- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Evidence of BVDV in Pigs from North Eastern Part of India- Genetic Profiling and Characterisation
Amit Kr Chakraborty1, 2, Priyanka Mukherjee1, 2, Amarjit Karam1, Samir Das1, Luit Barkalita3, Kekungo Puro1, Rajkumari Sanjukta1, Sandeep Ghatak1, Ingudam Sakuntala1, Ram Gopal Laha1, Prabodh Borah3, S.V. Ngachan1, Indu Sharma2, Arnab Sen1, *
Abstract
Introduction:
The work has been attempted to detect and genetically characterise the nature of Bovine Viral Diarrhea Virus (BVDV) isolates from the porcine population of the north east.
Methods and Material:
The samples have been collected over a two year period and are from areas where there is a mixed and integrated rearing of livestock in close proximity. The isolates were identified, cloned and sequenced using BVD specific genomic primers for two important domains viz., E-2 and 5’ UTR.
Results:
Porcine BVD Sequences were analysed phylogenetically. Divergence in 3 sequences is noted in the 5’ UTR region that are forming a clear outlier group while E-2 sequences are coming close to BVDV group but forming a separate cluster.
Article Information
Identifiers and Pagination:
Year: 2018Volume: 12
Issue: Suppl-2, M7
First Page: 110
Last Page: 120
Publisher Id: TOVJ-12-110
DOI: 10.2174/1874357901812010110
Article History:
Received Date: 16/4/2017Revision Received Date: 6/7/2018
Acceptance Date: 16/7/2018
Electronic publication date: 31/08/2018
Collection year: 2018
open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution 4.0 International Public License (CC-BY 4.0), a copy of which is available at: (https://creativecommons.org/licenses/by/4.0/legalcode). This license permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
* Address correspondence to this author at the Division of Animal Health, ICAR Research Complex for NEH, Barapani - 793103, India; E-mail: arnabsen123@gmail.com
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 16-4-2017 |
Original Manuscript | Evidence of BVDV in Pigs from North Eastern Part of India- Genetic Profiling and Characterisation | |
1. INTRODUCTION
Interspecies viral spill-over is responsible for most of the emerging infections. Emergence of new disease or new viral pathogens is often a result of species jumping or cross species transmission of viral pathogens.Successful cross species transmission of a virus is a relatively rare phenomenon as it must overcome the ecological and evolutionary barriers in the process to establish itself into a new host [1Geoghegan JL, Senior AM, Holmes EC. Pathogen population bottlenecks and adaptive landscapes: Overcoming the barriers to disease emergence. Proc R SocLond B 1837; 2016: 283.]. This phenomenon can play a major role in Virus evolution particularly in case of retroviruses which is related to their host cell morphology suggesting their long term Co divergence as per their phylogenetic tree [2Jackson AP, Charleston MA. A cophylogenetic perspective of RNA-virus evolution. Molecular biologyand evolution 2004; 21(1): 45-57.], while in case of Flaviviruses host switch over is more frequent among other RNA viruses as many of them transmitted by arthropod vectors and cause a short term infection [3Kitchen A, Shackelton LA, Holmes EC. Family level phylogenies reveal modes of macroevolution in RNA viruses 2011; 108(1): 238-43.
[http://dx.doi.org/10.1073/pnas.1011090108] ].
Bovine Viral Diarrhoea Virus (BVDV), a member of Flaviviridae family under genus Pestivirus, one of themost clinically and economically important ruminant pathogen. It was confirmed that BVDV and CSFV (another Pestivirus cause havoc mortality in pigs worldwide) shares cross reactive antigens [4Darbyshire JH. A serological relationship between swine fever andmucosal disease of cattle. Vet Rec 1960; 72: 331.]. Fernetius et al. in 1973 sucessfully isolated BVDV from pigs in Australia. In Asia BVDV in pigs was first reported in 1996 [5Wang XP, Tu CC, Li HW, et al. Pigs naturally infected by bovine diarrhea virus present signs resembling hog cholera. Chin J Vet Sci 1996; 16: 341-5.] in China. In India however the scenario is quite unknown. Pestivirus like BVDV, CSFV and BDV are antigenically close to each other and cross reaction or species spill over are often found [6Becher P, Avalos Ramirez R, Orlich M, et al. Genetic and antigenic characterization of novel pestivirus genotypes: Implications for classification. Virology 2003; 311(1): 96-104.
[http://dx.doi.org/10.1016/S0042-6822(03)00192-2] [PMID: 12832207] ].
Integrated farming systems are the mainstay of north eastern agriculture sector. Co-habitation of small livestock holds is quite common place. It is very common to see a herd of cattle co-existing with porcine in the near vicinity. This practice is unique to the region and could well serve in bridging niche-gaps leading to cross species transmission. We conducted this study here to observe if any such scenario is happening.
BVDV can be found in cattle, sheep, swine, goats, and other wild animals [7Ridpath JF. Bovine viral diarrhea virus: Global status. Vet Clin North Am Food Anim Pract 2010; 26(1): 105-21.
[http://dx.doi.org/10.1016/j.cvfa.2009.10.007] [PMID: 20117546] ]. BVDV in pigs can create smiler symptoms like classical swine fever and thus cause difficulty while differentiating from each other [8Passler T, Walz PH. Bovine viral diarrhea virus infections in heterologous species. Anim Health Res Rev 2010; 11(2): 191-205.
[http://dx.doi.org/10.1017/S1466252309990065] [PMID: 19883524] ]. But mainly BVDV in pigs shows no significant signs thus it has the advantage to spread without getting noticed [9Le Potier M, Mesplede A, Vanner P. Classical swine fever other pestiviruses.Diseases of Swine 9th ed. 2006; 309-11.], [10Liess B, Moennig V. Ruminant pestivirus infection in pigs. Rev - Off Int Epizoot 1990; 9(1): 151-61.
[http://dx.doi.org/10.20506/rst.9.1.484] [PMID: 1966720] ].
2. MATERIAL AND METHODS
2.1. Origin of Samples
Pig serum samples (n=206) were randomly collected from villages or from organised farms of Meghalaya (n=82), Assam (n=31), Mizoram (n=44), Nagaland (n=6), Manipur (n=12) and Arunachal Pradesh (n=31) for a period of two years (2013-15). Each sample was aseptically collected from ear veins in a vacutainer using sterile precaution and was transferred in -200C gel packs to the laboratory within 24 to 48 hours. The samples were collected over a two year period (2013 – 2015). These samples were collected from areas that had integrated farming operations and a mixed livestock population comprising of bovines, porcines and poultry. In the north eastern region small holder farms with livestock being reared in close proximity is quite common. Hence the samples were obtained from these farms or zones of integrated farming practice.
The protocols followed in the study were OIE approved as per genomic detections of BVD in are concerned. (OIE Manual, 2015). Standardised primers and validated methods of detection of BVD in porcines as reported by earlier workers were followed.
