- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Bioactive Compounds Journal
(Discontinued)
ISSN: 1874-8473 ― Volume 9, 2020
Non-Intercalative Triterpenoid Inhibitors of Topoisomerase II: A Molecular Docking Study
William N. Setzer*
Abstract
Theoretical flexible docking studies were carried out on a number of triterpenoids previously shown to be inhibitors of topoisomerase II in order to assess the nature of binding of these non-intercalative inhibitors to the enzyme. The molecular docking results suggest that most of the triterpenoids preferentially bind to the DNA binding site of topoisomerase II, while a few also bind to the ATP binding site. These results provide some insight into the mode of activity of these cytotoxic natural products.
Article Information
Identifiers and Pagination:
Year: 2008Volume: 1
First Page: 13
Last Page: 17
Publisher Id: TOBCJ-1-13
DOI: 10.2174/1874847300801010013
Article History:
Received Date: 17/4/2008Revision Received Date: 9/5/2008
Acceptance Date: 29/5/2008
Electronic publication date: 10/07/2008
Collection year: 2008
open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.5/), which permits unrestrictive use, distribution, and reproduction in any medium, provided the original work is properly cited.
* Address correspondence to this author at the Department of Chemistry, University of Alabama in Huntsville, Huntsville, AL 35899, USA; Tel: 256-824-6519; Fax: 256-824-6349; E-mail: wsetzer@chemistry.uah.edu
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 17-4-2008 |
Original Manuscript | Non-Intercalative Triterpenoid Inhibitors of Topoisomerase II: A Molecular Docking Study | |
INTRODUCTION
Topoisomerases are essential enzymes that catalyze modifications to the tertiary structure of DNA. There are two well-characterized classes of human topoisomerases. Topoisomerase I acts by breaking and religating one DNA strand [1Pommier Y, Pourquier P, Fan Y, Strumberg D. Biochim. Biophys Acta 1998; 1400: 83-105.
[http://dx.doi.org/10.1016/S0167-4781(98)00129-8] ], while topoisomerase II involves double-strand breaking [2Burden DA, Osheroff N. Biochim. Biophys Acta 1998; 1400: 139-54.
[http://dx.doi.org/10.1016/S0167-4781(98)00132-8] ]. These enzymes serve to relieve DNA twisting and supercoiling, playing key roles in replication, transcription, and recombinant repair. Topoisomerase II is highly expressed in rapidly proliferating cells [3Heck MM, Earnshaw WCJ. J. Cell Biol 1986; 103: 2569-81.
[http://dx.doi.org/10.1083/jcb.103.6.2569] ] and is therefore an attractive target for antitumor drugs.
There are two general classes of topoisomerase II targeting drugs: topoisomerase II poisons and topoisomerase II catalytic inhibitors. Topoisomerase II poisons include etoposide, doxorubicin, and mitoxantrone. These compounds serve to stabilize the enzyme-DNA complex (the “cleavable complex”) and prevent the enzyme from religating the cleaved DNA [4Wilstermann AM, Osheroff N. Curr Top Med Chem 2003; 3: 1349-64.
[http://dx.doi.org/10.2174/1568026033452519] ]. Both doxorubicin and mitoxantrone are DNA intercalating agents [5Hande KR. Update Cancer Ther 2006; 1: 3-15.
[http://dx.doi.org/10.1016/j.uct.2006.04.001] ] whereas etoposide does not bind DNA but rather apparently binds to the ATP binding site of the N-terminal domain of topoisomerase II [6Leroy D, Kajava AV, Frei C, Gasser SM. Biochemistry Source Journal 2001; 40: 1624-34.
[http://dx.doi.org/10.1021/bi0019141] [PMID: 11327821] , 7Sengupta T, Mukherjee M, Das A, et al. Biochem J 2005; 390: 419-26.
[http://dx.doi.org/10.1042/BJ20042128] [PMID: 15901238] [PMCID: PMC1198921] ].
The catalytic inhibitors, on the other hand, block the catalytic activity of DNA topoisomerase II but do not stabilize the DNA-topoisomerase II cleavable complex [5Hande KR. Update Cancer Ther 2006; 1: 3-15.
[http://dx.doi.org/10.1016/j.uct.2006.04.001] , 8Andoh T, Ishida R. Biochim Biophys Acta 1998; 1400: 155-71.
[http://dx.doi.org/10.1016/S0167-4781(98)00133-X] ]. Examples of catalytic topoisomerase II inhibitors include the anthracycline aclarubicin, the polyanionic compound surname, the coumarin novobiocin, and bisdioxopiperazines such as sobuzoxane and dexrazoxane [9Larsen AK, Escargueil AE, Skladanowski A. Pharmacol Ther 2003; 99: 167-81.
