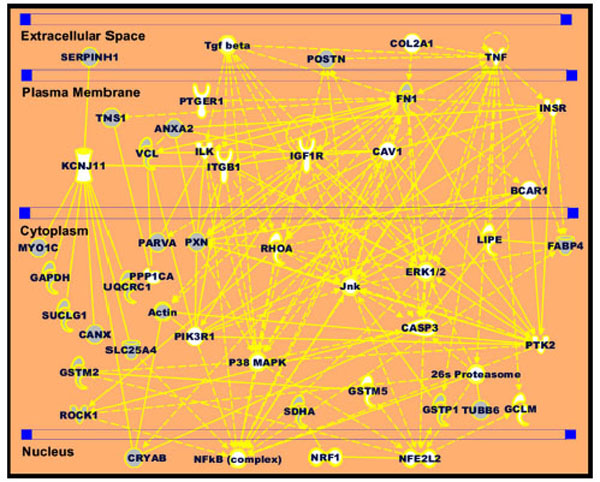
Fig. (10) IPA network analysis of proteins identified by iTRAQ in the aorta and their potential functional link to other proteins in human
cells (in various tissues, under various conditions). The six major IPA-provided networks were analyzed based on the data of proteins expressed
in rat aorta. The network (i) considered 14 proteins (Actin, BCAR1, CANX, CASP3, CAV1, CRYAB, GAPDH, GLMC, GCLM,
GSTM1, GSTM5, GSTP1, ILK, ITGB1, KCNJ11, MYO1C, NFE2L2, PARVA, PIK3R1, PPP1CA, PTK2, PXN, SERPINH1, SLC25A4,
SUCLG1, TNF, TNS1, UQCRC1, VCL) involved in necrosis/cell death, cell morphology. Network (ii) considered proteins (26S Proteasome,
TUBB6) involved in the cardiovascular system development and function. Network (iii) considered proteins (RHOA, ROCK1) involved
in cell morphology, cellular development, nervous system development and function. Network (iv) considered proteins (FABP4,
LIPE) in-volved genetic disorders and lipid metabolic diseases. Network (v) considered proteins (NRF1, SDHA) involved in lipid metabolism,
small molecule biochemistry, connective tissue development and function. Network (vi) considered proteins (ANXA2, IGF1R, INSR)
involved in organ morphology, cell-to-cell signaling and interaction and embryonic development. The solid lines show direct proteinprotein
interaction, while dotted lines show indirect interactions between the proteins. Proteins identified by iTRAQ, as shown in Supplemental
Table 1, are pre-sented in grey-color circles, while additional interaction proteins were added by the IPA network analysis in white
color.