2.2. Isolation of RNA and Detection by RT PCR
Viral RNA were extracted from serum samples using Qia Amp Viral RNA mini kit (Qiagen, Germany) following manufacturer’s instructions and subjected to the synthesis of cDNA using Revert Aid First strand cDNA synthesis kit (Thermoscientific, USA) with random hexamer primers. These cDNA’s were then checked with a set of BVDV specific nested primers namely Pan Pesti and then Pan BVDV and Pan CSFV (Table 1) to detect the presence of BVDV and CSFV respectively by amplifying the 5’UTR region of both the viruses, as both the specific primers are specific for a partial region of 5’UTR. The initial identity was checked again using PnPesti, subsequently PnBVDV and PnCSF were used to ascertain whether the nucleic acids were from BVD and/or CSFV. The cyclic conditions which are same for the three sets of primers are primary denaturation at 950C for 3 minutes followed by 40 cycles at 950C for 30 second, 550C for 30 seconds, 720C for 30seconds and final extension at 720C for 5 minutes. DreamTaq green from Thermoscientific, USA was used for PCR. The products were then observed in a 2% agarose gel. After detection partial E2 region of BVDV was amplified from PCR positive samples for further phylogenetic study.
2.3. Detection and Quantification of BVDV by RT qPCR
The positive samples from conventional PCR were subjected for absolute quantification using Anigen BVDV Real-Time Detection Kit from Bionote, Korea (cat no. PD65 050N) in a standard curve method following manufacturer’s instruction. qPCR was performed using Step One Plus Real Time PCR machine from Applied Biosystems, USA. The control positive BVDV RNA was supplied by the company with the kit. The experiment was cross checked using 10 RNA samples which were negative for BVDV and positive for CSFV as negative controls to determine kit’s specificity and confidence level. The negative controls were confirmed by conventional PCR using BVDV and CSFV 5’UTR detection primers listed in Table 1. The Taqman primer probe was directed against specific genomic regions of BVDV albeit the exact genetic sequences of the primer probe sets were not disclosed by the manufacturer.
2.4. Cloning, Nucleotide Sequencing and Phylogenetic Analysis
The positive RNA samples obtained in Taqman real time PCR kit were then further amplified with Pan Pesti forward and PnBVDV reverse primer in a hemi nested manner. A 232bp PCR product (5’NTR) and a 606bp PCR product (E2) then purified with TaKaRa PCR purification kit and purified fragment then ligated with pTZ57R/T (plasmid) vector (Thermoscientific, USA) and then DH5α competent cells (prepared with CaCl2treatment) were transformed with the plasmid containing insert fragment. Transformed colonies were isolated by blue white screening and plasmid were extracted with plasmid mini kit (Thermoscientific, USA) and then sequencing was done with ABI Big Dye Terminator v3.1 cycle sequencing kit (Applied Biosystems, USA) in ABI 3500xl genetic analyser. The sequences were assembled using Seqman software (DNASTAR Inc., Madison, USA). Additional pestiviral sequences were retrieved from NCBI GenBank and used in subsequent analysis. A total of eight E-2 partial gene sequences and eight 5’ UTR sequences were submitted to the Gen Bank during March. 2017 and provisional accession numbers were obtained. (KY852361-69/KY836194-200). These sequences were used for building the phylogenetic tree and derived protein data.
3. RESULTS
3.1. Conventional PCR
Out of 206 porcine serum samples 10 (Meghalaya (n=4), Mizoram (n=6)) samples were showing presence of BVDV positive bands that consistently amplified with PnBVDV primers that were indicative of amplification of 5’UTR. However 8 among those 10 positive samples were also showing BVDV 606bp E2 specific bands (Fig. 1a , b
, b ).
).
3.2. Real Time PCR
These 10 samples further were confirmed as positive in Real Time PCR with Taqman BVDV detection kit. These 10 BVDV positive samples were cross checked along with 10 known negative controls for testing the specificity of the kit. Those 10known negative controls were negative in the test (Fig. 2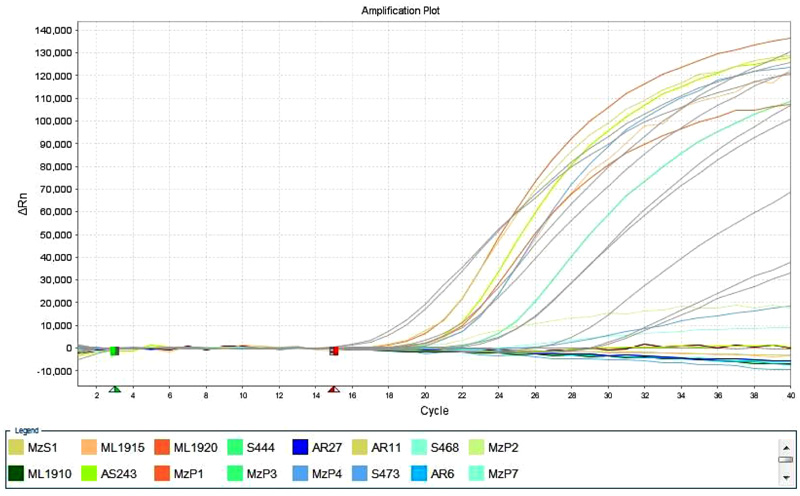 ).
).
 |
Fig. (2) Amplification plot of Taqman RT qPCR of BVDV detection kit from. |
3.3. Phylogenetic analysis
Sequences were aligned in Molecular Evolutionary Genetics Analysis version 7.0 software [13Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016; 33(7): 1870-4.
[http://dx.doi.org/10.1093/molbev/msw054] [PMID: 27004904] ] using clustal W method and tree were constructed using maximum parsimony [14Nei M, Kumar S. Molecular Evolution and Phylogenetics 2000.]. Partial E2 sequences were converted to protein sequence using online translation tools and they were further aligned to observe if there was any difference in protein levels due to species adaptation (Fig. 3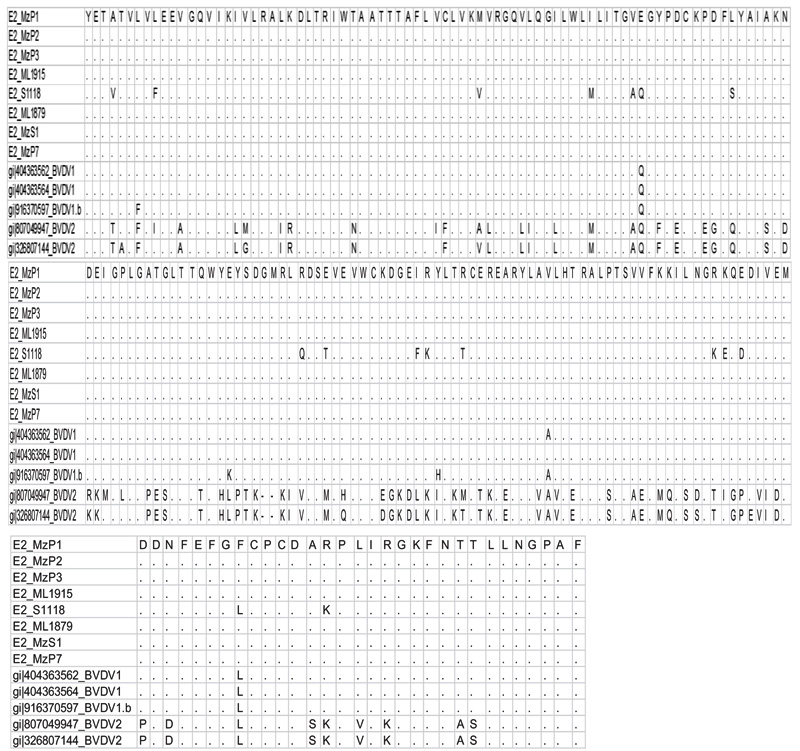 ).
