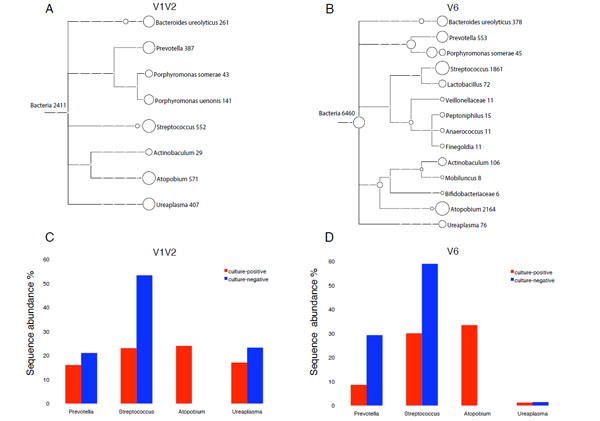
Fig. (1) Taxonomical classification of 16S rDNA found in reported samples.A and B: Tree view of 16S rDNA sequences from culture-positive urine (α-hemolytic Streptococcus >105 CFU/ml), assigned at different taxonomy levels (family, genus and species) as computed by MEGAN 3.4 for V1V2 (A) and V6 (B) amplicons. Each circle represents a taxon and is labeled by its name and the number of sequenced reads assigned. Taxonomical assignment was done by comparing the sequences to a curated version of the SSUrdp database, then assigning the sequences to the taxon of the best-matched reference sequence (lowest common ancestor). The size of the circles is scaled logarithmically to the number of reads assigned to the taxon.C and D: Comparison of bacterial communities from culture-positive urine positive for α-hemolytic green Streptococcus (red bars) and culture-negative urine (blue bars) from a case subject diagnosed with overactive bladder syndrome. Sequence analysis is based on V1V2 (C) and V6 (D) amplicons. Only major genera with a sequence abundance ≥10 % for either V1V2 or V6 analysis are shown. The calculation of sequence abundance for each genus was relative to the total number of sequence assigned to Bacteria in each dataset.