- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Environmental Research Journal
Formerly: The Open Ecology Journal
ISSN: 2590-2776 ― Volume 16, 2023
- New Journal Website
- Journal Home
- Editorial Board
- Board Recruitment Workflow
- Instructions for Authors
- Plagiarism Prevention
- Fabricating and Stating False Information
- Research Misconduct
- Post Publication Discussions and Corrections
- Allegations from Whistleblowers
- Advertising Policy
- Short Guide
- Editorial Policies
- Publishing Ethics and Rectitude
- Quick Track Option
- Reviewer Guidelines
- Guidelines for Guest Editors
- Publication Fee
- Publication Cycle - Process Flowchart
- Archiving Policies
- Special Issues
Spatial Variation of nosZ-communities in the Low Oxygen Waters of Prado Hydroelectric (South-West of Colombia)
Mauricio E. Pacheco Montealegre, Maribeb Castro González*
Abstract
Denitrification is a process of reduction of nitrate to dinitrogen by facultative anaerobic microorganisms, which have functional genes encoding denitrification enzymes (reductases). The nosZ gene encoding the enzyme that reduces nitrous oxide to dinitrogen was utilized in this research, as molecular marker for denitrifying communities into low oxygen waters of Prado reservoir. Our objective was to analyze and compare the composition (richness and abundance) of nosZ-type denitrifiers in relationship with physicochemical variables (oxygen, pH, temperature, nitrate, nitrite and ammonium) in three areas of this dam: Isla del Sol, Lozanía and Tomogó which are distant and have different anthropogenic influences. For this, we performed DNA extraction, amplification, 454 pyrosequencing and phylogenetic analysis of nosZ gene. The Chao1 estimator and Shannon index were used for compare richness and diversity of nosZ gene; and the relationship between compositions of operational taxonomic units (OTU) with physicochemical variables was established by canonical correspondence analysis (CCA). In the reservoir 45 nosZ-OTUs to species level (80% similarity) were detected. Lozanía had the highest number of OTUs (25) and greatest diversity (SCHAO1: 35; H: 2.0), compared to the other areas sampling. The phylogenetic analysis showed the presence of many OTUs (28 of 45) with low similarity to Proteobacteria group and high similarity with sequences of environmental clones reported previously. The CCA showed that the nosZ-community composition of Prado dam was related with low pH (6.2), oxygen (0.01mg / L) and nitrate (<0.25 mg / L) recorded in the water column.
Article Information
Identifiers and Pagination:
Year: 2016Volume: 9
First Page: 1
Last Page: 12
Publisher Id: TOECOLJ-9-1-1
DOI: 10.2174/1874213001609010001
Article History:
Received Date: 21/4/2015Revision Received Date: 17/5/2016
Acceptance Date: 30/06/2016
Electronic publication date: 15/09/2016
Collection year: 2016
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution-Non-Commercial 4.0 International Public License (CC BY-NC 4.0) (https://creativecommons.org/licenses/by-nc/4.0/legalcode), which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Facultad de Ciencias, Programa de Biología Aplicada, Universidad Militar Nueva Granada, Cajicá, Colombia; E-mail: maribeb.castro@unimilitar.edu.co
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 21-4-2015 |
Original Manuscript | Spatial Variation of nosZ-communities in the Low Oxygen Waters of Prado Hydroelectric (South-West of Colombia) | |
INTRODUCTION
In the natural environments, the nitrate and nitrite is removing by a microbial process known as denitrification, in which dissimilatory oxidized nitrogen is used as an alternative electron acceptor for energy production when oxygen is limited (Zumft and Kroneck 2007Matulich, KL (2013) Microbial Biodiversity. Encyclopedia of Biodiversity, 5, 252-58.). Denitrifying microorganisms are in environments where three conditions exist: (Alldredge and Cohen 1987Alldredge, AL & Cohen, Y (1987) Can microscale chemical patches persist in the sea? Microelectrode study of marine snow, fecal pellets. Science, 235(4789), 689-91.
[http://dx.doi.org/10.1126/science.235.4789.689] [PMID: 17833630] ) low oxygen levels, (American Public Health Association 1999American Public Health Association. (1999). >Standard Methods for the Examination of Water and Wastewater. (20nd). Baltimore, Maryland. ) availability of nitrate (NO3-) and nitrite (NO2-), and (Baxter et al. 2013Baxter, AM, Johnson, L, Royer, T & Leff, LG (2013) Spatial differences in denitrification and bacterial community structure of streams: relationships with environmental conditions. Aquatic Science, 75(2), 275-84.
[http://dx.doi.org/10.1007/s00027-012-0272-5] ) availability of organic matter (OM) (Correa-Galeote et al. 2013Correa-Galeote, D, Tortosa, G & Bedmar, EJ (2013) Determination of Denitrification Genes Abundance in Environmental Samples. Metagenomics, 2, 1-14.
[http://dx.doi.org/10.4303/mg/235702] ). The denitrifiers are known to belong to more than 50 genera of bacteria, including members of the groups: Proteobacteria, Firmicutes, Actinobacteria, Bacteroides, and Planctomyces, however, some Archaea and Fungi can denitrify too (Zumft and KroneckZhao, X, Wei, Z, Zhao, Y, Xi, B, Wang, X, Zhao, T, Zhang, X & Wei, Y (2015) Environmental factors influencing the distribution of ammonifying and denitrifying bacteria and water qualities in 10 lakes and reservoirs of the Northeast, China. Microbial Biotechnology, 8(3), 541-8.
[http://dx.doi.org/10.1111/1751-7915.12260] [PMID: 25756244] 2007).
Among the genes of the denitrification, the nosZ-gene encoding the nitrous oxide reductase is one of the molecular marker most widely implemented for phylogenetic and ecological analysis of denitrifying communities from natural environments (Braker et al. 2012Braker, G, Dörsch, P & Bakken, LR (2012) Genetic characterization of denitrifier communities with contrasting intrinsic functional traits. FEMS Microbiology Ecology, 79(2), 542-54.
[http://dx.doi.org/10.1111/j.1574-6941.2011.01237.x] [PMID: 22092293] , Jones and Hallin 2010Braker, G, Dörsch, P & Bakken, LR (2012) Genetic characterization of denitrifier communities with contrasting intrinsic functional traits. FEMS Microbiology Ecology, 79(2), 542-54.
[http://dx.doi.org/10.1111/j.1574-6941.2011.01237.x] [PMID: 22092293] ). It has recently been established that 80% and 86% similarity of DNA and amino acid sequences respectively of nosZ gene, allows detection of the OTUs in different habitats (Palmer et al. 2009Palmer, K & Horn, MA (2012) Actinobacterial nitrate reducers and proteobacterial denitrifiers are abundant in N2O-metabolizing palsa peat. Applied and Environmental Microbiology, 78(16), 5584-96.
[http://dx.doi.org/10.1128/AEM.00810-12] [PMID: 22660709] ). Many studies attribute the similarity of nosZ-gene sequences mostly to bacteria of Proteobacteria group (Jones et al. 2013, Wyman et al. 2013Wen, D, Bai, Y, Shi, Q & Li, Z (2012) Bacterial diversity in the polluted water of the Dianchi lakeshore in China. Annals of Microbiology, 62, 715-23.
[http://dx.doi.org/10.1007/s13213-011-0311-9] ). However, Jones et al. (2013) and Sanford et alSanford, RA, Wagner, DD, Wu, Q, Chee-Sanford, JC, Thomas, SH, Cruz-García, C, Rodríguez, G, Massol-Deyá, A, Krishnani, KK, Ritalahti, KM, Nissen, S, Konstantinidis, KT & Löffler, FE (2012) Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. Proceedings of the Natural Academy of Sciences of the United States of America, 109(48), 19709-14.