).
 |
Fig. (3) The representation of the alignment of protein sequence derived from the partial E2 sequences of BVDV from porcine isolates with the online available sequences. |
A total of eight sequences for E-2 and eight sequences for 5’UTR were aligned using Clustal W method and phylogenetic trees were constructed using maximum parsimony statistical method. In the E-2 tree (Fig. 5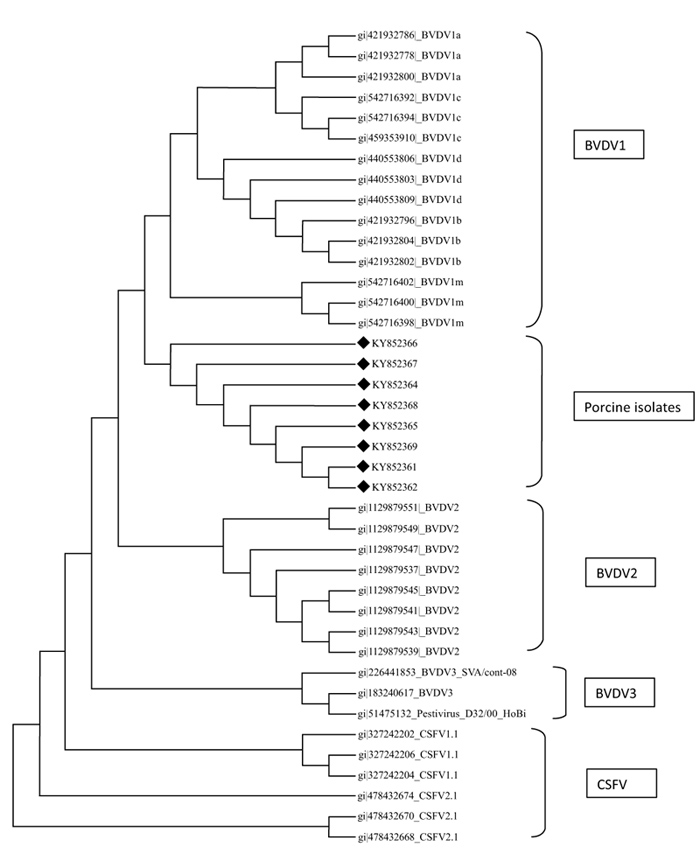 ) it was seen that all the eight porcine BVD sequences were tightly clustered and were located close to BVDV 1 group of cattle. A comparison was done with reference isolates of BVD1, 2 and 3 (Cattle), BVDV, CSFV and Giraffe pestivirus (Fig. 4
) it was seen that all the eight porcine BVD sequences were tightly clustered and were located close to BVDV 1 group of cattle. A comparison was done with reference isolates of BVD1, 2 and 3 (Cattle), BVDV, CSFV and Giraffe pestivirus (Fig. 4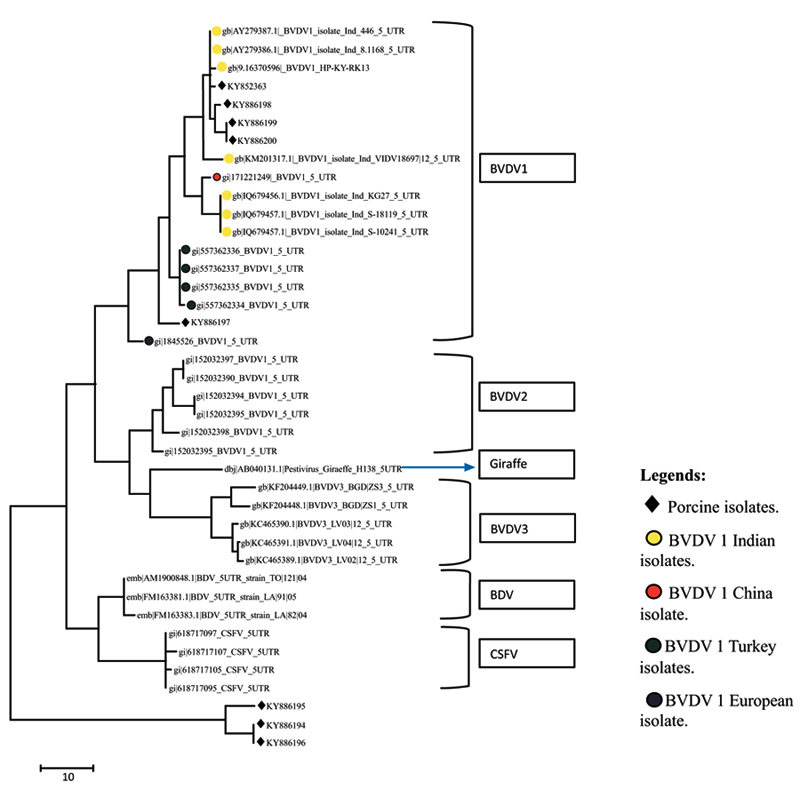 ). However the 5’ UTR tree revealed that the isolates were present in two distinct groups, i.e, five porcine BVD isolates were interspersed within BVDV type 1 cattle isolates and among them 4 porcine BVD isolates were close to previously reported Indian cattle isolates of BVDV 1and a single porcine BVD isolate was close to the Turkey isolates of cattle BVDV-1 and three porcine BVD isolates were present as an outlier cluster. The data clearly reveals that the porcine BVD isolates from Meghalaya show closeness to the cattle BVDV isolates of Indian origin.
). However the 5’ UTR tree revealed that the isolates were present in two distinct groups, i.e, five porcine BVD isolates were interspersed within BVDV type 1 cattle isolates and among them 4 porcine BVD isolates were close to previously reported Indian cattle isolates of BVDV 1and a single porcine BVD isolate was close to the Turkey isolates of cattle BVDV-1 and three porcine BVD isolates were present as an outlier cluster. The data clearly reveals that the porcine BVD isolates from Meghalaya show closeness to the cattle BVDV isolates of Indian origin.
4. DISCUSSION
Since 1960 there were a lots of work done related to pigs infected with BVDV. CSFV antibodies were found in pigs having no clinical signs raised questions and subsequently it was confirmed that the causative agent might be BVDV [15Flynn DM, Jones TE. The position regarding swine fever in Victoria. Aust Vet J 1964; 40: 131-7.
