- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

Current Chemical Genomics and Translational Medicine
(Discontinued)
ISSN: 2213-9885 ― Volume 12, 2018
Improved Dual-Luciferase Reporter Assays for Nuclear Receptors
Aileen Paguio, Pete Stecha, Keith V Wood , Frank Fan*
Abstract
Nuclear receptors play important roles in many cellular functions through control of gene transcription. It is also a large target class for drug discovery. Luciferase reporter assays are frequently used to study nuclear receptor function because of their wide dynamic range, low endogenous activity, and ease of use. Recent improvements of luciferase genes and vectors have further enhanced their utilities. Here we applied these improvements to two reporter formats for studying nuclear receptors. The first assay contains a Murine Mammary Tumor Virus promoter upstream of a destabilized luciferase. The presence of response elements for nuclear hormone receptor in this promoter allows the studies of endogenous and/or exogenous full length receptors. The second assay contains a ligand binding domain (LBD) of a nuclear receptor fused to the GAL4 DNA binding domain (DBD) on one vector and multiple Gal4 Upstream Activator Sequences (UAS) upstream of luciferase reporter on another vector. We showed that codon optimization of luciferase reporter genes increased expression levels in conjunction with the incorporation of protein destabilizing sequences into luciferase led to a larger assay dynamic range in both formats. The optimum number of UAS to generate the best response was determined. The expression vector for nuclear receptor LBD/GAL4 DBD fusion also constitutively expresses a Renilla luciferase-neoR fusion protein, which provides selection capability (G418 resistance, neoR) as well as an internal control (Renilla luciferase). This dual-luciferase format allowed detecting compound cytotoxicity or off-target change in expression during drug screening, therefore improved data quality. These luciferase reporter assays provided better research and drug discovery tools for studying the functions of full length nuclear receptors and ligand binding domains.
Article Information
Identifiers and Pagination:
Year: 2010Volume: 4
First Page: 43
Last Page: 49
Publisher Id: CCGTM-4-43
DOI: 10.2174/1875397301004010043
Article History:
Received Date: 19/2/2010Revision Received Date: 24/3/2010
Acceptance Date: 25/3/2010
Electronic publication date: 26/05/2010
Collection year: 2010
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Promega Corporation, Madison, Wisconsin, USA; Tel: 608-277-2531; Fax: 608-298-4818 E-mail: frank.fan@promega.com
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 19-2-2010 |
Original Manuscript | Improved Dual-Luciferase Reporter Assays for Nuclear Receptors | |
INTRODUCTION
The Nuclear Hormone Receptor (NHR) superfamily includes receptors for steroids, retinoids, thyroid hormone and vitamin D. These receptors are ligand-inducible transcription factors that regulate the expression of target genes involved in development, cell differentiation, and organ physiology [1Mangelsdorf DJ, Thummel C, Beato M, et al. The nuclear receptor superfamily: the second decade Cell 1995; 83: 835-9.]. Steroid hormones, since their first isolation in 1927, have been the targets of many drugs and the research of steroid receptors have advanced the treatment of cancer, osteoporosis, and endocrine and reproductive disorders [2Bai C, Schmidt A, Freedman LP. Steroid hormone receptors and drug discovery: therapeutic opportunities and assay designs Assay Drug Dev Technol 2003; 1(6): 843-52.]. The induction of steroid hormone pathways has also been increasingly important in the study of off-target drug effects and environmental toxicology [3Committee on Toxicity Testing and Assessment of Environmental Agents National Research Council Toxicity Testing in the 21st Century: A Vision and a Strategy. Washington DC: The National Academies Press 2007.].
NHRs have two major structural features, a highly conserved DNA binding domain (DBD), that targets the receptor to a specific DNA sequence upstream of a target gene, the hormone responsive element, and a C-terminal conserved ligand binding domain (LBD), which is involved in dimerization of the receptor and transactivation of target genes.