[http://dx.doi.org/10.1073/pnas.1211238109] [PMID: 23150571] . (2012) discuss the presence of nosZ gene into a wide range of groups of high abundance in different ecosystems with structural variations of gene. The nosZ-gene study has allowed to analyze denitrifying communities related with the reduction of nitrous oxide to dinitrogen in aquatic systems such as ocean (Zehr and Kudela 2011Wyman, M, Hodgson, S & Bird, C (2013) Denitrifying alphaproteobacteria from the Arabian Sea that express nosZ, the gene encoding nitrous oxide reductase, in oxic and suboxic waters. Applied and Environmental Microbiology, 79(8), 2670-81.
[http://dx.doi.org/10.1128/AEM.03705-12] [PMID: 23396348] ), oxygen minimum zones (Wyman et al. 2013, Castro-González et al. 2015), estuaries (Chon et al. 2011Chon, K, Chang, JS, Lee, E, Lee, J, Ryu, J & Cho, J (2011) Abundance of denitrifying genes coding for nitrate (narG), nitrite (nirS), and nitrous oxide (nosZ) reductases in estuarine versus wastewater effluent-fed constructed wetlands. Ecological Engineering, 37(1), 64-9.
[http://dx.doi.org/10.1016/j.ecoleng.2009.04.005] ), streams (Baxter et al. 2013) and wastewater (Hou et al. 2012Roldán, G (2003) Implicaciones ecológicas en la construcción de embalses en Colombia. Revista Universidad Católica de Oriente, 16, 71-83.). However, it is important to study these organisms in eutrophic lakes and reservoirs where the denitrification plays an important role in the balance of N2O production/reduction which can determine if such areas are source or sink of this greenhouse gas (Wang et al. 2013Wang, C, Zhu, G, Wang, Y, Wang, S & Yin, C (2013) Nitrous oxide reductase gene (nosZ) and N2O reduction along the littoral gradient of a eutrophic freshwater lake. Journal of Environmental Science (China), 25(1), 44-52.
[http://dx.doi.org/10.1016/S1001-0742(12)60005-9] [PMID: 23586298] , YuWetzel, RG (2001) Limnology Lake and River Ecosystems , 3rd ed, Academic Press, USA. et al. 2014).
In Colombia, there are currently 26 dam in operation, and among them the Prado Reservoir is the fourth largest. This reservoir is a multi-purpose (electricity, irrigation, aquaculture, fisheries, recreation and tourism) tropical dam constructed for water storage of the Cunday and Negro Rivers and other minor tributaries in the south west of Colombia (Guevara et al. 2009). Likewise, this reservoir provides drinking water for the population despite its water quality being strongly influenced by agricultural activities and domestic discharge. This reservoir was constructed in 1973, it has an average depth of ~45 m and a surface area of ~3.900 hectares covered with water during all the year. Because of the high organic load and the flooded vegetation not removed during its inundation the Prado Reservoir has high H2S and ammonia production in the hypolimnetic anoxic environment and it has been classified as hyper-eutrophic (DucharmeDucharme, A (1975) Informe técnico de biología pesquera (Limnología). Publicación #4. Proyecto para el Desarrollo de la Pesca Continental. INDERENA-FAO, Bogotá, Colombia. 1975, Márquez and Guillot 2001, Roldán 2003). In the surface waters Guevara et al. (2009) have reported alkalinity from 17.95 to 39.38mg/L, phosphates between 0.01–0.30mg/L, conductivity from 48.73 to 67.06 mS/cm and pH between 6.42 - 8.48. Recently, Castro-González and Torres et al. (2015)Ducharme, A (1975) Informe técnico de biología pesquera (Limnología). Publicación #4. Proyecto para el Desarrollo de la Pesca Continental. INDERENA-FAO, Bogotá, Colombia. reported a strong oxycline (151 - 4 µM O2) and variability of nutrients along the column water (between 0-10 m depth) with levels of nitrate and ammonium between 2.6 - 36 µM and of 2.8 to 50 µM respectively. Taking into account all these characteristics, the Prado dam is an ideal place to explore aspects about denitrifiers diversity, given its eutrophic condition, thermal stratification, and hypoxic/suboxic water column (Guevara et al. 2009, Perea and VillanuevaPerea, EM & Villanueva, E (2010) In: Reinoso-Flórez, G., Villa-Navarro, F.A., Garcia-Melo, J., (Eds.), Capacidad de carga, caracterización y evaluación de la actividad pesquera y acuícola en el Embalse de Prado 30-46. 2010). Until today, in this reservoir there have been studies about fishes (Villa and LozadaVilla, FA & Losada, S (1999) Aspectos tróficos de Petenia umbrifera (Pisces: Cichlidae) en la represa de Prado (Tolima). Revista de la Asociación Colombiana de Ciencias Biológicas, 11, 24-34. 1999, 2004), zooplankton, phytoplankton (Guevara et al. 2009) and bacterioplankton (Canosa and Pinilla 2007Canosa, A & Pinilla, G (2007) Relaciones entre las abundancias del bacterioplancton y del fitoplancton en tres ecosistemas lénticos de los Andes Colombianos. Biología Tropical, 55, 135-46.), but not related to the diversity of denitrifying microorganisms, excepting a recent study developed by Castro-GonzálezCastro-González, M & Torres, V (2015) Gases invernadero en aguas con bajo oxígeno en el reservorio eutrófico de Prado (Colombia). Revista de la Academia Colombiana de Ciencias Exactas. Físicas y Naturales, 39, 399-407. (2014) about the TRFLP diversity of nosZ-denitrifying communities between hypoxic and suboxic conditions in the water column at Isla del Sol. For this, the goal of this study was to determine and compare the composition (richness/abundance) of nosZ-type denitrifiers in three places of this dam, through PCR and 454 pyrosequencing of this functional gene. Also, we analyzed the relationship between denitrifying community composition with the physicochemical variables (oxygen, pH, temperature, nitrate, nitrite and ammonium) prevalent in the water column of this hydroelectric.
MATERIALS AND METHODS
Study Sites, Sample Collection and Total DNA Extraction
The sampling was done in the Prado dam in the south west of Tolima, which is one of the biggest reservoirs in Colombia (1.254 Ha), characterized by its eutrophic conditions, physical-chemical stratification, low oxygen and high levels of organic matter and nutrient in the water column in contrast with others Colombian hydroelectric (Canosa and Pinilla 2007, Guevara et al. 2009, Márquez and Guillot 2001, Perea and Villanueva 2010Roldán, G (2003) Implicaciones ecológicas en la construcción de embalses en Colombia. Revista Universidad Católica de Oriente, 16, 71-83., Roldán 2003Roldán, G (2003) Implicaciones ecológicas en la construcción de embalses en Colombia. Revista Universidad Católica de Oriente, 16, 71-83.).
The water samples were taken in September 2012 (dry season) from three places: Isla del sol (3°45’N - 74°51’W), Lozanía (3°52’N - 74°48’W) and Tomogó (3°43’N - 74°53’W), between 6-9 m depth where low oxygen levels were detected during sampling (Fig. 1 ). These sampling areas were selected taking into account previous reports about spatial heterogeneity of limnological variables caused by differences in the influence of rivers, small tributaries and anthropogenic activities related with changes in nutrient concentration, water temperature, transparency, conductivity and pH (SODEIC 1993SODEIC. (1993). Estudio de ordenamiento y manejo ambiental de la cuenca hidrográfica del Rio Prado Fase A: ordenamiento ambiental. Convenio CORTOLIMA-ICEL-ELECTROLIMA., Guevara 1993 et al. 2009 SODEIC. (1993). Estudio de ordenamiento y manejo ambiental de la cuenca hidrográfica del Rio Prado Fase A: ordenamiento ambiental. Convenio CORTOLIMA-ICEL-ELECTROLIMA.).