[http://dx.doi.org/10.1111/j.1751-0813.1964.tb01718.x] ]. After a long waiting BVDV could be succefully isolated from naturally infected pigs in 1973 by Fernelius et al but it was already reported back in 1968 by Snowdon and French. Detection of BVDV antibodies were used for classical swine fever quarantine [16Carbrey EA, Stewart WC, Kresse JI, Snyder ML. Natural infection of pigs with bovine viral diarrhea virus and its differential diagnosis from hog cholera. J Am Vet Med Assoc 1976; 169(11): 1217-9.
[PMID: 187565] ]. It was also confirmed that clinical cases showing symptoms of CSF were actually caused by natural BVDV infection [17Terpstra C, Wensvoort G. Bovine virus diarrhea virus infections in swine. Tijdschr Diergeneeskd 1991; 116(19): 943-8.
[PMID: 1656546] ]. Early reports suggests that BVDV infection caused in pigs due to hog cholera vaccines contaminated with BVDV [18Wensvoort G, Terpstra C. Bovine viral diarrhoea virus infections in piglets born to sows vaccinated against swine fever with contaminated vaccine. Res Vet Sci 1988; 45(2): 143-8.
[PMID: 2848299] ]. The presence of BVDV antibodies in pigs in Austria and Germany has been confirmed at 3–40% and in Holland at 15–20% [19Liess B, Moennig V. Ruminant pestivirus infection in pigs. Rev - Off Int Epizoot 1990; 9(1): 151-61.
[http://dx.doi.org/10.20506/rst.9.1.484] [PMID: 1966720] ], [20O’Connor M, Lenihan P, Dillon P. Pestivirus antibodies in pigs in Ireland. Vet Rec 1991; 129(12): 269.
[http://dx.doi.org/10.1136/vr.129.12.269] [PMID: 1660192] ], [21Terpstra C, Wensvoort G. Bovine virus diarrhea virus infections in swine. Tijdschr Diergeneeskd 1991; 116(19): 943-8.
[PMID: 1656546] ]. Seroprevalence of BVDV in domestic pigs was also recorded in Norway, Denmark, and Ireland respectively at 2.2%, 6.4% and 3.2% [22Løken T, Krogsrud J, Larsen IL. Pestivirus infections in Norway. Serological investigations in cattle, sheep and pigs. Acta Vet Scand 1991; 32(1): 27-34.
[PMID: 1659160] ], [23Jensen MH. Screening for neutralizing antibodies against hog cholera- and/or bovine viral diarrhea virus in Danish pigs. Acta Vet Scand 1985; 26(1): 72-80.
[PMID: 2994428] ], [24Graham DA, Calvert V, German A, McCullough SJ. Pestiviral infections in sheep and pigs in Northern Ireland. Vet Rec 2001; 148(3): 69-72.
[http://dx.doi.org/10.1136/vr.148.3.69] [PMID: 12503593] ]. The presence of BVDV antibodies during outbreaks of CSFV in the Netherlands caused havoc interference in the proper serological diagnosis of CSFV [25de Smit AJ, Eblé PL, de Kluijver EP, Bloemraad M, Bouma A. Laboratory decision-making during the classical swine fever epidemic of 1997-1998 in The Netherlands. Prev Vet Med 1999; 42(3-4): 185-99.
[http://dx.doi.org/10.1016/S0167-5877(99)00075-6] [PMID: 10619155] ]. Seroprevalence of BVDV in North American pig were ranging from 2% to 43%, with cattle being demonstrated as the most common source of BVDV infection in pigs [26O’Sullivan T, Friendship R, Carman S, Pearl DL, McEwen B, Dewey C. Seroprevalence of bovine viral diarrhea virus neutralizing antibodies in finisher hogs in Ontario swine herds and targeted diagnostic testing of 2 suspect herds. Can Vet J 2011; 52(12): 1342-4.
[PMID: 22654141] ]. Seroprevalance status of BVDV in pigs in China was considered threatening and it was also confirmed that BVDV1 was predominant strain in China [27Song YF, Zhang Z, Zhang YX, Wu FX, Li XC. The initial study on the prevalence of Bovine Viral Diarrhea Virus (BVDV) from swine. Chin J Anim Quarant 2008; 7: 25-7.].
 |
Fig. (4) Phylogenetic diversity of BVDV based on 5’UTR sequences. The tree constructed using maximum parsimony method in MEGA ver.7. |
 |
Fig. (5) Phylogenetic diversity of BVDV based on 5’UTR sequences. The tree constructed using maximum parsimony method in MEGA ver.7. |
The effects of intraspecies dynamics as a mediator of spill over is quite intriguing. Feral pigs can potentially become sources for disease transmission because persistent infection can occur in domestic pigs.Several experiments established the presence of BVDV in species other than cattle. Like in mountain goats [28Gaede W, Reiting R, Schirrmeier H, Depner KR, Beer M. Detection and species-specific differentiation of pestiviruses using real-time RT-PCR. Berl Munch Tierarztl Wochenschr 2005; 118(3-4): 113-20.
[PMID: 15803758] ], wild Alpine and Iberian ibex [29Fernández-Sirera L, Cabezón O, Rossi L, et al. Investigations of pestivirus infection in wild Caprinae in Europe. Vet Rec 2011; 169(1): 15.
[http://dx.doi.org/10.1136/vr.d1831] [PMID: 21709051] ]. Reports also shows that domestic goats infected with BVDV shows symptoms like abortion of production of PI kids [30Bachofen C, Vogt HR, Stalder H, et al. Persistent infections after natural transmission of bovine viral diarrhoea virus from cattle to goats and among goats. Vet Res (Faisalabad) 2013; 44: 32.
[http://dx.doi.org/10.1186/1297-9716-44-32] [PMID: 23675947] ]. Eland or Taurotragus orxy in Zimbabwe near cattle farms were found positive for BVDV antibody [31Vilcek S, Drew TW, McGoldrick A, Paton DJ. Genetic typing of bovine pestiviruses from England and Wales. Vet Microbiol 1999; 69(4): 227-37.
[http://dx.doi.org/10.1016/S0378-1135(99)00111-X] [PMID: 10535769] ]. BVDV was also confirmed in Mule deer by IHC and also suspected to be a spillover case [32Duncan C, Ridpath J, Palmer MV, Driskell E, Spraker T. Histopathologic and immunohistochemical findings in two white-tailed deer fawns persistently infected with Bovine viral diarrhea virus. J Vet Diagn Invest 2008; 20(3): 289-96.
[http://dx.doi.org/10.1177/104063870802000305] [PMID: 18460614] ]. Increasing level of BVDV infection in pigs in the last decade were recorded [26O’Sullivan T, Friendship R, Carman S, Pearl DL, McEwen B, Dewey C. Seroprevalence of bovine viral diarrhea virus neutralizing antibodies in finisher hogs in Ontario swine herds and targeted diagnostic testing of 2 suspect herds. Can Vet J 2011; 52(12): 1342-4.