[http://dx.doi.org/10.1016/S0163-7258(03)00058-5] ]. These agents inhibit the catalytic activity of topoisomerase II by preventing the binding of the enzyme to DNA. A number of natural and semisynthetic triterpenoids have shown topoisomerase II inhibitory activity. These include 3,4-seco-8βH-ferna-4(23),9(11)-dien-3-oic acid (1) and its corresponding alcohol derivative (2) [10Wada S, Tanaka R, Iida A, Matsunaga S. Bioorg Med Chem Lett 1998; 8: 2829-32.
[http://dx.doi.org/10.1016/S0960-894X(98)00515-0] ]; seco-3,4-friedelin (3), seco-3,4-taraxerone (4), lupeol (5) [11Setzer WN, Shen X, Bates RB, et al. Fitoterapia Source Journal 2000; 71: 195-8.]; fomitellic acids A and B (6 and 7) [12Mizushina Y, Iida A, Ohta K, Sugawara F, Sakaguchi K. Biochem J 2000; 350: 757-63.
[http://dx.doi.org/10.1042/0264-6021:3500757] [PMID: 10970789] [PMCID: PMC1221307] ]; ursolic acid (8), oleanolic acid (9), betulinic acid (10), acetyl α-boswellic acid (11) [13Syrovets T, Büchele B, Gedig E, Slupsky JR, Simmet T. Mol Pharmacol Source Journal 2000; 58: 71-81.
[PMID: 10860928] ]; demethylzeylasterone (12) [14Furbacher TR, Gunatilaka AAL. J Nat Prod Source Journal 2001; 64: 1294-6.
[http://dx.doi.org/10.1021/np010123c] ];
dihydrobetulinic acid (13) [15Chowdhury AR, Mandal S, Goswami A, et al. Mol Med 2003; 9: 26-36.
[PMID: 12765337] [PMCID: PMC1430381] ]; corosolic acid (14), 3α-corosolic acid (15), 3β-corosolic acid lactone (16) [16Mizushina Y, Ikuta A, Endoh K, et al. Biochem Biophys Res Commun 2003; 305: 365-73.
[http://dx.doi.org/10.1016/S0006-291X(03)00765-4] ]; celastrol (17), dihydrocelastrol (18) [17Nagase M, Oto J, Sugiyama S, Yube K, Takaishi Y, Sakato N. Biosci Biotech Biochem 2003; 67: 1883-7.
[http://dx.doi.org/10.1271/bbb.67.1883] [PMID: 14519971] ]; dehydrotrametononic acid (19), dehydroebriconic acid (20) [18Mizushina Y, Akihisa T, Ukiya M, et al. Cancer Sci 2004; 95: 354-60.
[http://dx.doi.org/10.1111/j.1349-7006.2004.tb03215.x] [PMID: 15072595] ]; ganoderic acid X (21) [19Li CH, Chen PY, Chang UM, et al. Life Sci 2005; 77: 252-65.
[http://dx.doi.org/10.1016/j.lfs.2004.09.045] [PMID: 15878354] ]; and the semisynthetic lanostane derivative (22) [20Wada S, Tanaka R. Bioorg Med Chem Lett 2005; 15: 2966-9.
[http://dx.doi.org/10.1016/j.bmcl.2005.04.052] [PMID: 15914002] ]. In this study, molecular docking techniques have been used to examine the potential binding sites of these known triterpenoid inhibitors of topoisomerase II in order to probe the possible mechanism of enzyme inhibition.
ATP is a required cofactor for topoisomerase II [2Burden DA, Osheroff N. Biochim. Biophys Acta 1998; 1400: 139-54.
[http://dx.doi.org/10.1016/S0167-4781(98)00132-8] , 8Andoh T, Ishida R. Biochim Biophys Acta 1998; 1400: 155-71.
[http://dx.doi.org/10.1016/S0167-4781(98)00133-X] , 21McClendon AK, Osheroff N. Mutat Res 2007; 623: 83-97.
[http://dx.doi.org/10.1016/j.mrfmmm.2007.06.009] [PMID: 17681352] [PMCID: PMC2679583] ]. Topoisomerase II uses the energy released by ATP hydrolysis to induce DNA strand passage. In addition, the binding of ATP causes a conformational change of the enzyme from an open form to a closed clamp form. Therefore, ATP binding and hydrolysis result in opening and closing of the topoisomerase II enzyme. Some topoisomerase II inhibitors (e.g., bisdioxopiperazines and coumarins) act by binding to the ATPase domain of the enzyme [8Andoh T, Ishida R. Biochim Biophys Acta 1998; 1400: 155-71.
[http://dx.doi.org/10.1016/S0167-4781(98)00133-X] , 9Larsen AK, Escargueil AE, Skladanowski A. Pharmacol Ther 2003; 99: 167-81.