). These sampling areas were selected taking into account previous reports about spatial heterogeneity of limnological variables caused by differences in the influence of rivers, small tributaries and anthropogenic activities related with changes in nutrient concentration, water temperature, transparency, conductivity and pH (SODEIC 1993SODEIC. (1993). Estudio de ordenamiento y manejo ambiental de la cuenca hidrográfica del Rio Prado Fase A: ordenamiento ambiental. Convenio CORTOLIMA-ICEL-ELECTROLIMA., Guevara 1993 et al. 2009 SODEIC. (1993). Estudio de ordenamiento y manejo ambiental de la cuenca hidrográfica del Rio Prado Fase A: ordenamiento ambiental. Convenio CORTOLIMA-ICEL-ELECTROLIMA.).
Temperature, pH and dissolved oxygen were measured on board with a waterproof Oakton® multi-parameter. Eight liters of water were maintained under refrigeration until collecting the microorganisms on membranes by vacuum filtration. On board, 350 mL of freshwater samples were sampled, filtered (through GF/F membranes) and frozen for nutrient analyses. The concentration of ammonium (NH4+) was determined by Kjeldahl method, and the dissolved nitrate (NO3-) and nitrite (NO2-) were measured to be 220-275 nm y 510 nm respectively, with spectrophotometric techniques following the Standard Methods for the Examination of Water and Wastewater (American Public Health Association 1999).
Water samples (8 L) were filtered sequentially through Millipore membrane filters of 20 µm, 8 µm and 0.45 µm pore size. The last one membrane was used for bacterial DNA extraction with the PowerWater® DNA isolation kit (MoBio Laboratories, Inc., CA, USA). The DNA extracts were visualized with etidium bromide on 0.9% agarose gels and were quantified by spectrophotometry 260/280 in a nanodrop.
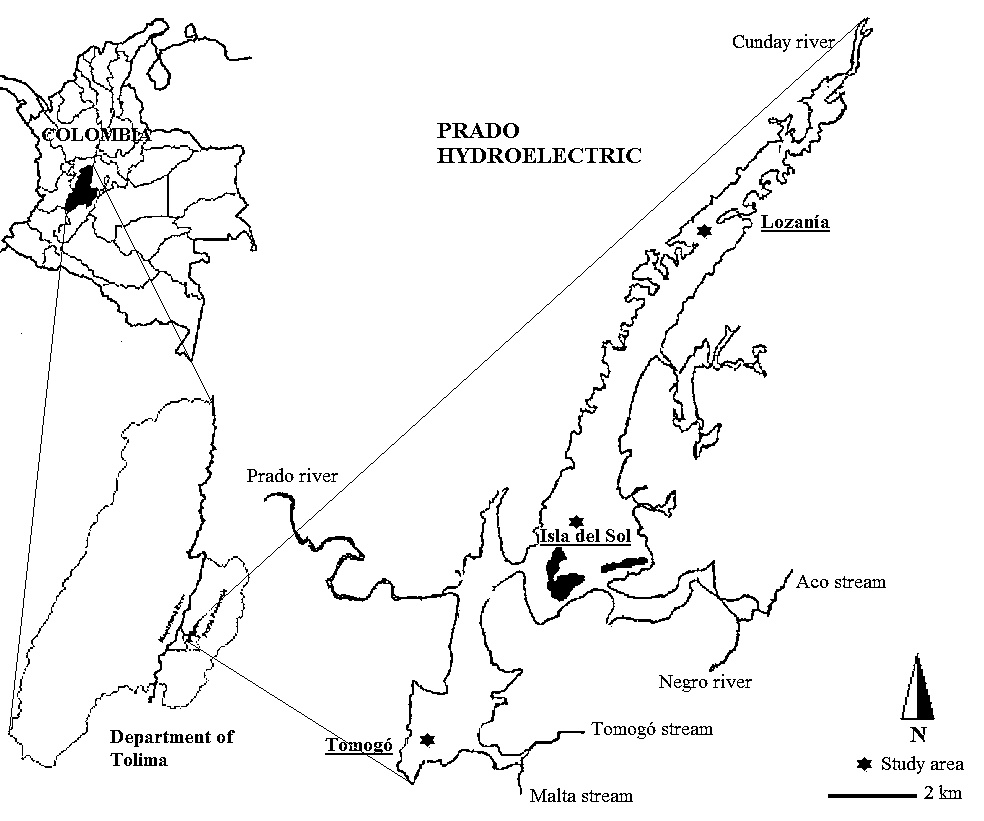 |
Fig. (1) Sampling areas in the Prado dam at south west of Tolima, Colombia. |
Pyrosequencing and Sequence Filtering
Tag pyrosequencing of nosZ genes was used to study the microbial communities with the potential for N2O reduction to N2. DNA (10 µl, 20 ng/µl) from each sample was sent to the Research and Testing (R&T) Laboratory (Lubbock, TX). PCR amplification was performed using primers nosZF and nosZ1622R (Kloos et al. 2001Kloos, K, Mergel, A, Rösch, C & Bothe, H (2001) Denitrification within the genus Azospirillum and other associative bacteria. Australian Journal of Plant Physiology, 28(8), 991-8.). Sequencing reactions utilized a Roche 454 GS-FLX+ instrument (Roche, Indianapolis, IN) with titanium reagents. Each study area had its specific barcode. The raw sequences processed and denoised by R & T Laboratory, where they were checked for elimination sequences with incorrect barcode and primers. After, a validation of sequences was performed according to Mao et al. (2013)Mao, Y, Yannarell, AC, Davis, SC & Mackie, RI (2013) Impact of different bioenergy crops on N-cycling bacterial and archaeal communities in soil. Environmental Microbiology, 15(3), 928-42.
[http://dx.doi.org/10.1111/j.1462-2920.2012.02844.x] [PMID: 22891790] and Palmer et al. (2012)Sanford, RA, Wagner, DD, Wu, Q, Chee-Sanford, JC, Thomas, SH, Cruz-García, C, Rodríguez, G, Massol-Deyá, A, Krishnani, KK, Ritalahti, KM, Nissen, S, Konstantinidis, KT & Löffler, FE (2012) Unexpected nondenitrifier nitrous oxide reductase gene diversity and abundance in soils. Proceedings of the Natural Academy of Sciences of the United States of America, 109(48), 19709-14.
[http://dx.doi.org/10.1073/pnas.1211238109] [PMID: 23150571] . As a result of this validation about 43% of sequences were eliminated and a total of 6697 sequences were used for the analysis (see Table 1). In this process chimeras and duplicates were eliminated with USEARCH/UCHIME (Edgar et al. 2011Edgar, RC, Haas, BJ, Clemente, JC, Quince, C & Knight, R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 27(16), 2194-200.
[http://dx.doi.org/10.1093/bioinformatics/btr381] [PMID: 21700674] ) and DEREPLICATOR using the Fun Gene Pipeline (Fish et al. 2013Fish, JA, Chai, B, Wang, Q, Sun, Y, Brown, CT, Tiedje, JM & Cole, JR (2013) FunGene: the functional gene pipeline and repository. Frontiers in Microbiology, 4, 291.
[http://dx.doi.org/10.3389/fmicb.2013.00291] [PMID: 24101916] ). Ambiguous sequences or low quality regions (with Q-values less than 20) and sequences <350 bp were eliminated with Bio Edit v 7.1.11 (Hall 1999Hall, TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium, 95-8.). Finally, no coding sequences for nitrous oxide reductase were eliminated with Blast2 go v2.6.6 (Conesa et al. 2005Conesa, A, Götz, S, García-Gómez, JM, Terol, J, Talón, M & Robles, M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 21(18), 3674-6.