[PMID: 22654141] ], [33Woods RD, Kunkle RA, Ridpath JE, Bolin SR. Bovine viral diarrhea virus isolated from fetal calf serum enhances pathogenicity of attenuated transmissible gastroenteritis virus in neonatal pigs. J Vet Diagn Invest 1999; 11(5): 400-7.
[http://dx.doi.org/10.1177/104063879901100503] [PMID: 12968752] ], [34Terpstra C, Wensvoort G. A congenital persistent infection of bovine virus diarrhoea virus in pigs: Clinical, virological and immunological observations. Vet Q 1997; 19(3): 97-101.
[http://dx.doi.org/10.1080/01652176.1997.9694750] [PMID: 9323848] ]. BVDV-1 strain ZM- 95 infections in swine population in China were first detected by Wang et al. in 1996.
The role of cattle and ruminants in harboring and disseminating BVDV is well documented in India. In the North eastern region of India the practice of composite farming is quite common, it is a matter of practice to rear both the species in close proximity hence facilitating the spread of multi host viruses.
In India, prevalence of BVDV were recorded in cattle, buffaloes, sheep and goats and all the genotypes of BVDV viz, BVDV-1, BVDV-2, and BVDV-3 have been detected [35Mishra N, Pattnaik B, Vilcek S, et al. Genetic typing of bovine viral diarrhoea virus isolates from India. Vet Microbiol 2004; 104(3-4): 207-12.
[http://dx.doi.org/10.1016/j.vetmic.2004.08.003] [PMID: 15564029] ], [36Mishra N, Rajukumar K, Vilcek S, Tiwari A, Satav JS, Dubey SC. Molecular characterization of bovine viral diarrhea virus type 2 isolate originating from a native Indian sheep (Ovies aries). Vet Microbiol 2008; 130(1-2): 88-98.
[http://dx.doi.org/10.1016/j.vetmic.2008.01.005] [PMID: 18308487] ], [37Mishra N, Rajukumar K, Pateriya A, et al. Identification and molecular characterization of novel and divergent HoBi-like pestiviruses from naturally infected cattle in India. Vet Microbiol 2014; 174(1-2): 239-46.
[http://dx.doi.org/10.1016/j.vetmic.2014.09.017] [PMID: 25301283] ]. Yaks (Bos grunniens) have been found positive for BVDV in Arunachal Pradesh state [38Mishra N, Dubey R, Rajukumar K, et al. Genetic and antigenic characterization of bovine viral diarrhea virus type 2 isolated from Indian goats (Capra hircus). Vet Microbiol 2007; 124(3-4): 340-7.
[http://dx.doi.org/10.1016/j.vetmic.2007.04.023] [PMID: 17509780] ] and Bos frontalis also known as mithun have been found positive for BVDV antibody from Nagaland, Mizoram and Arunachal Pradesh [39Singh V, Mishra N, Kalaiyarasu S, et al. First report on serological evidence of Bovine Viral Diarrhea Virus (BVDV) infection in farmed and free ranging mithuns (Bos frontalis). Trop Anim Health Prod 2017; 49(6): 1149-56.
[http://dx.doi.org/10.1007/s11250-017-1310-z] [PMID: 28504301] ]. There are earlier reports of BVDV in cattle (37.6%), buffaloes (30.76%), sheep (23.4%), and goats (16.9%) in India [40Bhatia S, Sood R, Mishra N, Pattnaik B, Pradhan HK. 2008.Development and evaluation of a MAb based competitive-ELISA using helicase domain of NS3 protein for sero-diagnosis of bovine viral diarrhea in cattle and buffaloes Research inVeterinary Science
[http://dx.doi.org/10.1016/j.rvsc.2007.09.013] ].
In a related study we have screened a total of 916 cattle serum originating in the north east India (especially from nearby places from where pig serum were collected) for the presence of BVDV with Ingezim BVDV compact ELISA for Antibody detection during the period of 2013 to 2015. 113 cattle serum were positive in Ab ELISA and 42 out of 113 cattle serum screened were showing positive result in RT-PCR technique as described by Sandvik et al in 1997 [12Couvreur B, Letellier C, Collard A, et al. Genetic and antigenic variability in Bovine Viral Diarrhea Virus (BVDV) isolates from Belgium. Virus Res 2002; 85(1): 17-28.
[http://dx.doi.org/10.1016/S0168-1702(02)00014-X] [PMID: 11955635] ] (Communicated). In the present study, a total of 10 (out of 206 porcine serum samples) genomic isolates of porcine origin- BVDV was detected by the amplification of partial 5’UTR region of the ORF.
As partial 5’UTR is the mos dominant gene fragment, it has been used widely for phylogenetic analysis of geographically diverse strains of BVDV from field samples [41Vilcek S, Paton DJ, Durkovic B, et al. Bovine viral diarrhoea virus genotype 1 can be separated into at least eleven genetic groups. Arch Virol 2001; 146(1): 99-115.
[http://dx.doi.org/10.1007/s007050170194] [PMID: 11266221] ], [42Nagai M, Hayashi M, Sugita S, et al. Phylogenetic analysis of bovine viral diarrhea viruses using five different genetic regions. Virus Res 2004; 99(2): 103-13.
[http://dx.doi.org/10.1016/j.virusres.2003.10.006] [PMID: 14749175] ], [43Wakeley PR, Turner JLE, Ibata G, et al. Characterisation of a type 2 bovine viral diarrhoea virus isolated from cattle in the UK. Vet Microbiol 2004; 102(1-2): 19-24.
[http://dx.doi.org/10.1016/j.vetmic.2004.05.005] [PMID: 15288923] ], [36Mishra N, Rajukumar K, Vilcek S, Tiwari A, Satav JS, Dubey SC. Molecular characterization of bovine viral diarrhea virus type 2 isolate originating from a native Indian sheep (Ovies aries). Vet Microbiol 2008; 130(1-2): 88-98.
[http://dx.doi.org/10.1016/j.vetmic.2008.01.005] [PMID: 18308487] ], [44Hornberg A, Fernández SR, Vogl C, et al. Genetic diversity of pestivirus isolates in cattle from Western Austria. Vet Microbiol 2009; 135(3-4): 205-13.
[http://dx.doi.org/10.1016/j.vetmic.2008.09.068] [PMID: 19019571] ].
Additionally, E2 have been found useful for more accurate phylogenetic analysis in segregating BVDV into its subgenotypes [41Vilcek S, Paton DJ, Durkovic B, et al. Bovine viral diarrhoea virus genotype 1 can be separated into at least eleven genetic groups. Arch Virol 2001; 146(1): 99-115.
[http://dx.doi.org/10.1007/s007050170194] [PMID: 11266221] ], [42Nagai M, Hayashi M, Sugita S, et al. Phylogenetic analysis of bovine viral diarrhea viruses using five different genetic regions. Virus Res 2004; 99(2): 103-13.
[http://dx.doi.org/10.1016/j.virusres.2003.10.006] [PMID: 14749175] ], [44Hornberg A, Fernández SR, Vogl C, et al. Genetic diversity of pestivirus isolates in cattle from Western Austria. Vet Microbiol 2009; 135(3-4): 205-13.