[http://dx.doi.org/10.1016/S0163-7258(03)00058-5] ]. Potential binding of triterpenoid topoisomerase II inhibitors was also investigated by docking the compounds into the ATP binding sites of the N-terminal domain of topoisomerase II.
RESULTS AND DISCUSSION
The binding energies of the lowest-energy poses for each of the triterpenoid topoisomerase II inhibitors for the DNA binding site (PDB: 1bjt [22Fass D, Bogden CE, Berger JM. Nature Struct Biol 1999; 6: 322-6.
[http://dx.doi.org/10.1038/7556] [PMID: 10201398] ] and 2rgr [23Dong KC, Berger JM. Nature 2007; 450: 1201-5.
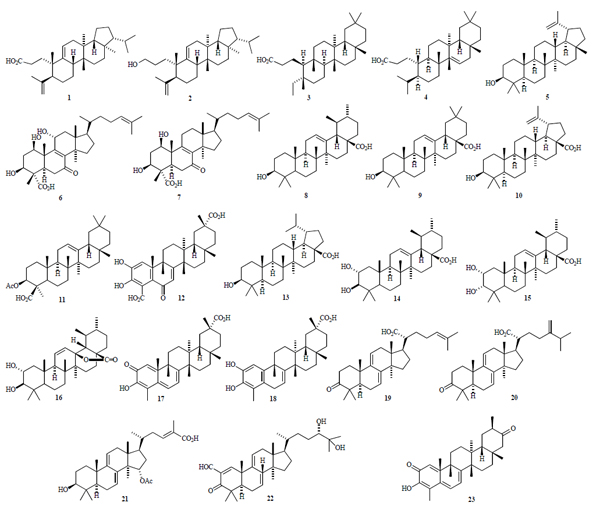
[http://dx.doi.org/10.1038/nature06396] [PMID: 18097402] ]) and the ATP binding sites (PDB: 1qzr, 1pvg, and 1zxm) are summarized in Table 1. The lowest-energy docking poses for most of the triterpenoids is the DNA binding site of topoisomerase II (see Figs. 1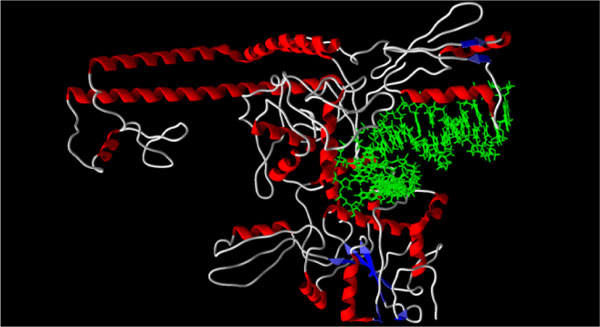 and 2
and 2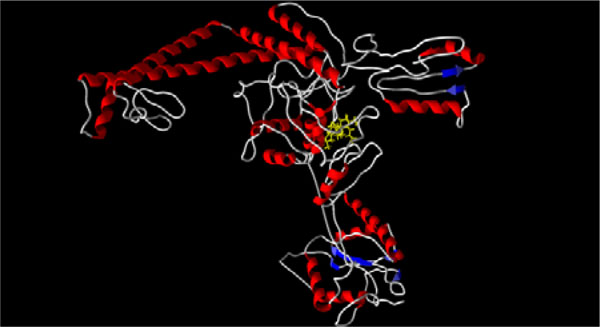 ), including 1-3, 5-14, 16,and 19-22. The key amino acid residues at this binding site are Arg 690, Asp 687. Gln 599, Gln 739, Gln 743, Glu 738, Glu 831, Gly 737, Gly 832, Ile 833, Lys 598, Lys 700, Phe 595, Ser 691, Thr 596, and Trp 597 (Fig. 3
), including 1-3, 5-14, 16,and 19-22. The key amino acid residues at this binding site are Arg 690, Asp 687. Gln 599, Gln 739, Gln 743, Glu 738, Glu 831, Gly 737, Gly 832, Ile 833, Lys 598, Lys 700, Phe 595, Ser 691, Thr 596, and Trp 597 (Fig. 3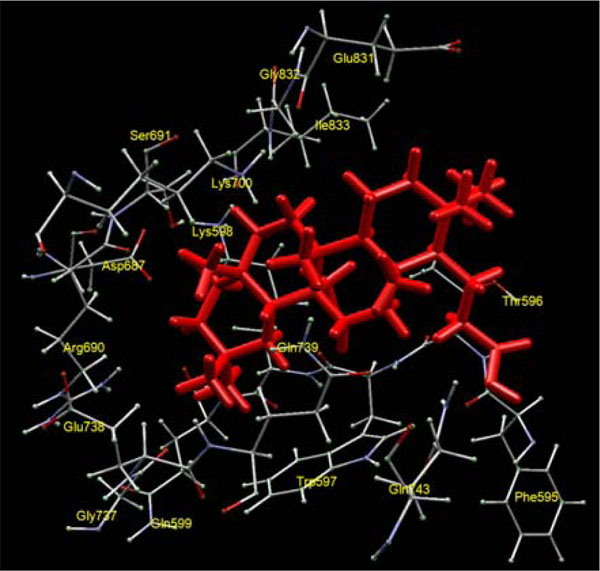 ). Mizushina and co-workers [24Mizushina Y, Sugawara F, Iida A. J Mol Biol 2000; 304: 385-95.