Reporter gene assays are uniquely suited to studying NHR because activating gene expression is a direct result of its action. In particular, luciferase reporter assays are frequently used because of their wide dynamic range, low endogenous activity, and ease of use [2Bai C, Schmidt A, Freedman LP. Steroid hormone receptors and drug discovery: therapeutic opportunities and assay designs Assay Drug Dev Technol 2003; 1(6): 843-52., 4Chen T, Xie W, Agler M, Banks M. Coactivators in assay design for nuclear hormone receptor drug discovery Assay Drug Dev Technol 2003; 1: 835-42.]. Recent improvements of luciferase genes and vectors represented by the pGL4 vector series have further enhanced the utilities of reporter assays for research and drug discovery. Most notably, codon optimization of luciferase reporter genes has increased expression levels and the incorporation of PEST- amino acid sequences that signal the protein for degradation by the proteosome- into luciferase has led to a larger assay dynamic range up to several hundred-fold [5Fan F, Wood KV. Bioluminescent assays for high-throughput screening Assay Drug Dev Technol 2007; 5(1): 127-36., 6Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis Science 1986; 234(4774): 364-8.].
Two reporter formats are commonly utilized for assaying nuclear receptor function (Fig. 1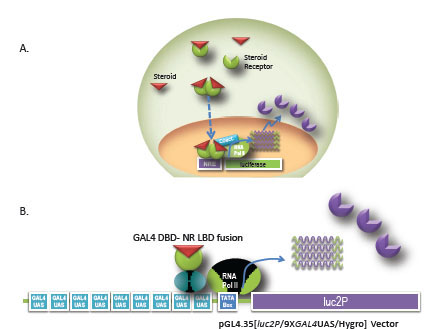 ). The first assay format incorporates a response element known to induce expression of target genes upstream of a reporter gene. This format enables the study of full length endogenous or exogenous receptors. Transfection of the response element and reporter gene into a cell line containing the endogenous receptor of interest has the advantage of analyzing the receptor signaling pathway in a native biological context. However, signal induction from endogenous receptors can be low. In addition, multiple receptors are known to bind to the same response element, which can complicate data interpretation. To improve assay quality, receptors can be introduced into the cell exogenously. In this case, the receptor is commonly expressed from a strong viral promoter such as Cytomegalovirus or SV40 promoter for high levels of expression. The MMTV promoter contains hormone response elements that are known to stimulate the androgen (AR), mineralcorticoid (MR), Glucocorticoid (GR), and progesterone receptors (PR) and has been used successfully in drug discovery compound screening and profiling [2Bai C, Schmidt A, Freedman LP. Steroid hormone receptors and drug discovery: therapeutic opportunities and assay designs Assay Drug Dev Technol 2003; 1(6): 843-52., 7Katso RM, Parham JH, Caivano M, et al. Evaluation of cell-based assays for steroid nuclear receptors delivered by recombinant baculoviruses J Biomol Scr 2005; 10: 715-24., 8Truss M, Chalepakis G, Beato M. Interplay of steroid hormone receptors and transcription factors on the mouse mammary tumor virus promoter J Steroid Biochem Mol Biol 1992; 43: 365-78.]. The second assay format utilizes the ligand binding domain of a nuclear receptor fused to the GAL4 DNA binding domain on one vector and multiple GAL4 Upstream Activator Sequences (UAS) upstream of luciferase reporter on another vector to monitor ligand binding and transactivation. The advantages of this format include low cross-reactivity with other nuclear receptors and the fact that the chimeric receptor is generally less toxic to the cells when overexpressed [2Bai C, Schmidt A, Freedman LP. Steroid hormone receptors and drug discovery: therapeutic opportunities and assay designs Assay Drug Dev Technol 2003; 1(6): 843-52., 4Chen T, Xie W, Agler M, Banks M. Coactivators in assay design for nuclear hormone receptor drug discovery Assay Drug Dev Technol 2003; 1: 835-42.]. Often, researchers will use both assay formats. For example, during drug discovery, the GAL4 hybrid system is frequently used to screen compounds for agonist or antagonist hit identification. The response element- luciferase reporter assay in a biologically relevant cell line is then used for hit to lead verification during secondary screening.