[http://dx.doi.org/10.1093/bioinformatics/bti610] [PMID: 16081474] ), using the non-redundant protein sequence database with an E-value cutoff of 0.001 (Mao et al. 2013Mao, Y, Yannarell, AC, Davis, SC & Mackie, RI (2013) Impact of different bioenergy crops on N-cycling bacterial and archaeal communities in soil. Environmental Microbiology, 15(3), 928-42.
[http://dx.doi.org/10.1111/j.1462-2920.2012.02844.x] [PMID: 22891790] ).
Composition and Diversity of NosZ-type Denitrifiers
The valid sequences were clustered as operational taxonomic units at species-level threshold distances of 20% (Palmer et al. 2009) based on DNA sequences. The OTUs were defined with DOTUR program [Distance-Based OTU and Richness (Schloss and HandelsmanSchloss, PD & Handelsman, J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Applied and Environmental Microbiology, 71(3), 1501-6.
[http://dx.doi.org/10.1128/AEM.71.3.1501-1506.2005] [PMID: 15746353] 2005)] using furthest neighbor assignment algorithm with 1.000 iterations. Later, nosZ sequences within OTUs that started with the barcode assigned to Tomogó, Isla del Sol and Lozanía were selected for recognizing the OTUs generated for each area. The coverage percent (%) of OTUs for the number of valid sequences, was calculated according to Good (1953) in Mao et al. (2013)Mao, Y, Yannarell, AC, Davis, SC & Mackie, RI (2013) Impact of different bioenergy crops on N-cycling bacterial and archaeal communities in soil. Environmental Microbiology, 15(3), 928-42.
[http://dx.doi.org/10.1111/j.1462-2920.2012.02844.x] [PMID: 22891790] . The CHAO1 richness estimator, Shannon weaver diversity index and the Bray-Curtis similarity index were calculated with PAST v 3.0 (Hammer et al. 2001Hammer, Ø, Harper, DA & Ryan, PD (2001) PAST : Paleontological statistics software package for education and data analysis. Palaeontologia Electronica, 4(1), 1-9.).
The phylogenetic tree was constructed with MEGA v 5.2.1 (Tamura et al. 2011Tamura, K, Peterson, D, Peterson, N, Stecher, G, Nei, M & Kumar, S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28(10), 2731-9.
[http://dx.doi.org/10.1093/molbev/msr121] [PMID: 21546353] ). We selected 45 in silico-translated amino acid OTUs representative sequences, and multiple alignments were performed among OTU-sequences and nosZ-sequences from several environment (soil, sediment and water column) and bacterial cultures reported in the GenBank (http//:www.ncbi.nlm.nih.gov). The tree was reconstructed by the neighbor-joining algorithm using p-distances and bootstrap resampling based on 10.000 replicates, according to Palmer et al. (2012). The Haloarcula marismortui strain ATCC 43049 (AY596297) was used as the out-group sequence. All original 454 sequences were archived at NCBI Sequence Read Archive under accession SAMN03402215, SAMN03402334, SAMN03402335.
Statistical Analysis
The Shapiro-Wilk W test was used to estimate the normal distribution of OTUs abundances data in each study area, and the Kruskal-wallis test was used to estimate the statistical difference of nosZ genes abundances among study areas; using the PAST v 3.0 (Hammer et al. 2001). To evaluate the effect of physical-chemical variables in nosZ-OTU distribution, we used the canonical correspondence analysis (CCA) using the software Canoco for Windows 4.5 (Microcomputer Power, USA) (Ter Braak et al. 2002 Tamura, K, Peterson, D, Peterson, N, Stecher, G, Nei, M & Kumar, S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28(10), 2731-9.
[http://dx.doi.org/10.1093/molbev/msr121] [PMID: 21546353] ).
RESULTS
Physicochemical Variables
Among study areas the water average temperature was 26,4ºC, Isla del Sol registered the highest value (26,8°C) when compared with the others areas. The average pH in the sampling areas was 6,37 and Tomogó registered the highest value (6, 66) when compared with the others areas (with a pH of 6.2). The contents of O2, NH4+, NO3- and NO2- showed great spatial differences among study areas. In the tree areas low levels of O2 were recorded, but Tomogó had a high O2 level (3,4mg/L) and Lozanía the lowest (0,01mg/L). Highest values of NH4+ (0,88mg/L) were reported in Tomogó, and the highest NO3- (0,78mg/L) and NO2- (0,079mg/L) levels were registered in Isla del Sol and Lozanía, respectively (Table 2).
Distribution, Abundance and Diversity of NosZ OTUs
The sequences analysis showed that Lozanía had the highest number of sequences (3,184), followed by Tomogó (2,890) and Isla del Sol (623). From them 45 species-level OTUs were obtained by grouping to 80% of similarity. The greatest OTUs richness was observed in Tomogó (28) of which 18 were area-specific, followed by Lozanía (25) of which 10 were area-specific and finally Isla del Sol (12) of which two were area-specific. The data indicated that among study areas 5 OTUs were shared, except between Isla del Sol and Tomogó that did not share OTUs (Fig. 2A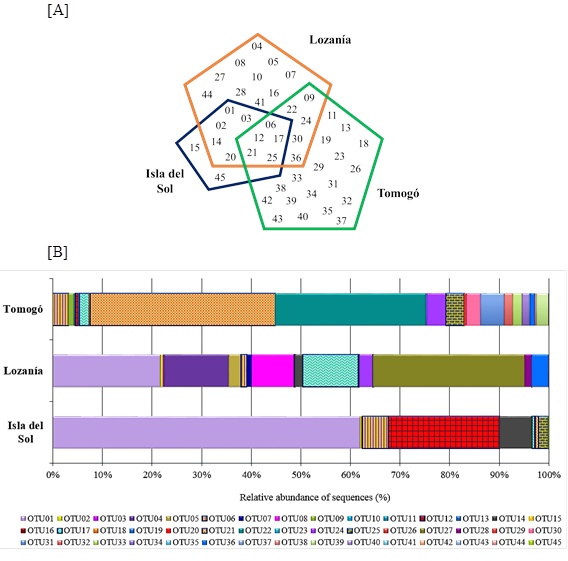 ). The average percentage of coverage, estimated with the number of OTUs (represented by one sequences) and the total number of sequences was 99,6%, indicating that the number of valid sequences used in this study was sufficient for the diversity, phylogeny and statistical analysis.
). The average percentage of coverage, estimated with the number of OTUs (represented by one sequences) and the total number of sequences was 99,6%, indicating that the number of valid sequences used in this study was sufficient for the diversity, phylogeny and statistical analysis.
Approximately 65% of the nosZ sequences of Isla del Sol, Lozanía and Tomogó, were grouped into 2 or 3 OTUs (Fig. 2B ). The OTU01 was the most abundant (62%) in Isla Del Sol, the OTU27 was the most abundant (30%) in Lozanía and OTU21 was the most abundant (40%) in Tomogó.
). The OTU01 was the most abundant (62%) in Isla Del Sol, the OTU27 was the most abundant (30%) in Lozanía and OTU21 was the most abundant (40%) in Tomogó.