[http://dx.doi.org/10.1016/j.vetmic.2008.09.068] [PMID: 19019571] ], [45Becher P, Orlich M, Shannon AD, Horner G, König M, Thiel HJ. Phylogenetic analysis of pestiviruses from domestic and wild ruminants. J Gen Virol 1997; 78(Pt 6): 1357-66.
[http://dx.doi.org/10.1099/0022-1317-78-6-1357] [PMID: 9191930] ], [6Becher P, Avalos Ramirez R, Orlich M, et al. Genetic and antigenic characterization of novel pestivirus genotypes: Implications for classification. Virology 2003; 311(1): 96-104.
[http://dx.doi.org/10.1016/S0042-6822(03)00192-2] [PMID: 12832207] ]. Main circulating genotype of BVDV in cattle of India is BVDV-1b [35Mishra N, Pattnaik B, Vilcek S, et al. Genetic typing of bovine viral diarrhoea virus isolates from India. Vet Microbiol 2004; 104(3-4): 207-12.
[http://dx.doi.org/10.1016/j.vetmic.2004.08.003] [PMID: 15564029] ] while BVDV type 2 occurred sporadically in India [35Mishra N, Pattnaik B, Vilcek S, et al. Genetic typing of bovine viral diarrhoea virus isolates from India. Vet Microbiol 2004; 104(3-4): 207-12.
[http://dx.doi.org/10.1016/j.vetmic.2004.08.003] [PMID: 15564029] ]. Identification of BVDV-2 in sheep and goats [46Mishra N, Dubey R, Rajukumar K, et al. Genetic and antigenic characterization of bovine viral diarrhea virus type 2 isolated from Indian goats (Capra hircus). Vet Microbiol 2007; 124(3-4): 340-7.
[http://dx.doi.org/10.1016/j.vetmic.2007.04.023] [PMID: 17509780] ], [36Mishra N, Rajukumar K, Vilcek S, Tiwari A, Satav JS, Dubey SC. Molecular characterization of bovine viral diarrhea virus type 2 isolate originating from a native Indian sheep (Ovies aries). Vet Microbiol 2008; 130(1-2): 88-98.
[http://dx.doi.org/10.1016/j.vetmic.2008.01.005] [PMID: 18308487] ] demanded a search in cattle considering migration and trading in ruminants from the porous borders.HoBi like pestivirus or BVDV type 3 also detected from cattle serum in India in 2014 [37Mishra N, Rajukumar K, Pateriya A, et al. Identification and molecular characterization of novel and divergent HoBi-like pestiviruses from naturally infected cattle in India. Vet Microbiol 2014; 174(1-2): 239-46.
[http://dx.doi.org/10.1016/j.vetmic.2014.09.017] [PMID: 25301283] ].
In phylogenetic analysis it could be seen that on the basis of E-2 sequence alignment all the 8 isolates clustered in one group that was close to BVDV type 1 (Fig. 5 ). However the same isolates on the basis of 5’- UTR alignment clustered in two groups viz, five isolates with BVDV type 1 and three isolates as a distinct outlier group (Fig. 4
). However the same isolates on the basis of 5’- UTR alignment clustered in two groups viz, five isolates with BVDV type 1 and three isolates as a distinct outlier group (Fig. 4 ). The E-2 alignment indicated closeness to Cattle BVDV type 1. However, further studies on subtyping the isolates on the basis of full length 5’-UTR are underway. Regarding molecular phylogenetic studies of pestiviruses in the north eastern part of India, several reports have been published stating prevalence of CSFV subgenotype 1.1 and emergence of subgenotype 2.2 in the region [47Patil SS, Hemadri D, Veeresh H, Sreekala K, Gajendragad MR, Prabhudas K. Phylogenetic analysis of NS5B gene of classical swine fever virus isolates indicates plausible Chinese origin of Indian subgroup 2.2 viruses. Virus Genes 2012; 44(1): 104-8.
). The E-2 alignment indicated closeness to Cattle BVDV type 1. However, further studies on subtyping the isolates on the basis of full length 5’-UTR are underway. Regarding molecular phylogenetic studies of pestiviruses in the north eastern part of India, several reports have been published stating prevalence of CSFV subgenotype 1.1 and emergence of subgenotype 2.2 in the region [47Patil SS, Hemadri D, Veeresh H, Sreekala K, Gajendragad MR, Prabhudas K. Phylogenetic analysis of NS5B gene of classical swine fever virus isolates indicates plausible Chinese origin of Indian subgroup 2.2 viruses. Virus Genes 2012; 44(1): 104-8.
[http://dx.doi.org/10.1007/s11262-011-0572-1] [PMID: 21246270] ]. Recently, CSFV of genotype 2.2 from wild hog has also been reported from the region [48Barman NN, Bora DP, Khatoon E, et al. Classical swine fever in wild hog: Report of its prevalence in Northeast India. Transbound Emerg Dis 2014.
[http://dx.doi.org/10.1111/tbed.12298] [PMID: 25430917] ]. Overall results in our laboratory showed that the CSFV isolates from India, Nepal and China might have some common ancestor and subgenotype 2.2 is wide spread in the north- eastern region of India.
The occurrence of BVDV in pigs in a single cluster with closeness to BVDV-2 and 1 of cattle on the basis of E-2 sequencing and closeness to BVDV-1 of cattle on the basis of 5’UTR sequencing could indicate emergence and niche adaptation of the virus, as Type 1 BVDV is more genetically variable than type 2 [12Couvreur B, Letellier C, Collard A, et al. Genetic and antigenic variability in Bovine Viral Diarrhea Virus (BVDV) isolates from Belgium. Virus Res 2002; 85(1): 17-28.
[http://dx.doi.org/10.1016/S0168-1702(02)00014-X] [PMID: 11955635] ]. However if the phylogenetic analysis of 5’-UTR sequences are considered exclusively then the presence of an outlier group closer to CSFV1.1 genogroup could indicate an emergence of a genetically divergent type of BVDV that is establishing itself in the swine population of the north east. This could well be the isolates that would be encountered in the future. As the 5’UTR is relatively stable when compared to E-2; such a change could very well signal an initial adaptation event that could subsequently translate to presumable changes in the E-2 domain after a more prolonged adaptation of BVDV in pigs the north eastern part of India.
CONCLUSION
The study is the first of its kind regarding identification and preliminary molecular epidemiology of BVDV isolates from porcine in the north eastern part of India, under conditions of integrated and mixed livestock farming. Our study indicates a very minute prevalence of such infections (<5%) from nearly 200 samples analysed over a period of two years. However it can be seen that all the partial E2 gene sequences (8 nos) are close to the BVDV type 1 but forming a distinct cluster. The same isolates show a difference/variation on the basis of 5’ UTR gene sequences (10 nos) and are close to the BVDV type 1. Sentinel surveillance for both BVDV type 1 and 2 isolates need to be established to study the emergence of these isolates in the swine population of the north east. There has to be a heightened surveillance for atypical CSF cases that could indicate emergence of a new strain of BVDV (most likely type 1) that is stabilising itself in the porcine population of the north east.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
the reported experiments were in accordance with the standards set forth in the 8th Edition of Guide for the Care and Use of Laboratory Animals (http://grants.nih.gov/grants/olaw/Guide-for-the-care-and-use-of-laboratory-animals.pdf) published by the National Academy of Sciences, The National Academies Press, Washington DC, United States of America.