). The first assay format incorporates a response element known to induce expression of target genes upstream of a reporter gene. This format enables the study of full length endogenous or exogenous receptors. Transfection of the response element and reporter gene into a cell line containing the endogenous receptor of interest has the advantage of analyzing the receptor signaling pathway in a native biological context. However, signal induction from endogenous receptors can be low. In addition, multiple receptors are known to bind to the same response element, which can complicate data interpretation. To improve assay quality, receptors can be introduced into the cell exogenously. In this case, the receptor is commonly expressed from a strong viral promoter such as Cytomegalovirus or SV40 promoter for high levels of expression. The MMTV promoter contains hormone response elements that are known to stimulate the androgen (AR), mineralcorticoid (MR), Glucocorticoid (GR), and progesterone receptors (PR) and has been used successfully in drug discovery compound screening and profiling [2Bai C, Schmidt A, Freedman LP. Steroid hormone receptors and drug discovery: therapeutic opportunities and assay designs Assay Drug Dev Technol 2003; 1(6): 843-52., 7Katso RM, Parham JH, Caivano M, et al. Evaluation of cell-based assays for steroid nuclear receptors delivered by recombinant baculoviruses J Biomol Scr 2005; 10: 715-24., 8Truss M, Chalepakis G, Beato M. Interplay of steroid hormone receptors and transcription factors on the mouse mammary tumor virus promoter J Steroid Biochem Mol Biol 1992; 43: 365-78.]. The second assay format utilizes the ligand binding domain of a nuclear receptor fused to the GAL4 DNA binding domain on one vector and multiple GAL4 Upstream Activator Sequences (UAS) upstream of luciferase reporter on another vector to monitor ligand binding and transactivation. The advantages of this format include low cross-reactivity with other nuclear receptors and the fact that the chimeric receptor is generally less toxic to the cells when overexpressed [2Bai C, Schmidt A, Freedman LP. Steroid hormone receptors and drug discovery: therapeutic opportunities and assay designs Assay Drug Dev Technol 2003; 1(6): 843-52., 4Chen T, Xie W, Agler M, Banks M. Coactivators in assay design for nuclear hormone receptor drug discovery Assay Drug Dev Technol 2003; 1: 835-42.]. Often, researchers will use both assay formats. For example, during drug discovery, the GAL4 hybrid system is frequently used to screen compounds for agonist or antagonist hit identification. The response element- luciferase reporter assay in a biologically relevant cell line is then used for hit to lead verification during secondary screening.
In this report, we applied the advancement of luciferase gene, vector and genetic engineering to current assays to create better tools for studying nuclear receptor research and drug discovery.
MATERIALS AND METHODOLOGY
Chemicals
17β- estradiol (E2), estrone, ICI 182,780, tamoxifen, raloxifene, dihydrotestosterone (DHT), dexamethasone (Dex), hydrocortisone (HC), aldosterone, RU486, progesterone, prednisone, were purchased from Sigma Chemical Company (St. Louis, MO).
Plasmid Construction
Either 5 or 9 copies of the GAL4 UAS was inserted into pGL4.15[luc2P /Hygro] or pGL4.14 [luc2/Hygro] (Promega, Madison, WI), by PCR, utilizing the Acc65I and HindIII restriction enzyme sites and standard molecular cloning techniques. The clone containing 9 copies of the GAL4 UAS was named pGL4.35[luc2P/9XGAL4UAS/Hygro]. A DNA sequence of ERα-LBD (amino acids 303-595; Genbank NM_000125) was synthesized by DNA 2.0 (Menlo Park, CA), and cloned into pFN26A (BIND) hRluc-neo (Promega) by SgfI/PmeI so that the SgfI site yields an in-frame protein fusion with GAL4-DBD. A DNA sequence of GR-LBD (amino acids 500-777; Genbank NM_000176) was synthesized and cloned into pFN26A (BIND) hRluc-neo by the same method as ERα. These constructs were named pBIND-ERα and pBIND-GR respectively. The MMTV promoter (GenBank: U13861.1) was synthesized and cloned into pGL4.15 [luc2P/Hygro] and pGL4.14 [luc2/Hygro] using two SfiI sites each with unique internal sequences, yielding unique 3’ overhangs for unidirectional ligation. Full length AR (Genbank: NM_000044) driven by the CMV promoter was purchased from Origene (Rockville, MD). Constructions were verified by restriction digest and sequencing. One amino acid difference relative to the Genbank sequence (amino acid 240 of Genbank sequence) was introduced in the ERα-LBD by accident. It led to significantly increased induction (100- Fold) and right shifted the EC50’s of agonists (supplementary material Fig. 1), but did not affect potency ranking (data not shown).