Measurements of nosZ-OTUs diversity (CHAO1 estimator and Shannon index) from Lozanía and Tomogó were consistently higher than those of Isla del Sol; and the Bray Curtis similarity index was 0, 03 and 0,07 between Tomogó-Isla del Sol and Tomogó-Lozanía, respectively, suggesting differences among stations (Table 2). The Shapiro-Wilk W test and the Kruskal-Wallis anova established significant differences between Isla del Sol and Lozanía (p-value: 0,003) and between Isla del Sol and Tomogó (p-value: 0, 0002).
 |
Fig. (2) Composition of nosZ-type OTUs from study areas of Prado reservoir (A): Number of nosZ OTUs founded in the study areas. (B): Relative abundance of nosZ OTUs between study areas. |
Phylogenetic Analysis of nosZ OTUs
Phylogenetic analysis was done with 45 representative sequences of OTUs. In the phylogenetic tree two clusters were generated (Fig. 3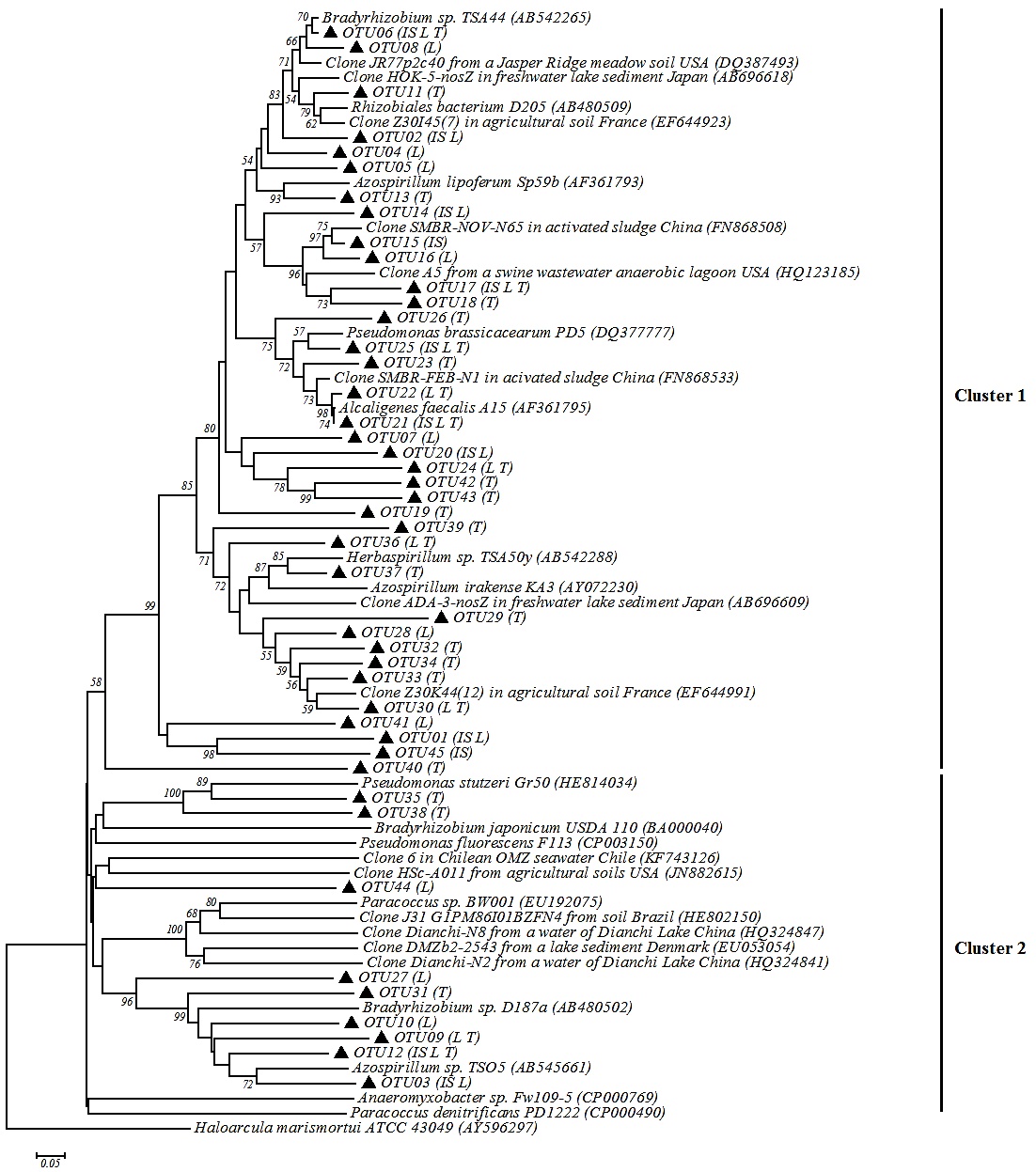 ): the cluster one included 53% of the nosZ-OTUs sequences (OTU02, OTU6, OTU15, OTU33 and OTU34) which shared 60%-100% similarity with sequences of clones from soils, sediments, sewage and activated sludge. The 47% of remaining representative sequences had 80%-100% similarity with sequences of microorganisms isolated and cultured. Some of these OTUs were 80%-100% similar with Proteobacteria nosZ sequences, such as OTU21 with Alcaligenes faecalis A15 (100%); OTU04, OTU08 and OTU11 with Bradyrhizobium sp. TSA44 (80%, 90% and 87%); and OTU21, OTU22 and OTU25 with Pseudomonas brassicacearum PD5 (90% or 88%).
): the cluster one included 53% of the nosZ-OTUs sequences (OTU02, OTU6, OTU15, OTU33 and OTU34) which shared 60%-100% similarity with sequences of clones from soils, sediments, sewage and activated sludge. The 47% of remaining representative sequences had 80%-100% similarity with sequences of microorganisms isolated and cultured. Some of these OTUs were 80%-100% similar with Proteobacteria nosZ sequences, such as OTU21 with Alcaligenes faecalis A15 (100%); OTU04, OTU08 and OTU11 with Bradyrhizobium sp. TSA44 (80%, 90% and 87%); and OTU21, OTU22 and OTU25 with Pseudomonas brassicacearum PD5 (90% or 88%).
The cluster 2 grouped 9 OTUs the most of which had low similarity with Proteobacteria group (<50% similarity). Some OTUs (OTU03, OTU09, OTU10, OTU12 and OTU31) had a 40% and 60% similarity with Azospirillum sp TSO5. All OTUs showed <45% similarity with sequences of clones isolated from soil, seawater, freshwater and sediment.
Canonical Correspondence Analysis
In the analysis of the relationship among the composition of the nosZ community and the physicochemical variables in each study area, we observed that the studied environmental variables (oxygen, pH, nitrite and nitrate) explained the structure of the nosZ-community (Fig. 4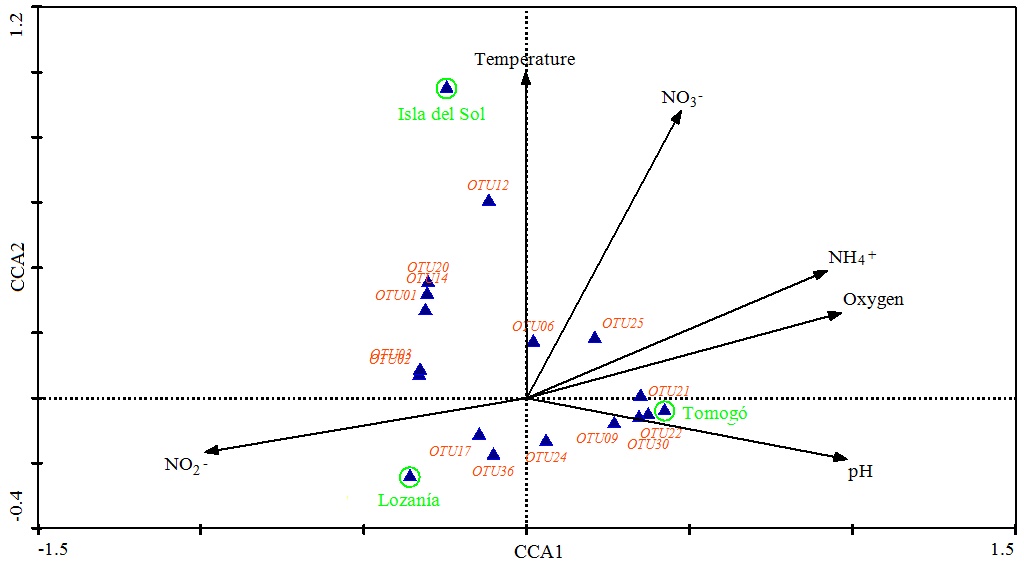 ). Both axes explained 100% of total variation. The first axis explained 67, 3% of variation and was dominated by pH (weight correlation: 0, 98), O2 (weight correlation: 0, 96) and NO2- (weight correlation: -0, 98). The second axis explained 32, 7% of variation and was dominated by NO3- (weight correlation: 0, 88) and temperature (weight correlation: 0,8).