HUMAN AND ANIMAL RIGHTS
The protocols followed in the study were OIE approved as per genomic detections of BVD in are concerned. (OIE Manual, 2015). Standardised primers and validated methods of detection of BVD in porcines as reported by earlier workers were followed.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
We want to thank DBT GOI for providing nessesary funding for the work through project title "NER DBT ADMaC" (DBT-NER/LIVS/11/2012 dated: 24th April 2014).
REFERENCES
| [1] | Geoghegan JL, Senior AM, Holmes EC. Pathogen population bottlenecks and adaptive landscapes: Overcoming the barriers to disease emergence. Proc R SocLond B 1837; 2016: 283. |
| [2] | Jackson AP, Charleston MA. A cophylogenetic perspective of RNA-virus evolution. Molecular biologyand evolution 2004; 21(1): 45-57. |
| [3] | Kitchen A, Shackelton LA, Holmes EC. Family level phylogenies reveal modes of macroevolution in RNA viruses 2011; 108(1): 238-43. [http://dx.doi.org/10.1073/pnas.1011090108] |
| [4] | Darbyshire JH. A serological relationship between swine fever andmucosal disease of cattle. Vet Rec 1960; 72: 331. |
| [5] | Wang XP, Tu CC, Li HW, et al. Pigs naturally infected by bovine diarrhea virus present signs resembling hog cholera. Chin J Vet Sci 1996; 16: 341-5. |
| [6] | Becher P, Avalos Ramirez R, Orlich M, et al. Genetic and antigenic characterization of novel pestivirus genotypes: Implications for classification. Virology 2003; 311(1): 96-104. [http://dx.doi.org/10.1016/S0042-6822(03)00192-2] [PMID: 12832207] |
| [7] | Ridpath JF. Bovine viral diarrhea virus: Global status. Vet Clin North Am Food Anim Pract 2010; 26(1): 105-21. [http://dx.doi.org/10.1016/j.cvfa.2009.10.007] [PMID: 20117546] |
| [8] | Passler T, Walz PH. Bovine viral diarrhea virus infections in heterologous species. Anim Health Res Rev 2010; 11(2): 191-205. [http://dx.doi.org/10.1017/S1466252309990065] [PMID: 19883524] |
| [9] | Le Potier M, Mesplede A, Vanner P. Classical swine fever other pestiviruses.Diseases of Swine 9th ed. 2006; 309-11. |
| [10] | Liess B, Moennig V. Ruminant pestivirus infection in pigs. Rev - Off Int Epizoot 1990; 9(1): 151-61. [http://dx.doi.org/10.20506/rst.9.1.484] [PMID: 1966720] |
| [11] | Sandvik T, Paton DJ, Lowings PJ. Detection and identification of ruminant and porcine pestiviruses by nested amplification of 5′ untranslated cDNA regions. J Virol Methods 1997; 64(1): 43-56. [http://dx.doi.org/10.1016/S0166-0934(96)02136-2] [PMID: 9029529] |
| [12] | Couvreur B, Letellier C, Collard A, et al. Genetic and antigenic variability in Bovine Viral Diarrhea Virus (BVDV) isolates from Belgium. Virus Res 2002; 85(1): 17-28. [http://dx.doi.org/10.1016/S0168-1702(02)00014-X] [PMID: 11955635] |
| [13] | Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016; 33(7): 1870-4. [http://dx.doi.org/10.1093/molbev/msw054] [PMID: 27004904] |
| [14] | Nei M, Kumar S. Molecular Evolution and Phylogenetics 2000. |
| [15] | Flynn DM, Jones TE. The position regarding swine fever in Victoria. Aust Vet J 1964; 40: 131-7. [http://dx.doi.org/10.1111/j.1751-0813.1964.tb01718.x] |
| [16] | Carbrey EA, Stewart WC, Kresse JI, Snyder ML. Natural infection of pigs with bovine viral diarrhea virus and its differential diagnosis from hog cholera. J Am Vet Med Assoc 1976; 169(11): 1217-9. [PMID: 187565] |
| [17] | Terpstra C, Wensvoort G. Bovine virus diarrhea virus infections in swine. Tijdschr Diergeneeskd 1991; 116(19): 943-8. [PMID: 1656546] |
| [18] | Wensvoort G, Terpstra C. Bovine viral diarrhoea virus infections in piglets born to sows vaccinated against swine fever with contaminated vaccine. Res Vet Sci 1988; 45(2): 143-8. [PMID: 2848299] |
| [19] | Liess B, Moennig V. Ruminant pestivirus infection in pigs. Rev - Off Int Epizoot 1990; 9(1): 151-61. [http://dx.doi.org/10.20506/rst.9.1.484] [PMID: 1966720] |
| [20] | O’Connor M, Lenihan P, Dillon P. Pestivirus antibodies in pigs in Ireland. Vet Rec 1991; 129(12): 269. [http://dx.doi.org/10.1136/vr.129.12.269] [PMID: 1660192] |
| [21] | Terpstra C, Wensvoort G. Bovine virus diarrhea virus infections in swine. Tijdschr Diergeneeskd 1991; 116(19): 943-8. [PMID: 1656546] |
| [22] | Løken T, Krogsrud J, Larsen IL. Pestivirus infections in Norway. Serological investigations in cattle, sheep and pigs. Acta Vet Scand 1991; 32(1): 27-34. [PMID: 1659160] |
| [23] | Jensen MH. Screening for neutralizing antibodies against hog cholera- and/or bovine viral diarrhea virus in Danish pigs. Acta Vet Scand 1985; 26(1): 72-80. [PMID: 2994428] |
| [24] | Graham DA, Calvert V, German A, McCullough SJ. Pestiviral infections in sheep and pigs in Northern Ireland. Vet Rec 2001; 148(3): 69-72. [http://dx.doi.org/10.1136/vr.148.3.69] [PMID: 12503593] |
| [25] | de Smit AJ, Eblé PL, de Kluijver EP, Bloemraad M, Bouma A. Laboratory decision-making during the classical swine fever epidemic of 1997-1998 in The Netherlands. Prev Vet Med 1999; 42(3-4): 185-99. [http://dx.doi.org/10.1016/S0167-5877(99)00075-6] [PMID: 10619155] |
| [26] | O’Sullivan T, Friendship R, Carman S, Pearl DL, McEwen B, Dewey C. Seroprevalence of bovine viral diarrhea virus neutralizing antibodies in finisher hogs in Ontario swine herds and targeted diagnostic testing of 2 suspect herds. Can Vet J 2011; 52(12): 1342-4. [PMID: 22654141] |
| [27] | Song YF, Zhang Z, Zhang YX, Wu FX, Li XC. The initial study on the prevalence of Bovine Viral Diarrhea Virus (BVDV) from swine. Chin J Anim Quarant 2008; 7: 25-7. |