Cell Culture and Generation of GloResponse™ 9XGAL4UAS-luc2P HEK293 Cell Line
HEK293 cells were cultured in DMEM (Invitrogen, Carlsbad CA) supplemented with 10% fetal bovine serum at 37oC with a humidified atmosphere at 5% CO2.
A stable cell line containing 9XGAL4UAS-luc2P was generated by lipid transfection of HEK293 with pGL4.35 luc2P/9XGAL4UAS/Hygro]. 2 x 106 cells were seeded in a 10 cm dish. The next day plasmid DNA was transfected using TransIT®-LT1 Transfection Reagent (Mirus Bio, Madison WI) following manufacturer’s protocol. A hygromycin resistant population was selected at 500µg/ml after two weeks. Clones were isolated by limiting dilution plating in 96-well plates. Individual clones were ranked by level of activation of ER by E2 or GR by dex after transient transfection of the receptor. The best clone was the clone with the highest induction of ER and GR and above background basal luciferase expression, which was subsequently named the GloResponse™ 9XGAL4UAS-luc2P HEK293 Cell Line.
Luciferase Reporter Assays
96-wells assays were performed by seeding 10,000 cell/ well and transeintly transfecting pGL4.35 luc2P/9XGAL4 UAS/Hygro] and pBIND-ERα or pBIND-GR. For assays in the GloResponseTM 9XGAL4UAS-luc2P, either the pBIND-ERα or pBIND-GR was transfected. 20-24 hours after transfection cells were treated with agonist or mock treated.
For assays in 384-well plates, cells were first seeded at 2 million cells/ 10 cm dish and transfected. 20-24 hours later, cells were transferred to 384-well plates containing compound at 10,000 cells/well, 20 µl per well.
For GR antagonist assays, each antagonist was serially diluted and added to cells in the presence of 100 nM Dex. For ER antagonist assays, the antagonist was serially diluted and added to cells in the presence of 1.0 nM E2.
Transfected cells were assayed 20-24 hours post- treatment with Dual-Glo® Luciferase Assay System (Promega, Madison WI). Assays in 96-wells plates were read with the Turner Biosystems (Sunnyvale, CA) GloMax® 96 Microplate Luminometer, 0.5 second integration/ well. Assays in 384-well plates were read with the Thermo Fisher Scientific (Waltham, MA) VarioSkan multimode reader, 0.5 second integration/well.
Inductions were calculated as the relative light units of the treated sample/ average relative light units of the untreated samples and plotted against the log of the compound concentration using Prism 4 software (GraphPad Software, San Diego CA). The sigmoidal does-response equation with varying slope was used to fit the data to generate the EC50 or IC50 values. Z’ values were calculated using the following formula: Z´= 1–[(3*SDtreatd + 3*SDuntreated)/ (averagetreated – averageuntreated)].
