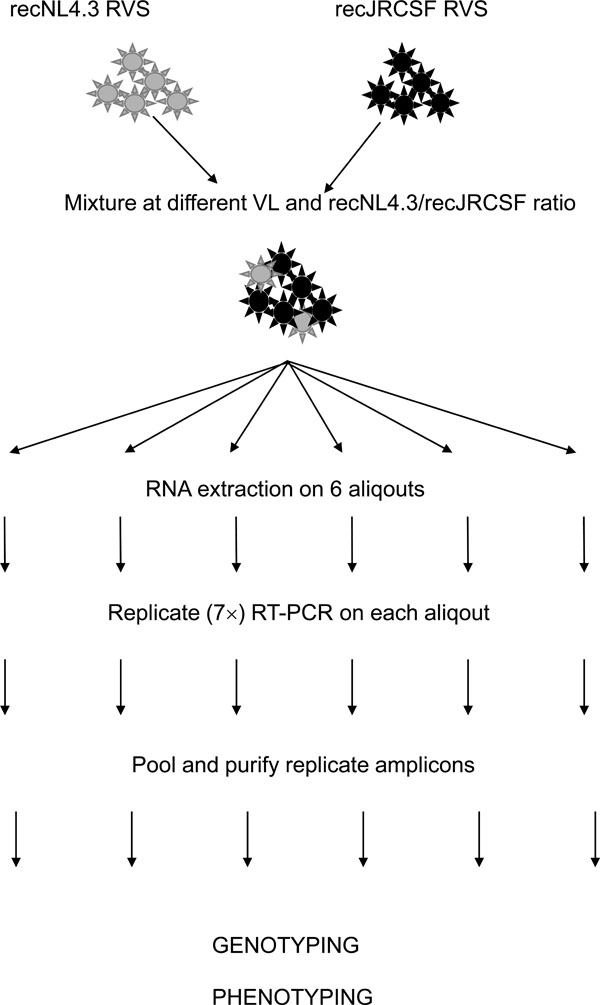
Fig. (2) Workflow for genotypic and phenotypic analysis of the recJRCSF/recNL4.3 mixtures. Two RVS were mixed at different ratios and viral loads. Each mixture was split into 6 aliquots and processed in parallel. RNA was extracted of each aliquot, followed by 7 replicate amplifications of the NH2-V4 region. PCR products were pooled, purified and cloned. Colonies were picked, the insert was PCR amplified, purified and analyzed by AluI restriction analysis. Tropism was assessed by population-based phenotyping as described [7]. Infection of CCR5- and CXCR4-expressing U87-cells was scored using a fluorescent microscope and FACS.