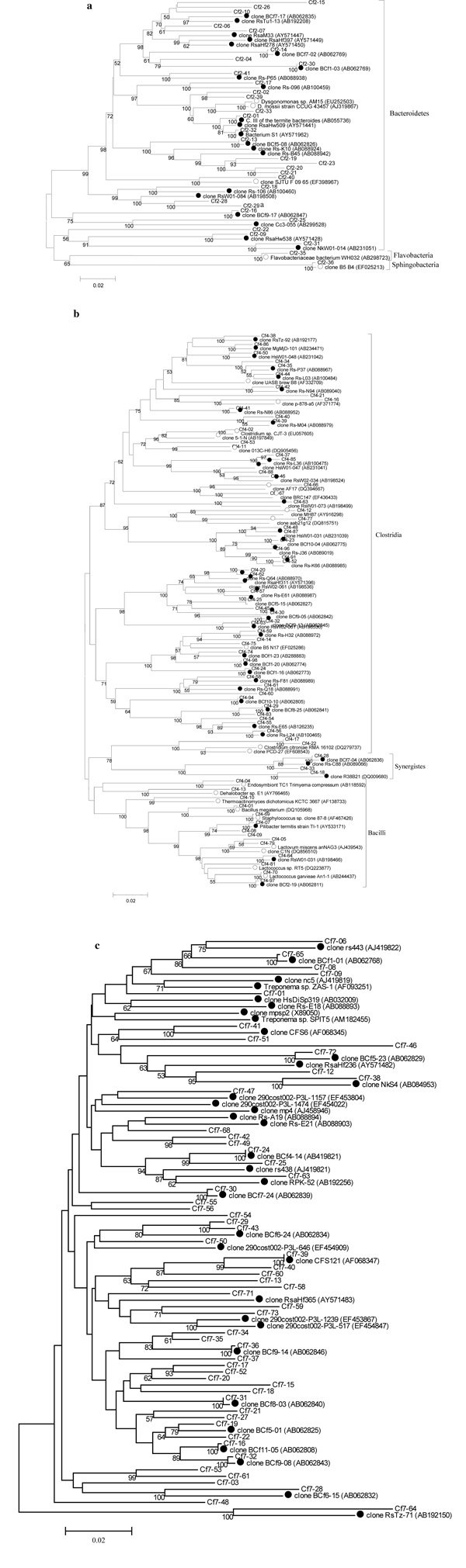
Fig. (1) Phylogenetic trees constructed from the 16S rRNA gene sequences of gut bacteria of Formosan subterranean termite (FST) samples
from Louisiana and China (Cf) and their highest match from the DDBJ/EMBL/GenBank databases. A. Bacteroidetes, B. Firmicutes, C.
Spirochaetes, D. Actinobacteria, E. Proteobacteria, F. TM7, G. Verrucomicrobia and Planctomycetes. The phylogenetic trees were
constructed using the neighbor joining method with 1000 bootstrap replicates. Only bootstrap values of ≥50 are indicated on the branch
nodes. The scale bars represent 2% difference in nucleotide sequence. Closely related ribotype sequences from other studies were included for
comparison and marked with circles (● termite-specific bacteria, ○ environmental bacteria). A. Bacteroidetes. B. Firmicutes & Synergistes.
C. Spirochaetes. D. Proteobacteria. E. Actinobacteria. F. TM7. G. Planctomycetes & Verrucomicrobia.