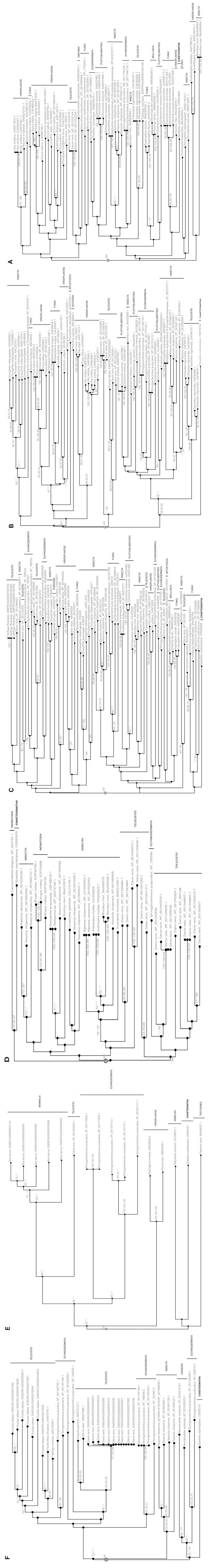
Fig. (1) Phylogenetic trees from predicted translations of coding regions of reverse transcriptase domain of Gypsy/Ty3 LTR-retrotransposons in (A), (B) and (C), of Bel-Pao LTR-retrotransposons in (D) and LINE elements in (E) and (F). The chaetognath EST accession numbers CR953949, (CR953634, CR953418, CR953554), CR952896, (CR950076, CR950075), CR950197 and CR950101 in respectively (A), (B), (C), (D), (E) and (F) phylogenetic analyses. Each tree is the fusion of three phylogenetic trees built on Neighbor Joining (NJ), Maximum Parsimony (MP), and Maximum Likelihood (ML) methods using FIGENIX software platform. For each node, bootstrap values are reported for each method. *, means that the bootstrap value was inferior to 50% (e.g. a bootstrap 69,*,57 means that the node exists in NJ tree with a bootstrap value equal to 69, it exists in ML tree with a bootstrap value equal to 57, but does not exists in the MP tree). Branch lengths are correlated to the sequence evolution rate. Many of the same genes are mentioned more than once because they represent different GenBank sequences; since the trees are generated automatically, no discrimination was made.