). Mizushina and co-workers [24Mizushina Y, Sugawara F, Iida A. J Mol Biol 2000; 304: 385-95.
[http://dx.doi.org/10.1006/jmbi.2000.4223] [PMID: 11090281] ] found this to be the preferred binding site for unsaturated fatty acids with yeast topoisomerase II. Not surprisingly, the nature of binding of these lipophilic triterpenoids is largely hydrophobic, and the triterpenoids can dock in various orientations in this binding pocket. There are some trends, however. The lowest-energy pose of lupeol (5) is such that it forms hydrogen bonds between the C(3) hydroxyl group of the ligand and the carboxylate of Asp 687 and the guanidinium of Arg 690.
Ursolic acid (8) and ganoderic acid X (21) occupy analogous orientations. Both betulinic acid and ursolic acid orient themselves in the binding site to form a salt bridge between the carboxylates of the ligands and the ammonium of Lys 700. Fernane 19 and seco-3,4-friedelin (3) have very similar orientations, but no obvious interactions between the carboxylates and nearby amino acid residues. Fomitellic acid A (6), 3α-corosolic acid (15), and dihydrocelastrol (18), dock into the DNA binding site of topoisomerase II, but preferentially occupy different locations than the other triterpenoid ligands (see Fig. 4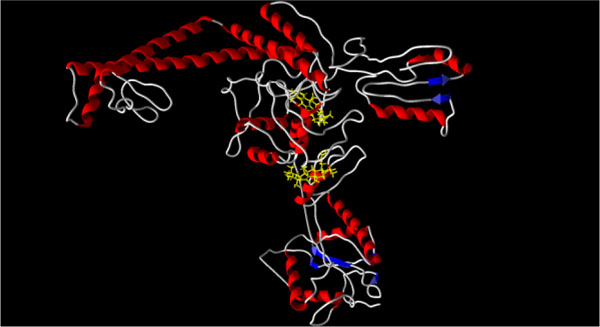 ). This alternative binding site is defined by Ala 830, Asn 756, Asn 828, Asp 697, Gln 703, Gln 739, Gln 750, Glu 831, Gly 698, Gly 747, Gly 829, Gly 832, Ile 758, Ile 825, Leu 748, Leu 760, Lys 700, Met 824, and Phe 699. The key interactions involved in docking dihydrocelastrol are a salt bridge between the carboxylate of the ligand and the ammonium moiety of Lys 700, hydrogen bonding between the C(2) hydroxyl group of the ligand and the carbonyl of Gly 829. Interestingly, seco-3,4-taraxerone (4) preferentially docks into an altogether different site (Fig. 4
). This alternative binding site is defined by Ala 830, Asn 756, Asn 828, Asp 697, Gln 703, Gln 739, Gln 750, Glu 831, Gly 698, Gly 747, Gly 829, Gly 832, Ile 758, Ile 825, Leu 748, Leu 760, Lys 700, Met 824, and Phe 699. The key interactions involved in docking dihydrocelastrol are a salt bridge between the carboxylate of the ligand and the ammonium moiety of Lys 700, hydrogen bonding between the C(2) hydroxyl group of the ligand and the carbonyl of Gly 829. Interestingly, seco-3,4-taraxerone (4) preferentially docks into an altogether different site (Fig. 4 ) in the DNA binding region of topoisomerase II, in contrast to the other seco-3,4-triter-penoids, 1-3. This binding site is defined by Ala 722, Ala 725, Ala 742, Ala 777, Ala 778, Ala 779, Ala 780, Arg 781, Gln 743, Glu 589, Glu 590, His 593, Ile 746, Pro 726, Ser 740, and Val 721. The key interaction in the docked pose is a salt bridge between the carboxylate of the ligand and His 593.