RESULTS AND DISCUSSION
Destabilized Luciferase Enhances Assay Performance
The codon optimized luciferase, luc2 and a version of the luc2 gene containing the PEST protein degradation sequence, luc2P, were compared in the two reporter formats. MMTV promoter contains multiple copies of a hormone response element, which is known to induce several NHR’s. We tested this MMTV promoter- luciferase construct for activation of two different receptors, an exogoneously expressed androgen receptor and an endogenously expressed glucocorticoid receptor. The activation of an exogenously introduced androgen receptor by DHT was compared with MMTV promoter driving either luc2 or luc2P. HEK293 cells were transiently co-transfected with pGL4.36[luc2P/MMTV/ hygro] or the same vector with luc2 and a plasmid expressing the full length androgen receptor (AR) driven by the CMV promoter. Titrating amounts of DHT were added to cells. Luciferase expression was induced 140-fold with an EC50 of 7.9 nM for the luc2P gene whereas the luc2 gene was only induced 64-fold, with an EC50= 9.9 nM. The presence of the PEST degradations sequence improves the dynamic range. A negative control of pGL4.36 [luc2P/MMTv/ hygro] co-transfected with pUC19 shows no induction (Fig. 2A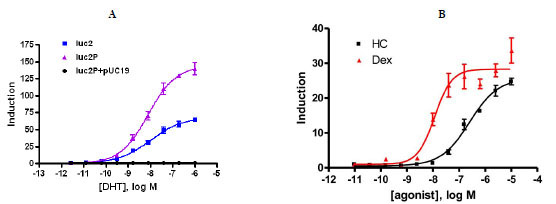 ). HeLa, which express endogenous glucocorticoid receptor were transiently transfected with pGL4.36 [luc2P/MMTV/hygro] or the same vector with luc2. Cells were induced with titrating amounts of dex. In this case, the difference between the two genes was minor, around 20% greater induction with luc2P (data not shown). To demonstrate the expected potency ranking, dex and HC were titrated and added to cells after transfecting with pGL4.36[luc2P/MMTV/hygro]. Luciferase expression was induced 35-fold with dex and 28-fold with HC (Fig. 2B
). HeLa, which express endogenous glucocorticoid receptor were transiently transfected with pGL4.36 [luc2P/MMTV/hygro] or the same vector with luc2. Cells were induced with titrating amounts of dex. In this case, the difference between the two genes was minor, around 20% greater induction with luc2P (data not shown). To demonstrate the expected potency ranking, dex and HC were titrated and added to cells after transfecting with pGL4.36[luc2P/MMTV/hygro]. Luciferase expression was induced 35-fold with dex and 28-fold with HC (Fig. 2B ) which is in agreement with published literature [9].
) which is in agreement with published literature [9].
Using the GAL4 hybrid format, luc2 and luc2P gene were compared for reporter dynamics with either the estrogen receptor alpha (ERα) LBD or the glucocorticoid receptor (GR) LBD fused to the GAL4 DBD. HEK293 cells were transiently co-transfected with each combination. With the ER-LBD, the expression from the luc2 gene was 170-fold over that of untreated. The luc2P gene induced with E2 expressed 270- fold over untreated. The Rapid Response luc2P had a 1.7-fold increased induction over luc2 for ER. With the GR-LBD induced with dex, the luc2 gene was induced 30-fold over untreated and the luc2P gene induced 62-fold over untreated cells. This corresponds to a 2.3 fold increased response of luc2P compared to luc2 at 24 hours post ligand exposure (Fig. 3A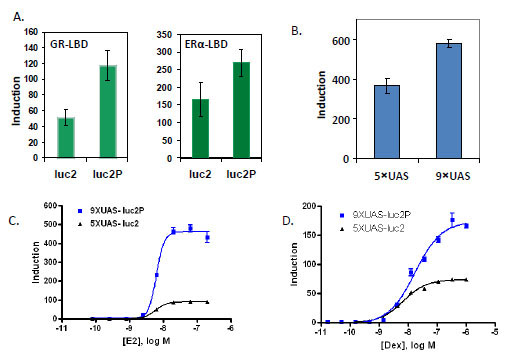 ).
).
Utilizing destabilized reporters to better reflect transcriptional dynamics have been reported with several popular reporters including luciferase and green fluorescent protein [10, 11]. The basic rationale for this approach is that destabilized reporters have lower steady-state levels so that changes in the promoter activities can be quickly represented. We have compared the performance of destabilized luciferase to its regular counterpart in several biological systems including GPCR induction (Fig.6 in ref. [5Fan F, Wood KV. Bioluminescent assays for high-throughput screening Assay Drug Dev Technol 2007; 5(1): 127-36.]), nuclear receptor studies as shown in this report and pathway analysis (manuscript in preparation). In general, the improved dynamic is observed. In time course experiments, destabilized luciferases often reach highest induction at earlier time points and gives higher maximum signal to background ratio over native luciferase. However, the details vary case by case, perhaps due to the complexity of each model system.