| [28] | Gaede W, Reiting R, Schirrmeier H, Depner KR, Beer M. Detection and species-specific differentiation of pestiviruses using real-time RT-PCR. Berl Munch Tierarztl Wochenschr 2005; 118(3-4): 113-20. [PMID: 15803758] |
| [29] | Fernández-Sirera L, Cabezón O, Rossi L, et al. Investigations of pestivirus infection in wild Caprinae in Europe. Vet Rec 2011; 169(1): 15. [http://dx.doi.org/10.1136/vr.d1831] [PMID: 21709051] |
| [30] | Bachofen C, Vogt HR, Stalder H, et al. Persistent infections after natural transmission of bovine viral diarrhoea virus from cattle to goats and among goats. Vet Res (Faisalabad) 2013; 44: 32. [http://dx.doi.org/10.1186/1297-9716-44-32] [PMID: 23675947] |
| [31] | Vilcek S, Drew TW, McGoldrick A, Paton DJ. Genetic typing of bovine pestiviruses from England and Wales. Vet Microbiol 1999; 69(4): 227-37. [http://dx.doi.org/10.1016/S0378-1135(99)00111-X] [PMID: 10535769] |
| [32] | Duncan C, Ridpath J, Palmer MV, Driskell E, Spraker T. Histopathologic and immunohistochemical findings in two white-tailed deer fawns persistently infected with Bovine viral diarrhea virus. J Vet Diagn Invest 2008; 20(3): 289-96. [http://dx.doi.org/10.1177/104063870802000305] [PMID: 18460614] |
| [33] | Woods RD, Kunkle RA, Ridpath JE, Bolin SR. Bovine viral diarrhea virus isolated from fetal calf serum enhances pathogenicity of attenuated transmissible gastroenteritis virus in neonatal pigs. J Vet Diagn Invest 1999; 11(5): 400-7. [http://dx.doi.org/10.1177/104063879901100503] [PMID: 12968752] |
| [34] | Terpstra C, Wensvoort G. A congenital persistent infection of bovine virus diarrhoea virus in pigs: Clinical, virological and immunological observations. Vet Q 1997; 19(3): 97-101. [http://dx.doi.org/10.1080/01652176.1997.9694750] [PMID: 9323848] |
| [35] | Mishra N, Pattnaik B, Vilcek S, et al. Genetic typing of bovine viral diarrhoea virus isolates from India. Vet Microbiol 2004; 104(3-4): 207-12. [http://dx.doi.org/10.1016/j.vetmic.2004.08.003] [PMID: 15564029] |
| [36] | Mishra N, Rajukumar K, Vilcek S, Tiwari A, Satav JS, Dubey SC. Molecular characterization of bovine viral diarrhea virus type 2 isolate originating from a native Indian sheep (Ovies aries). Vet Microbiol 2008; 130(1-2): 88-98. [http://dx.doi.org/10.1016/j.vetmic.2008.01.005] [PMID: 18308487] |
| [37] | Mishra N, Rajukumar K, Pateriya A, et al. Identification and molecular characterization of novel and divergent HoBi-like pestiviruses from naturally infected cattle in India. Vet Microbiol 2014; 174(1-2): 239-46. [http://dx.doi.org/10.1016/j.vetmic.2014.09.017] [PMID: 25301283] |
| [38] | Mishra N, Dubey R, Rajukumar K, et al. Genetic and antigenic characterization of bovine viral diarrhea virus type 2 isolated from Indian goats (Capra hircus). Vet Microbiol 2007; 124(3-4): 340-7. [http://dx.doi.org/10.1016/j.vetmic.2007.04.023] [PMID: 17509780] |
| [39] | Singh V, Mishra N, Kalaiyarasu S, et al. First report on serological evidence of Bovine Viral Diarrhea Virus (BVDV) infection in farmed and free ranging mithuns (Bos frontalis). Trop Anim Health Prod 2017; 49(6): 1149-56. [http://dx.doi.org/10.1007/s11250-017-1310-z] [PMID: 28504301] |
| [40] | Bhatia S, Sood R, Mishra N, Pattnaik B, Pradhan HK. 2008.Development and evaluation of a MAb based competitive-ELISA using helicase domain of NS3 protein for sero-diagnosis of bovine viral diarrhea in cattle and buffaloes Research inVeterinary Science [http://dx.doi.org/10.1016/j.rvsc.2007.09.013] |
| [41] | Vilcek S, Paton DJ, Durkovic B, et al. Bovine viral diarrhoea virus genotype 1 can be separated into at least eleven genetic groups. Arch Virol 2001; 146(1): 99-115. [http://dx.doi.org/10.1007/s007050170194] [PMID: 11266221] |
| [42] | Nagai M, Hayashi M, Sugita S, et al. Phylogenetic analysis of bovine viral diarrhea viruses using five different genetic regions. Virus Res 2004; 99(2): 103-13. [http://dx.doi.org/10.1016/j.virusres.2003.10.006] [PMID: 14749175] |
| [43] | Wakeley PR, Turner JLE, Ibata G, et al. Characterisation of a type 2 bovine viral diarrhoea virus isolated from cattle in the UK. Vet Microbiol 2004; 102(1-2): 19-24. [http://dx.doi.org/10.1016/j.vetmic.2004.05.005] [PMID: 15288923] |
| [44] | Hornberg A, Fernández SR, Vogl C, et al. Genetic diversity of pestivirus isolates in cattle from Western Austria. Vet Microbiol 2009; 135(3-4): 205-13. [http://dx.doi.org/10.1016/j.vetmic.2008.09.068] [PMID: 19019571] |
| [45] | Becher P, Orlich M, Shannon AD, Horner G, König M, Thiel HJ. Phylogenetic analysis of pestiviruses from domestic and wild ruminants. J Gen Virol 1997; 78(Pt 6): 1357-66. [http://dx.doi.org/10.1099/0022-1317-78-6-1357] [PMID: 9191930] |
| [46] | Mishra N, Dubey R, Rajukumar K, et al. Genetic and antigenic characterization of bovine viral diarrhea virus type 2 isolated from Indian goats (Capra hircus). Vet Microbiol 2007; 124(3-4): 340-7. [http://dx.doi.org/10.1016/j.vetmic.2007.04.023] [PMID: 17509780] |
| [47] | Patil SS, Hemadri D, Veeresh H, Sreekala K, Gajendragad MR, Prabhudas K. Phylogenetic analysis of NS5B gene of classical swine fever virus isolates indicates plausible Chinese origin of Indian subgroup 2.2 viruses. Virus Genes 2012; 44(1): 104-8. [http://dx.doi.org/10.1007/s11262-011-0572-1] [PMID: 21246270] |
| [48] | Barman NN, Bora DP, Khatoon E, et al. Classical swine fever in wild hog: Report of its prevalence in Northeast India. Transbound Emerg Dis 2014. [http://dx.doi.org/10.1111/tbed.12298] [PMID: 25430917] |