) in the DNA binding region of topoisomerase II, in contrast to the other seco-3,4-triter-penoids, 1-3. This binding site is defined by Ala 722, Ala 725, Ala 742, Ala 777, Ala 778, Ala 779, Ala 780, Arg 781, Gln 743, Glu 589, Glu 590, His 593, Ile 746, Pro 726, Ser 740, and Val 721. The key interaction in the docked pose is a salt bridge between the carboxylate of the ligand and His 593.
 |
Fig. (1) X-ray crystal structure of human topoisomerase II bound
to DNA (PDB: 2rgr) [23Dong KC, Berger JM. Nature 2007; 450: 1201-5. |
 |
Fig. (2) X-ray crystal structure of Saccharomyces cerevisiae topoisomerase
II (PDB: 1bjt) [22Fass D, Bogden CE, Berger JM. Nature Struct Biol 1999; 6: 322-6. |
 |
Fig. (3) Preferred triterpenoid binding site of yeast topoisomerase II with seco-3,4-friedelin as ligand. |
 |
Fig. (4) Structure of yeast topoisomerase II (PDB: 1bjt) with docked ligands, fomitellic acid A, 6 (lower pose) and dihydrocelastrol, 18 (upper pose). |
The triterpenoid ligands were docked into the ATP binding sites of both Saccharomycescerevisiae topoisomerase II (two different structures, PDB: 1qzr and 1pvg [25Classen S, Olland S, Berger JM. Proc Natl Acad Sci USA 2003; 100: 10629-34.
[http://dx.doi.org/10.1073/pnas.1832879100] [PMID: 12963818] [PMCID: PMC196855] ]) and human topoisomerase II (PDB: 1zxm [26Wei H, Ruthenburg AJ, Bechis SK, Verdine GL. J Biol Chem 2005; 280: 37041-7.
[http://dx.doi.org/10.1074/jbc.M506520200] [PMID: 16100112] ]) (see Fig. 5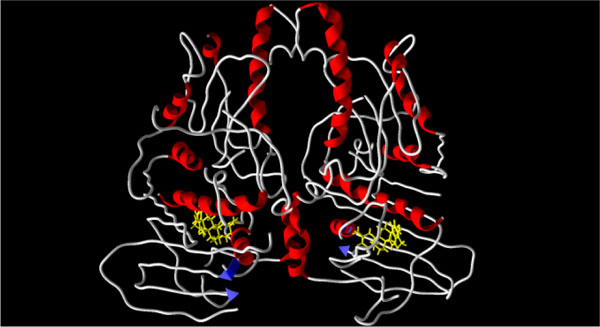 ). Most of the triterpenoid ligands showed lower binding (or no binding) affinity for the ATP binding sites. Four triterpenoids, however, seco-3,4-friedelin (3), demethylzeylasterone (12), celastrol (17), and dihydrocelastrol (18), showed stronger binding for the ATP binding sites than for the DNA binding site.
). Most of the triterpenoid ligands showed lower binding (or no binding) affinity for the ATP binding sites. Four triterpenoids, however, seco-3,4-friedelin (3), demethylzeylasterone (12), celastrol (17), and dihydrocelastrol (18), showed stronger binding for the ATP binding sites than for the DNA binding site.
 |
Fig. (5) X-ray crystal structure of Saccharomyces cerevisiae ATPase
region of topoisomerase II (PDB: 1pvg) [25Classen S, Olland S, Berger JM. Proc Natl Acad Sci USA 2003; 100: 10629-34. |
In the ATPase domain of yeast topoisomerase II, demethylzeylasterone (12) forms salt bridges between the C(23) carboxylate and the ammonium group of Lys 11 and the guanidinium group of Arg 77, as well as a hydrogen bond with Ser 128; hydrogen bonds between the C(29) carboxylate with the amide hydrogens of Arg 141, Gln 365, Gly 145, and Tyr 144; and a hydrogen bond between the C(6) ketone and the hydroxyl of Tyr 12. Both celastrol (16) and dihydrocelastrol (17) dock in the same orientation as demethylzeylasterone, with the same interactions between the C(29) carboxylate and Arg 141, Gln 365, Gly 145, and Tyr 144. Tingenone (23), known to be a cytotoxic agent [27Setzer WN, Setzer MC, Hopper AL, et al. Planta Med 1998; 64: 583.], but not yet shown to be a topoisomerase II inhibitor, also docks into the same site with the same orientation (see Fig. 6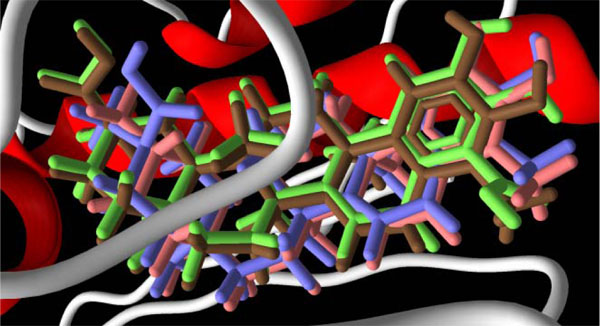 ). Tingenone, therefore, would be expected to be a topoisomerase II inhibitor. Friedelane 3 does not have planar rings and therefore binds to the ATP binding site differently than the quinone-methide triterpenoids 12, 17, 18, and 23. Key hydrogen bonding interactions between seco-3,4-friedelin (3) and the protein are between the C(3) carboxylate of the ligand and the amide of Asn 129 as well as the hydroxyl of Ser 128.