Increased GAL4 UAS Repeats Enhances Assay Performance
We tested 5 to 10 copies of the GAL4 Upstream Activator Sequence (UAS) and found that increasing the number of UAS up to 9 copies also increased induction response with the ERα-LBD in the pG5luc (Promega) backbone by around 1.5 fold (data not shown). We then transferred either 5 or 9 copies of the GAL4 UAS to the pGL4 backbone with luc2P and co-transfected with pBIND-ERα. Nine copies of the GAL4 UAS also increased the induction response in the pGL4 vector backbone, 310-fold for 5X compared to 490- fold for 9X, or an increase of 1.7- fold (Fig. 3B ). When combined, these two elements, nine copies of the GAL4 UAS and use of the destabilized and codon optimized, luc2P, contribute to a 5-fold increase in induction with ER and 2.2-fold increase with GR. The EC50’s were not changed. For ER, 5XUAS-luc2 the EC50 was 6.2 nM and 6.7 nM for 9XUAS-luc2P. For GR, the EC50’s were 6.5 nM and 17 nM for 5XUAS-luc2 and 9XUAS-luc2P respectively (Figs. 3C
). When combined, these two elements, nine copies of the GAL4 UAS and use of the destabilized and codon optimized, luc2P, contribute to a 5-fold increase in induction with ER and 2.2-fold increase with GR. The EC50’s were not changed. For ER, 5XUAS-luc2 the EC50 was 6.2 nM and 6.7 nM for 9XUAS-luc2P. For GR, the EC50’s were 6.5 nM and 17 nM for 5XUAS-luc2 and 9XUAS-luc2P respectively (Figs. 3C and D
and D ).
).
Improved Luciferase Nuclear Receptor Assays for Drug Discovery
The GloResponseTM 9XGAL4UASluc2P HEK293 cell line was transiently transfected with either pBIND-ER or pBIND-GR and were evaluated for their performance in high throughput screening (HTS) assays by determining the Z´-factor value for an agonist assay in 384-well plate format. The ER assay induced with E2 had a Z’ of 0.8 and the GR assay had a Z’ of 0.79 when induced with dex. These Z´-factor values significantly higher than 0.5, an acceptance criterion commonly used for high-throughput screening [12]. These assays also maintained a Z’ greater than 0.5 in the presence of up to 1% DMSO or ETOH (data not shown).
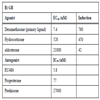
To evaluate the compound specificity of the GAL4 hybrid assay the EC50 and IC50 of known agonists and antagonist for the estrogen receptor and glucocorticoid receptor were determined. Fig. (4A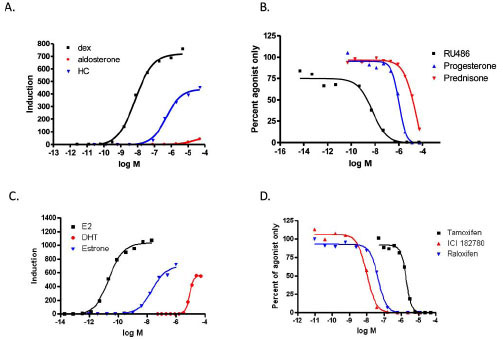 ) shows the EC50 generated in the GAL4 hybrid format for GR agonists. The potency rankings for GR agonists were dex<HC< aldosterone. The IC50’s for GR antagonists ranked as follows (Fig. 4B
) shows the EC50 generated in the GAL4 hybrid format for GR agonists. The potency rankings for GR agonists were dex<HC< aldosterone. The IC50’s for GR antagonists ranked as follows (Fig. 4B ): RU486 <Progesterone<Prednisone. Both results are in agreement with published literature [9Wilkinson JM, Hayes S, Thompson D, Whitney P Bi K. Com-pound profiling using a panel of steroid hormone receptor cell-based assays J Biomol Scr 2008; 13: 755-65.]. The rank order potency for ER agonists of E2<estrone<DHT is comparable with other transactivation assays (Fig. 4C
