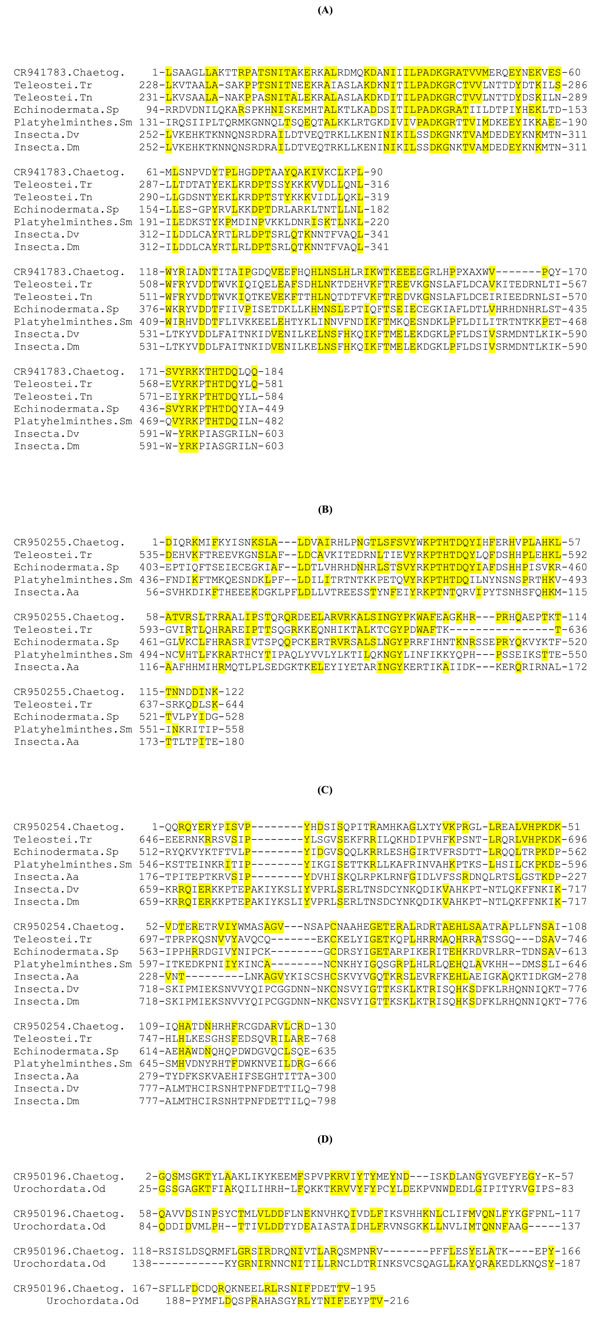
Fig. (2) Alignments of predicted amino acid sequence of chaetognath ESTs with the homologous regions of various retrotransposons. (A), (B), (C) and (D) correspond respectively to alignments with CR941783, CR950255, CR950254, CR950196 chaetognath ESTs. In each alignment, the first sequence corresponds to the predicted amino acid sequence of a chaetognath EST. In (A), (B) and (C) the reverse transcriptase homologous sequences belong to Penelope-like elements, whereas in (D), only one putative integrase has been found partially homologous to chaetognath sequence. Sequences were aligned using program CLUSTAL W; positions of nucleotides are shown. The abbreviations of the other sequences and their accession numbers are: (Teleostei.Tr) Takifugu rubripes, AAK58879.1; (Telostei.tn) Tetraodon nigroviridis, CAF94542; (Echinodermata.Sp) Strongylocentrotus purpuratus, XP_001200565; (Platyhelminthes.Sm) Schistosoma mansoni, DAA00890; (Insecta.Aa) Aedes aegypti, AAZ15235; (Insecta.Dv) Drosophila virilis AAL14979; (Insecta.Dm) Drosophila melanogaster, AAR86938. Amino acid residues which are identical to those of the chaetognath sequences are underlined in yellow. Dashes represent gaps introduced to improve the sequence alignment. Stop codons appear as a result of unknown amino acid residue marked as X.