). Tingenone, therefore, would be expected to be a topoisomerase II inhibitor. Friedelane 3 does not have planar rings and therefore binds to the ATP binding site differently than the quinone-methide triterpenoids 12, 17, 18, and 23. Key hydrogen bonding interactions between seco-3,4-friedelin (3) and the protein are between the C(3) carboxylate of the ligand and the amide of Asn 129 as well as the hydroxyl of Ser 128.
The binding of the quinone-methide triterpenoids to the ATPase domain of human topoisomerase II is similar to that observed in the yeast. Demethylzeylasterone (12) interacts with Arg 98 and Ser 149, through the C(23) carboxylate. The C(29) carboxylate interacts with Arg 162, Gln 376, Gly 166, and Tyr 165; and the C(6) ketone hydrogen bonds with Tyr 34. Similarly, seco-3,4-friedelin (3) and seco-3,4-taraxerone (4) bind to the ATP binding site of human topoisomerase II through hydrogen bonding between the C(3) carboxylates of the ligands and Asn 150 and Ser 149. The binding energies of the quinone-methide triterpenoids to the ATP binding sites of topoisomerase II are comparable to those calculated (Molegro) for known ATP binders salvicine [28Hu CX, Zuo ZL, Xiong B, et al. Mol Pharmacol 2006; 70: 1593-601.
[http://dx.doi.org/10.1124/mol.106.027714] [PMID: 16914642] ] (average binding energy = -25.2 kcal/mol) or etoposide [6Leroy D, Kajava AV, Frei C, Gasser SM. Biochemistry Source Journal 2001; 40: 1624-34.
[http://dx.doi.org/10.1021/bi0019141] [PMID: 11327821] , 7Sengupta T, Mukherjee M, Das A, et al. Biochem J 2005; 390: 419-26.
[http://dx.doi.org/10.1042/BJ20042128] [PMID: 15901238] [PMCID: PMC1198921] ] (average binding energy = -22.4 kcal/mol).
There is no discernable trend between the calculated binding energies in this study and the reported topoisomerase II inhibitory activities. This may be due to the different sources of topoisomerase II used (e.g., yeast, human, parasite), or the fact that enzyme inhibitory concentrations have large differences. Thus, for example, ursolic acid has been reported to have IC50 values of 20 μM [13Syrovets T, Büchele B, Gedig E, Slupsky JR, Simmet T. Mol Pharmacol Source Journal 2000; 58: 71-81.
[PMID: 10860928] ], 36 μM [16Mizushina Y, Ikuta A, Endoh K, et al. Biochem Biophys Res Commun 2003; 305: 365-73.
[http://dx.doi.org/10.1016/S0006-291X(03)00765-4] ], 150 μM [12Mizushina Y, Iida A, Ohta K, Sugawara F, Sakaguchi K. Biochem J 2000; 350: 757-63.
[http://dx.doi.org/10.1042/0264-6021:3500757] [PMID: 10970789] [PMCID: PMC1221307] ]; etoposide had MIC values of 20 μM [15Chowdhury AR, Mandal S, Goswami A, et al. Mol Med 2003; 9: 26-36.
[PMID: 12765337] [PMCID: PMC1430381] ] and 25 μM [11Setzer WN, Shen X, Bates RB, et al. Fitoterapia Source Journal 2000; 71: 195-8.]; and acetyl α-boswellic acid had MIC values of 3 μM [13Syrovets T, Büchele B, Gedig E, Slupsky JR, Simmet T. Mol Pharmacol Source Journal 2000; 58: 71-81.
[PMID: 10860928] ] and > 50 μM [29Zhao W, Entschladen F, Liu H, et al. Cancer Detec Prev 2003; 27: 67-75.
[http://dx.doi.org/10.1016/S0361-090X(02)00170-8] ].
COMPUTATIONAL METHODS
Molecular structures for the triterpenoids were built using Spartan ’06 for Windows [30SPARTAN ’06 for Windows Wavefunction Inc Irvine California 2006.] and geometries optimized using the MMFF 94 force field [31Halgren TA. J Comp Chem 1996; 17: 490-519.