): RU486 <Progesterone<Prednisone. Both results are in agreement with published literature [9Wilkinson JM, Hayes S, Thompson D, Whitney P Bi K. Com-pound profiling using a panel of steroid hormone receptor cell-based assays J Biomol Scr 2008; 13: 755-65.]. The rank order potency for ER agonists of E2<estrone<DHT is comparable with other transactivation assays (Fig. 4C ; [8Truss M, Chalepakis G, Beato M. Interplay of steroid hormone receptors and transcription factors on the mouse mammary tumor virus promoter J Steroid Biochem Mol Biol 1992; 43: 365-78., 9Wilkinson JM, Hayes S, Thompson D, Whitney P Bi K. Com-pound profiling using a panel of steroid hormone receptor cell-based assays J Biomol Scr 2008; 13: 755-65.]). The rank order potency for ER antagonist in this format was ICI,182,780 <raloxifene< tamoxifen (Fig. 4D
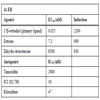
; [8Truss M, Chalepakis G, Beato M. Interplay of steroid hormone receptors and transcription factors on the mouse mammary tumor virus promoter J Steroid Biochem Mol Biol 1992; 43: 365-78., 9Wilkinson JM, Hayes S, Thompson D, Whitney P Bi K. Com-pound profiling using a panel of steroid hormone receptor cell-based assays J Biomol Scr 2008; 13: 755-65.]). The rank order potency for ER antagonist in this format was ICI,182,780 <raloxifene< tamoxifen (Fig. 4D ). The IC50 for ICI 182,780 was 10 nM and raloxifene, 47 nM in our tests. Published literature for other transactivation or binding assays show these two antagonists to have IC50’s within one log, but reports conflict as to their ranking [9Wilkinson JM, Hayes S, Thompson D, Whitney P Bi K. Com-pound profiling using a panel of steroid hormone receptor cell-based assays J Biomol Scr 2008; 13: 755-65., 13Wilson VS, Bobseine K, Gray LE LE Jr. Development and characterization of a cell line that stably expresses an estrogen-responsive luciferase reporter for the detection of estrogen receptor agonist and antagonists Toxicol Sci 2004; 81: 69-77.-15Shi X, Zheng W, Schneeweis JE, Fischer PA, Strulovici B, Peekhaus NT. A short-incubation reporter-gene assay for high-throughput screening of estrogen receptor-alpha antagonists Assay Drug Dev Technol 2005; 3: 393-400.]. Our results are consistent with the similarity of the IC50’s. This similarity may cause alternating rankings depending on assay and cell type. Table 1 summarizes the EC50’s, IC50’s and inductions.
). The IC50 for ICI 182,780 was 10 nM and raloxifene, 47 nM in our tests. Published literature for other transactivation or binding assays show these two antagonists to have IC50’s within one log, but reports conflict as to their ranking [9Wilkinson JM, Hayes S, Thompson D, Whitney P Bi K. Com-pound profiling using a panel of steroid hormone receptor cell-based assays J Biomol Scr 2008; 13: 755-65., 13Wilson VS, Bobseine K, Gray LE LE Jr. Development and characterization of a cell line that stably expresses an estrogen-responsive luciferase reporter for the detection of estrogen receptor agonist and antagonists Toxicol Sci 2004; 81: 69-77.-15Shi X, Zheng W, Schneeweis JE, Fischer PA, Strulovici B, Peekhaus NT. A short-incubation reporter-gene assay for high-throughput screening of estrogen receptor-alpha antagonists Assay Drug Dev Technol 2005; 3: 393-400.]. Our results are consistent with the similarity of the IC50’s. This similarity may cause alternating rankings depending on assay and cell type. Table 1 summarizes the EC50’s, IC50’s and inductions.
