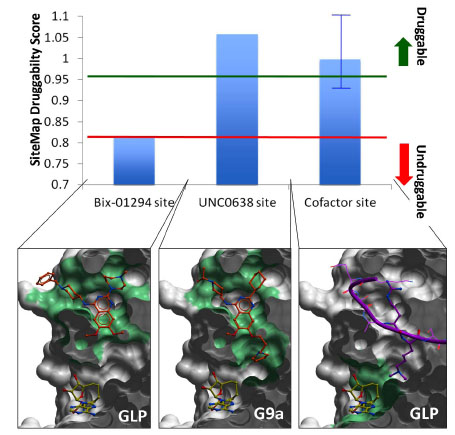
Fig. (8) Druggability of the peptide and cofactor binding sites. A druggability score was calculated with SiteMap (Schrodinger, NY) for
the binding sites of the GLP/G9a inhibitors Bix-01294, UNC0638, and the cofactor (top) [41]. The peptide-binding site (center) is druggable,
and can accommodate highly potent compounds, such as UNC0638. When the lysine channel is ignored, the druggability index drops sharply
(left). The cofactor site (right) is also predicted to be druggable (blue bar represents the mean value across all available co-crystallized human
SET domain PMT structures), but with varying druggability index from one enzyme to another (blue error bar), which reflects a variation in
hydrophilicity and enclosure of the site. Green: pocket used to calculate the druggability score. The molecular surface of the enzyme is
clipped across the Z-axis for better visualization.