[http://dx.doi.org/10.1002/(SICI)1096-987X(199604)17:5/6<490::AID-JCC1>3.0.CO;2-P] ]. Protein-ligand docking studies were carried out based on the crystal structure of the DNA-binding and cleavage core (residues 409-1201) of yeast topoisomerase II (PDB: 1bjt) [22Fass D, Bogden CE, Berger JM. Nature Struct Biol 1999; 6: 322-6.
[http://dx.doi.org/10.1038/7556] [PMID: 10201398] ], the crystal structure of the DNA-binding and cleavage domain (residues 419-1177) of human topoisomerase IIα bound to G-segment DNA (PDB: 2rgr) [23Dong KC, Berger JM. Nature 2007; 450: 1201-5.
[http://dx.doi.org/10.1038/nature06396] [PMID: 18097402] ], the crystal structure of the N-terminal ATPase region of yeast topoisomerase II bound to dexrazoxane (PDB: 1qzr) and imino-ATP (PDB: 1pvg) [25Classen S, Olland S, Berger JM. Proc Natl Acad Sci USA 2003; 100: 10629-34.
[http://dx.doi.org/10.1073/pnas.1832879100] [PMID: 12963818] [PMCID: PMC196855] ], and the crystal structure of the ATPase region of human topoisomerase IIα bound to imino-ATP (PDB: 1zxm) [26Wei H, Ruthenburg AJ, Bechis SK, Verdine GL. J Biol Chem 2005; 280: 37041-7.
[http://dx.doi.org/10.1074/jbc.M506520200] [PMID: 16100112] ]. All solvent molecules, cofactors, and co-crystallized ligands were removed from the structures. Molecular docking calculations for all compounds were undertaken using Molegro Virtual Docker 2.3 [32Molegro Virtual Docker 2 Molegro ApS Aarhus Denmark 2007., 33Thompsen R, Christensen MH. J Med Chem 2006; 49: 3315-21.
[http://dx.doi.org/10.1021/jm051197e] [PMID: 16722650] ]. Because it is unknown how and where triterpenoids might bind to topoisomerase II, many different sites were examined in order to probe the entire protein structure for 1bjt. For the 2rgr structure, the DNA was removed from the structure and the triterpenoid ligands were docked in the DNA binding site of the protein. In the case of the ATPase region, only the ATP binding pockets of 1qzr, 1pvg, and 1zxm, were modeled. A sphere of radius 15 Å was centered on the binding site in order to allow each ligand to search. Different orientations of the ligands were searched and ranked based on their energy scores.
SUMMARY
A number of triterpenoid natural products have shown potential antitumor activity by inhibition of topoisomerase II. These enzyme inhibitors are not planar molecules and clearly, then, do not intercalate DNA to form stable cleavable complexes with topoisomerase II. The mode of inhibition as revealed by this study may be either to bind to the enzyme at the DNA binding site, preventing DNA binding, or binding to the ATP binding site, conformationally locking the enzyme and thus preventing DNA binding. The calculated binding energies of the triterpenoid inhibitors, however, do not correlate well with experimental inhibitory concentrations.
REFERENCES
| [1] | Pommier Y, Pourquier P, Fan Y, Strumberg D. Biochim. Biophys Acta 1998; 1400: 83-105. [http://dx.doi.org/10.1016/S0167-4781(98)00129-8] |
| [2] | Burden DA, Osheroff N. Biochim. Biophys Acta 1998; 1400: 139-54. [http://dx.doi.org/10.1016/S0167-4781(98)00132-8] |
| [3] | Heck MM, Earnshaw WCJ. J. Cell Biol 1986; 103: 2569-81. [http://dx.doi.org/10.1083/jcb.103.6.2569] |
| [4] | Wilstermann AM, Osheroff N. Curr Top Med Chem 2003; 3: 1349-64. [http://dx.doi.org/10.2174/1568026033452519] |
| [5] | Hande KR. Update Cancer Ther 2006; 1: 3-15. [http://dx.doi.org/10.1016/j.uct.2006.04.001] |
| [6] | Leroy D, Kajava AV, Frei C, Gasser SM. Biochemistry Source Journal 2001; 40: 1624-34. [http://dx.doi.org/10.1021/bi0019141] [PMID: 11327821] |