). Both axes explained 100% of total variation. The first axis explained 67, 3% of variation and was dominated by pH (weight correlation: 0, 98), O2 (weight correlation: 0, 96) and NO2- (weight correlation: -0, 98). The second axis explained 32, 7% of variation and was dominated by NO3- (weight correlation: 0, 88) and temperature (weight correlation: 0,8).
 |
Fig. (4) Canonical correspondence analysis (CCA) of OTUs data and physicochemical variables in the three areas. Eigenvalues (500 runs), 0.703 and 0.342 for dimensions CCA1 and CCA2, respectively. |
We observed that nosZ-communities composition from Isla del Sol were related with high NO3- and temperature levels, the communities composition in Tomogó were related with high pH and NH4+ levels, and intermediate oxygen level and the nosZ-community composition from Lozanía was associated with low O2 and NO3-and higher NO2- level.
DISCUSSION
This is the first study in one Colombian’s dam whose goal was explore the spatial variation of denitrifying communities using the nosZ functional gene that codes for nitrous oxide reductase. The values of temperature, pH and nitrate reported in the surface water at the study areas (Isla del Sol, Lozanía and Tomogó) are slightly lower than those reported previously (Canosa and Pinilla 2007, Castro-González 2014, Perea and Villanueva 2010) supporting the temporal variability recorded in the area (Guevara Guevara, G, Lozano, P, Reinoso, G & Villa, F (2009) Horizontal and seasonal patterns of tropical zooplankton from the eutrophic Prado Reservoir (Colombia). Limnologica - Ecology and Management of Inland Waters, 39(2), 128-39.et al. 2009). Although profiles along water column of O2 and nutrients are very limited for this dam, the oxygen, nitrate and ammonium levels quantified during this study support the hypoxic (even suboxic) and eutrophic condition of this reservoir which has been reported several decades ago (Ducharme 1975, SODEIC 1993, Roldán 2003)
The molecular analysis indicated that Lozanía contributed with a high number of sequences (3, 184), followed by Tomogó (2, 890) and Isla del Sol (623). Likewise, the richness and diversity analysis showed high values at Lozanía and Tomogó, and low values at Isla del Sol. Both results suggested spatial variation of nosZ denitrifying communities in the Prado dam. This data are related with the previous report of Canosa and Pinilla (2007) whose quantified low number of heterotrophs in the center of the dam in comparison with other stations, which was associated with lower chlorophyll level. Our results are interesting if we consider that previous studies had also reported spatial variability in dissolved organic carbon (DOC) availability which is a key substrate for the heterotrophic activity (Guevara et al. 2009Guevara, G, Lozano, P, Reinoso, G & Villa, F (2009) Horizontal and seasonal patterns of tropical zooplankton from the eutrophic Prado Reservoir (Colombia). Limnologica - Ecology and Management of Inland Waters, 39(2), 128-39.). In this sense it is notorious that Guevara et al. (2009) recorded the highest DOC values at Tomogó (up to 11 mg/L), followed by Lozanía (9 mg/L) and by Isla del Sol (8mg/L) which are in agreement with our observations about a higher nosZ - diversity in the first two places where low O2 levels plus enough organic substrate might be facilitating its metabolic activity.
Also, the turbidity could be another key factor in the study area because denitrifying microorganisms usually are associated to particles in suspension, where they found reduced niches required to sustain suboxic processes such as denitrification (Alldredge and Cohen 1987Alldredge, AL & Cohen, Y (1987) Can microscale chemical patches persist in the sea? Microelectrode study of marine snow, fecal pellets. Science, 235(4789), 689-91.
[http://dx.doi.org/10.1126/science.235.4789.689] [PMID: 17833630] ). In this sense, it is possible that the high values of turbidity reported for Lozanía and Tomogó by influence of tributaries (Guevara et al. 2009) could also be related with the higher values of diversity registered in both areas by particle-attached communities in comparison with Isla del Sol where the low levels of particles in suspension could be related with a free-living nosZ-denitrifying community less rich and abundant. In this scenario, the formation of microhabitats as well as the development of segregated niche in the water column of this reservoir could be taking place such as has been proposed for other aquatic environments (Jones et al. 2013, Wang et al. 2013Wang, C, Zhu, G, Wang, Y, Wang, S & Yin, C (2013) Nitrous oxide reductase gene (nosZ) and N2O reduction along the littoral gradient of a eutrophic freshwater lake. Journal of Environmental Science (China), 25(1), 44-52.
[http://dx.doi.org/10.1016/S1001-0742(12)60005-9] [PMID: 23586298] ).
For other hand, although our analysis suggested that the number of sequences used in this study were optimal for the number of OTUs detected in a 99,6% (Palmer and Horn 2012), one has to bear in mind the biases associated with the nosZ-gene amplification and sequencing because they may be influencing the differential amplification of some OTUs in each study area as has been reported by Magalhães et al. (2008) and Throbäck et al. (2004)Throbäck, IN, Enwall, K, Jarvis, A & Hallin, S (2004) Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiology Ecology, 49(3), 401-17.
[http://dx.doi.org/10.1016/j.femsec.2004.04.011] [PMID: 19712290] . Another possible explanation depends on how many nosZ-sequences are detected by our primers, in each study area; and whether the denitrifying genes are present or not with enough abundance. However, the low diversity detected at Isla del Sol in this study is consistent with previous results where a similar number of OTUs were detected to 9 m depth through nosZ-TRFLP analysis with other primers (Castro-González 2014).
This is one of the first studies which reports nosZ-OTUs generated with a 80% similarity between sequences obtained from water column of a dam and although this distance has been recommended for soil studies (Palmer et al. 2009) in this case it was successful to give us a good idea of nosZ-denitrifying diversity. Our results indicated that the OTUs richness founded in the Prado reservoir was similar to these reported from soils, where between 10 to 53 OTUs had been generated (Depkat-Jakob et al. 2012Depkat-Jakob, PS, Brown, GG, Tsai, SM, Horn, MA & Drake, HL (2013) Emission of nitrous oxide and dinitrogen by diverse earthworm families from Brazil and resolution of associated denitrifying and nitrate-dissimilating taxa. FEMS Microbiology Ecology, 83(2), 375-91.
[http://dx.doi.org/10.1111/j.1574-6941.2012.01476.x] [PMID: 22928867] , Palmer et al. 2012Palmer, K, Biasi, C & Horn, MA (2012) Contrasting denitrifier communities relate to contrasting N2O emission patterns from acidic peat soils in arctic tundra. The ISME Journal, 6(5), 1058-77.