Potency Ranking. GloResponse 9XGAL4-luc2P HEK293 cell line was transiently transfected with vector containing receptor LBD in a 10-cm dish. The day after transfection, cells were transferred to 384-well plates and a titration of each compound was added to cells. The fold-induction was plotted using GraphPad Prism 4 software. The EC50/IC50 was determined from the sigmoidal dose-response equation with varying slope which fit the data. A) ER was tranfected. B) pBIND-GR was transfected
Dual Luciferase Format Improves Data Quality
The expression vector for nuclear receptor LBD/GAL4 DBD fusion (pFN26A (BIND) hRluc-neo) also constitutively expresses a Renilla (Rluc) luciferase-neoR fusion, which can be used for selection (neoR) of stable clones as well as an internal control reporter (Renilla luciferase). We observed a reduction in firefly (Fluc) stimulation at high E2 concentrations. To determine if this drop is caused by cytotoxicity of the compound and if the Renilla reporter could be used to detect compound cytotoxicity or any other off-target change in expression in a dual luciferase assay, HEK293 cells were transiently co-transfected in 96-well plates with pBIND-ERα-LBD and pGL4.35 [luc2P/9XGAL4UAS/Hygro] vectors and treated with titrated concentrations of E2. Firefly and Renilla luciferase activities were measured using the Dual-Glo™ Assay System. Another plate was prepared in parallel and measured with CellTiter-Glo® Luminescent Cell Viability Assay. Fig. (5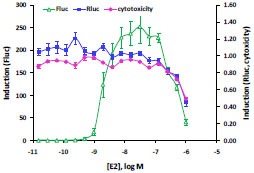 ) shows that at highest concentrations of E2, the expression of the Firefly reporter still shows induction of 50 fold over basal, though reduce from a peak of 250-fold. The Renilla reporter is not induced (induction around 1.0) by E2. However, for the three highest concentrations of E2, Renilla expression drops below basal, resulting in declining induction ratios from 1.0 to 0.5. Decreasing cells viability at these three highest points of E2 is verified by decreasing values in the CellTiter-Glo® Luminescent Cell Viability Assay, paralleling the Renilla reporter expression. In one assay, the Dual-Glo™ Assay System, two results can be obtained—the primary firefly reporter expression and a second control reporter Renilla reporter, which can detect off-target signal perturbations and cytotoxicity.
) shows that at highest concentrations of E2, the expression of the Firefly reporter still shows induction of 50 fold over basal, though reduce from a peak of 250-fold. The Renilla reporter is not induced (induction around 1.0) by E2. However, for the three highest concentrations of E2, Renilla expression drops below basal, resulting in declining induction ratios from 1.0 to 0.5. Decreasing cells viability at these three highest points of E2 is verified by decreasing values in the CellTiter-Glo® Luminescent Cell Viability Assay, paralleling the Renilla reporter expression. In one assay, the Dual-Glo™ Assay System, two results can be obtained—the primary firefly reporter expression and a second control reporter Renilla reporter, which can detect off-target signal perturbations and cytotoxicity.
Dual luciferase system has been widely used to study promoters and transcriptional response elements. In that application, firefly luciferase is used as functional reporter and Renilla as a control reporter to normalize transfection efficiency. For the application in HTS, Renilla luciferase can be used as an internal control to detect cytotoxicity, therefore improving data quality by identifying potential false hit as shown in this report. However, since Fluc and Rluc are unrelated enzymes, it will not be able to detect false hits due to the interference (inhibition or activation) with Fluc. In that aspect, a green and red click beetle luciferases system offers advantages since the two reporters are essentially the same except difference in emission spectrum rendered by several amino acids in the active site. With the appropriate instrument and data analysis, the click beetle system showed great utilities in HTS as demonstrated by Davis et al. [16Davis RE, Zhang YQ, Southall N, et al. A cell-based assay for IkappaBalpha stabilization using a two-color dual luciferase-based sensor Assay Drug Dev Technol 2007; 5(1): 85-103.]. Alternatively, Renilla luciferase can be used as a second functional reporter to detect another cellular event. The signal strength of Renilla luciferase can be adjusted depend on which constitutive promoter (e.g. CMV, SV40, TK) was chosen to drive Rluc expression.
CONCLUSION
We have described improved reporter performance in two reporter formats for assaying nuclear receptors by utilizing the codon optimized and destablized Rapid Response luciferase reporter gene, luc2P in the pGL4 vector backbone. The MMTV promoter driving luc2P is a single reporter that can be used to assay several different full length NR’s, endogenous or exogenously expressed. In the GAL4 hybrid format, nine copies of the GAL4 UAS and the luc2P gene both improved the dynamic range of the response to appropriate stimuli. The improved dynamic range in both these reporter formats will improve their use for high-throughput screening or profiling.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publishers Web site along with the published article Download Supplementary Material.
ACKNOWLEDGEMENTS
We would like to thank Kevin Kopish and Zhijie Cheng for their valuable discussion.