| [7] | Sengupta T, Mukherjee M, Das A, et al. Biochem J 2005; 390: 419-26. [http://dx.doi.org/10.1042/BJ20042128] [PMID: 15901238] [PMCID: PMC1198921] |
| [8] | Andoh T, Ishida R. Biochim Biophys Acta 1998; 1400: 155-71. [http://dx.doi.org/10.1016/S0167-4781(98)00133-X] |
| [9] | Larsen AK, Escargueil AE, Skladanowski A. Pharmacol Ther 2003; 99: 167-81. [http://dx.doi.org/10.1016/S0163-7258(03)00058-5] |
| [10] | Wada S, Tanaka R, Iida A, Matsunaga S. Bioorg Med Chem Lett 1998; 8: 2829-32. [http://dx.doi.org/10.1016/S0960-894X(98)00515-0] |
| [11] | Setzer WN, Shen X, Bates RB, et al. Fitoterapia Source Journal 2000; 71: 195-8. |
| [12] | Mizushina Y, Iida A, Ohta K, Sugawara F, Sakaguchi K. Biochem J 2000; 350: 757-63. [http://dx.doi.org/10.1042/0264-6021:3500757] [PMID: 10970789] [PMCID: PMC1221307] |
| [13] | Syrovets T, Büchele B, Gedig E, Slupsky JR, Simmet T. Mol Pharmacol Source Journal 2000; 58: 71-81. [PMID: 10860928] |
| [14] | Furbacher TR, Gunatilaka AAL. J Nat Prod Source Journal 2001; 64: 1294-6. [http://dx.doi.org/10.1021/np010123c] |
| [15] | Chowdhury AR, Mandal S, Goswami A, et al. Mol Med 2003; 9: 26-36. [PMID: 12765337] [PMCID: PMC1430381] |
| [16] | Mizushina Y, Ikuta A, Endoh K, et al. Biochem Biophys Res Commun 2003; 305: 365-73. [http://dx.doi.org/10.1016/S0006-291X(03)00765-4] |
| [17] | Nagase M, Oto J, Sugiyama S, Yube K, Takaishi Y, Sakato N. Biosci Biotech Biochem 2003; 67: 1883-7. [http://dx.doi.org/10.1271/bbb.67.1883] [PMID: 14519971] |
| [18] | Mizushina Y, Akihisa T, Ukiya M, et al. Cancer Sci 2004; 95: 354-60. [http://dx.doi.org/10.1111/j.1349-7006.2004.tb03215.x] [PMID: 15072595] |
| [19] | Li CH, Chen PY, Chang UM, et al. Life Sci 2005; 77: 252-65. [http://dx.doi.org/10.1016/j.lfs.2004.09.045] [PMID: 15878354] |
| [20] | Wada S, Tanaka R. Bioorg Med Chem Lett 2005; 15: 2966-9. [http://dx.doi.org/10.1016/j.bmcl.2005.04.052] [PMID: 15914002] |
| [21] | McClendon AK, Osheroff N. Mutat Res 2007; 623: 83-97. [http://dx.doi.org/10.1016/j.mrfmmm.2007.06.009] [PMID: 17681352] [PMCID: PMC2679583] |
| [22] | Fass D, Bogden CE, Berger JM. Nature Struct Biol 1999; 6: 322-6. [http://dx.doi.org/10.1038/7556] [PMID: 10201398] |
| [23] | Dong KC, Berger JM. Nature 2007; 450: 1201-5. [http://dx.doi.org/10.1038/nature06396] [PMID: 18097402] |
| [24] | Mizushina Y, Sugawara F, Iida A. J Mol Biol 2000; 304: 385-95. [http://dx.doi.org/10.1006/jmbi.2000.4223] [PMID: 11090281] |
| [25] | Classen S, Olland S, Berger JM. Proc Natl Acad Sci USA 2003; 100: 10629-34. [http://dx.doi.org/10.1073/pnas.1832879100] [PMID: 12963818] [PMCID: PMC196855] |
| [26] | Wei H, Ruthenburg AJ, Bechis SK, Verdine GL. J Biol Chem 2005; 280: 37041-7. [http://dx.doi.org/10.1074/jbc.M506520200] [PMID: 16100112] |
| [27] | Setzer WN, Setzer MC, Hopper AL, et al. Planta Med 1998; 64: 583. |
| [28] | Hu CX, Zuo ZL, Xiong B, et al. Mol Pharmacol 2006; 70: 1593-601. [http://dx.doi.org/10.1124/mol.106.027714] [PMID: 16914642] |
| [29] | Zhao W, Entschladen F, Liu H, et al. Cancer Detec Prev 2003; 27: 67-75. [http://dx.doi.org/10.1016/S0361-090X(02)00170-8] |
| [30] | SPARTAN ’06 for Windows Wavefunction Inc Irvine California 2006. |
| [31] | Halgren TA. J Comp Chem 1996; 17: 490-519. [http://dx.doi.org/10.1002/(SICI)1096-987X(199604)17:5/6<490::AID-JCC1>3.0.CO;2-P] |
| [32] | Molegro Virtual Docker 2 Molegro ApS Aarhus Denmark 2007. |
| [33] | Thompsen R, Christensen MH. J Med Chem 2006; 49: 3315-21. [http://dx.doi.org/10.1021/jm051197e] [PMID: 16722650] |