[http://dx.doi.org/10.1038/ismej.2011.172] [PMID: 22134649] ); which suggest that in the column water of eutrophic environments can also inhabit a high diversity of organisms. In comparison with reports about nosZ-diversity in freshwater sediments (Huang et al. 2011Huang, S, Chen, C, Wu, Q, Zhang, R & Yang, X (2011) Distribution of typical denitrifying functional genes and diversity of the nirS-encoding bacterial community related to environmental characteristics of river sediments. Biogeosciences Discussions, 8(3), 5251-80.
[http://dx.doi.org/10.5194/bgd-8-5251-2011] , Jones and Hallin 2010, Magalhães et al. 2008, Palmer and Horn 2012Villa, FA & Losada, S (2004) Aspectos bioecológicos del Caloche Sternopygus macrurus (Gymnotiformes: Sternopygidae), en la represa de Prado, Tolima, Colombia. Dahlia-Revista de la Asociación Colombiana de Ictiología, 7, 49-56.) we detect a similar diversity, however, there are no studies about diversity of denitrifying communities in the water column of other dams or reservoirs for comparison. Likewise, our data suggest the coexistence of a variety of nosZ-communities to suboxic depths (6-9 m depth) in the Prado dam, which have the gene that code for reductase of nitrous oxide with the potential to develop N2O reduction; however, it is necessary to develop more studies to determine the active transcription and functionality of this gene in the sampling areas.
Among the study areas, 5 OTUs (06, 12, 17, 21 and 25) were present in all areas and others OTUs were specific of each area (30 OTUs). This data suggest that in this dam we can observe organisms generalists and specialists such as has been recently proposed for denitrifiers by Bowen et al. (2013Bowen, JL, Byrnes, JE, Weisman, D, Colaneri, C & Jones, C (2013) Functional gene pyrosequencing and network analysis: an approach to examine the response of denitrifying bacteria to increased nitrogen supply in salt marsh sediments. Frontiers in Microbiology, 4(342), 1-12.
[http://dx.doi.org/10.3389/fmicb.2013.00342] [PMID: 24348464] ) where the generalists can inhabit in a wide range of habitats unlike specialists. We observe the presence of many OTUs with low number of sequences (36 of 45 OTUs) which also has been reported in other studies as “rare biosphere” (Bowen et al. 2013, Huse et al. 2010Huse, SM, Welch, DM, Morrison, HG & Sogin, ML (2010) Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environmental Microbiology, 12(7), 1889-98.
[http://dx.doi.org/10.1111/j.1462-2920.2010.02193.x] [PMID: 20236171] , Kalscheur et al. 2012Kalscheur, KN, Rojas, M, Peterson, CG, Kelly, JJ & Gray, KA (2012) Algal exudates and stream organic matter influence the structure and function of denitrifying bacterial communities. Microbial Ecology, 64(4), 881-92.
[http://dx.doi.org/10.1007/s00248-012-0091-1] [PMID: 22828897] ). This information suggest that several factor prevalent in the reservoir could favor the differentiation of ecological niches for different types of microorganisms, such as a wide range of substrates available, an intricate functional and ecological network between denitrifying bacteria, greater biological effectiveness (present only in specialists) and a great adaptive and evolutionary capacity, as has been proposed by others authors (Koeppel and Wu 2012Koeppel, AF & Wu, M (2012) Lineage-dependent ecological coherence in bacteria. FEMS Microbiology Ecology, 81(3), 574-82.
[http://dx.doi.org/10.1111/j.1574-6941.2012.01387.x] [PMID: 22486161] , Logares et al. 2013Logares, R, Lindström, ES, Langenheder, S, Logue, JB, Paterson, H, Laybourn-Parry, J, Rengefors, K, Tranvik, L & Bertilsson, S (2013) Biogeography of bacterial communities exposed to progressive long-term environmental change. The ISME Journal, 7(5), 937-48.
[http://dx.doi.org/10.1038/ismej.2012.168] [PMID: 23254515] , Matulich 2013Matulich, KL (2013) Microbial Biodiversity. Encyclopedia of Biodiversity, 5, 252-58.).
The phylogenetic analysis showed that a few OTUs were 100% similar to previously reported sequences of Alcaligenes faecalis A15, Bradyrhizobium sp. TSA44 and Pseudomonas brassicacearum PD5. Our data are in accordance with preliminary reports about of the Proteobacteria that are ubiquitous and widely distributed in reservoirs, lakes, hot springs, activated sludge and soils (Hou et al. 2012Hou, J, Li, L, Zhang, S, Wang, P & Wang, C (2012) Diversity of NosZ gene in three municipal wastewater treatment plants located in different geographic regions. African Journal of Microbiology Research, 6(15), 3574-81., Wang et al. 2013Wang, C, Zhu, G, Wang, Y, Wang, S & Yin, C (2013) Nitrous oxide reductase gene (nosZ) and N2O reduction along the littoral gradient of a eutrophic freshwater lake. Journal of Environmental Science (China), 25(1), 44-52.
[http://dx.doi.org/10.1016/S1001-0742(12)60005-9] [PMID: 23586298] , Yu et al. 2014). In this regard, in the Baiyangdian lake sediments and at Dianchi lake water column, affiliations of OTUs to Rhizobiales group (Wang et al. 2013Wang, C, Zhu, G, Wang, Y, Wang, S & Yin, C (2013) Nitrous oxide reductase gene (nosZ) and N2O reduction along the littoral gradient of a eutrophic freshwater lake. Journal of Environmental Science (China), 25(1), 44-52.
[http://dx.doi.org/10.1016/S1001-0742(12)60005-9] [PMID: 23586298] , Wen et al. 2012Villa, FA & Losada, S (2004) Aspectos bioecológicos del Caloche Sternopygus macrurus (Gymnotiformes: Sternopygidae), en la represa de Prado, Tolima, Colombia. Dahlia-Revista de la Asociación Colombiana de Ictiología, 7, 49-56.) were recorded. Likewise, in the water column of Lake Dianchi (Wen et al. 2012) and wastewater (Hou et al. 2012) has been reported affiliation of OTUs with Burkholderiales and Pseudomonadales groups, respectively.
The presence of several OTUs (for example OTUs 45, 41, 15) with low similarity to Proteobacteria and high similarity to sequences of environmental clones reported previously, suggest that in the Prado dam a nosZ-community exists which is phylogenetically different and unique such as that been recorded in other natural environments (Bowen et al. 2013Bowen, JL, Byrnes, JE, Weisman, D, Colaneri, C & Jones, C (2013) Functional gene pyrosequencing and network analysis: an approach to examine the response of denitrifying bacteria to increased nitrogen supply in salt marsh sediments. Frontiers in Microbiology, 4(342), 1-12.
[http://dx.doi.org/10.3389/fmicb.2013.00342] [PMID: 24348464] , Jones et al. 2013Jones, CM, Graf, DR, Bru, D, Philippot, L & Hallin, S (2013) The unaccounted yet abundant nitrous oxide-reducing microbial community: a potential nitrous oxide sink. The ISME Journal, 7(2), 417-26.
[http://dx.doi.org/10.1038/ismej.2012.125] [PMID: 23151640] , Jones and Hallin 2010Jones, CM & Hallin, S (2010) Ecological and evolutionary factors underlying global and local assembly of denitrifier communities. The ISME Journal, 4(5), 633-41.
[http://dx.doi.org/10.1038/ismej.2009.152] [PMID: 20090785] ). This data indicate the coexistence of different bacteria in the reservoir with the potential to perform the reduction of N2O to N2, probably generating a functional redundancy that stabilizes the denitrification process under changing conditions (fluctuations of O2 and nutrients), like that suggested for other habitats (Hou et al. 2012).
The environmental parameters selected as predictor variables (O2, pH, temperature, NO3-, NH4+ and NO2-) explained the differences in nosZ-denitrifying community composition among sampling sites at the Prado dam. The nosZ-community composition was associated with hypoxic (~3.4mg/L O2) and suboxic (0.01mg/L O2) conditions recorded at Tomogó and Lozanía respectively, which support previous data about the presence of denitrifiers in the water column to O2 levels between 1.1 - 4.1 mg/L (Castro-González 2014Castro-González, M (2014) Evaluación de la comunidad desnitrificante tipo nosZ en la columna de agua de Isla del Sol, embalse de Prado, Tolima Revista de La Academia Colombiana de Ciencias Exactas, Físicas Naturales, 38(149), 385-92.). In general, our results corroborate that the composition of nosZ-denitrifying microorganisms is related to low oxygen environments such as those founded in rivers (Baxter et al. 2013Baxter, AM, Johnson, L, Royer, T & Leff, LG (2013) Spatial differences in denitrification and bacterial community structure of streams: relationships with environmental conditions. Aquatic Science, 75(2), 275-84.
[http://dx.doi.org/10.1007/s00027-012-0272-5] ), estuaries (Magalhães et al. 2008Magalhães, C, Bano, N, Wiebe, WJ, Bordalo, AA & Hollibaugh, JT (2008) Dynamics of nitrous oxide reductase genes (nosZ) in intertidal rocky biofilms and sediments of the Douro River estuary (Portugal), and their relation to N-biogeochemistry. Microbial Ecology, 55(2), 259-69.
[http://dx.doi.org/10.1007/s00248-007-9273-7] [PMID: 17604988] ), wastewater (Hou et al. 2012), lakes (Wen et al. 2012) and reservoirs (Yu et al. 2014).
At Isla del Sol was founded a nosZ-community less diverse and rich whose composition was related with higher levels of NO3-, contrarily to observed at Lozanía were the nosZ community composition showing an inverse relationship with the nitrate levels. It is well known that the nitrate (NO3-) is required by denitrifying communities in estuaries, rivers, lakes and reservoirs (Zhao et alZehr, JP & Kudela, RM (2011) Nitrogen cycle of the open ocean: from genes to ecosystems. Annual Review of Marine Science, 3(1), 197-225.
[http://dx.doi.org/10.1146/annurev-marine-120709-142819] [PMID: 21329204] . 2015, Baxter et al.Baxter, AM, Johnson, L, Royer, T & Leff, LG (2013) Spatial differences in denitrification and bacterial community structure of streams: relationships with environmental conditions. Aquatic Science, 75(2), 275-84.
[http://dx.doi.org/10.1007/s00027-012-0272-5] 2013, Magalhães et al. 2008, Wen et al. 2012) and for this case the results suggest that the denitrifying community at Lozanía could be using the nitrate actively because was evident that this ion decreased to 0.25 mg/L in comparison with the recorded at Isla del Sol (0.78 mg/L NO3-). Our data are similar to those reported by Magalhães et al. (2008) in the Douro estuary (Portugal) and corroborate arguments about different nosZ-groups could have different competitive advantages for denitrification when nitrate levels fluctuate in a system (Jones and Hallin 2010, Magalhães et al. 2008).
For the reservoir, nitrate levels are related to the planktonic and benthic activities (Márquez and Guillot 2001), and the increase in nitrate level observed at Isla del Sol may be due to a higher nitrifying (to 1.2mg/L O2) than denitrifying activity, to difference that observed at Lozanía where the lower O2 level (0.01mg/L) favored the denitrification by a great variety of nosZ-OTUs. Our assessment is supported by studies showing as both ammonia and nitrite oxidation occurred at very low oxygen concentrations (even < 50 µM) in marine (Peng et al.Peng, X, Fuchsman, CA, Jayakumar, A, Oleynik, S, Martens-Habbena, W, Devol, AH & Ward, BB (2015) Ammonia and nitrite oxidation in the Eastern Tropical North Pacific. Global Biogeochemical Cycles, 29
[http://dx.doi.org/10.1002/2015GB005278] 2015) and freshwater ecosystems (Wetzel 2001Wang, C, Zhu, G, Wang, Y, Wang, S & Yin, C (2013) Nitrous oxide reductase gene (nosZ) and N2O reduction along the littoral gradient of a eutrophic freshwater lake. Journal of Environmental Science (China), 25(1), 44-52.
[http://dx.doi.org/10.1016/S1001-0742(12)60005-9] [PMID: 23586298] , Castro-González and Torres 2015Chon, K, Chang, JS, Lee, E, Lee, J, Ryu, J & Cho, J (2011) Abundance of denitrifying genes coding for nitrate (narG), nitrite (nirS), and nitrous oxide (nosZ) reductases in estuarine versus wastewater effluent-fed constructed wetlands. Ecological Engineering, 37(1), 64-9.
[http://dx.doi.org/10.1016/j.ecoleng.2009.04.005] ).
For other hand, the nosZ genes composition -in Tomogó- was associated with a pH level of 6,7 which are in accordance with a previous report where nosZ community was active and diverse in a wide range of pH (6.1-8) (Palmer et al. 2012Palmer, K, Biasi, C & Horn, MA (2012) Contrasting denitrifier communities relate to contrasting N2O emission patterns from acidic peat soils in arctic tundra. The ISME Journal, 6(5), 1058-77.
[http://dx.doi.org/10.1038/ismej.2011.172] [PMID: 22134649] ). The effect of pH on denitrification enzyme activity and diversity of denitrifying communities has also been reported for bacterial cultures (Saleh-lakha et al. 2009Saleh-Lakha, S, Shannon, KE, Henderson, SL, Goyer, C, Trevors, JT, Zebarth, BJ & Burton, DL (2009) Effect of pH and temperature on denitrification gene expression and activity in Pseudomonas mandelii. Applied and Environmental Microbiology, 75(12), 3903-11.
[http://dx.doi.org/10.1128/AEM.00080-09] [PMID: 19376915] ) and soils respectively (Enwall et al. 2005Enwall, K, Philippot, L & Hallin, S (2005) Activity and composition of the denitrifying bacterial community respond differently to long-term fertilization. Applied and Environmental Microbiology, 71(12), 8335-43.
[http://dx.doi.org/10.1128/AEM.71.12.8335-8343.2005] [PMID: 16332820] ).
The bacterial diversity and composition between Isla del Sol and Tomogó differed significantly (β-diversity), although both localities are not so far between them. This suggest that differences in the studied physical-chemical variables and other factors not analyzed in this report such as availability of organic matter and dissolved organic carbon, particulate carbon/dissolved carbon ratio, oxygen and nutrients dial fluctuations and interaction with other micro-communities could be contributing to the differences founded in the structure of the microbial community between study areas.
CONCLUSION
This study is the first report that analyzes the microbial composition of a nitrogen functional community related with the reduction of the greenhouse gas, nitrous oxide to N2. Our data demonstrate that such community is present, is diverse, rich and that its composition varies spatially in relationship with the environmental factors. This information is the base for future studies about of the activity and importance of this communities in the cycling of N2O from this dam that will permit to determine if this reservoir is a source or sink of this greenhouse gas.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
ACKNOWLEDGEMENTS
The Office of Research and Scientific Development of University of Tolima, funded this research. We are grateful to the Colombian Ministry of Environment who authorized the access and use of environmental bacterial DNA and CORTOLIMA who authorized this research in biological diversity. We thank A. Castro, J.C. Duque and A. Meneses for their help in the field sampling and Dr. J.F. Alzate for his advice about the bioinformatics analysis.







