- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Conference Proceedings Journal
(Biological Sciences, Chemical Sciences, Physical Sciences, Medicine, Engineering & Technology)
(Discontinued)
ISSN: 2210-2892 ― Volume 10, 2020
Whole Cell Biocatalyst for Soyasapogenol B Production from Soybean Saponin
Hala A. Amin1, *, Mostafa M. Abo Elsoud2, 3, Ahmed. F. Sahab4
Abstract
Soyasapogenol B (SB), aglycone of soybean saponin, is known to have hepatoprotective, antimutagenic, antivirus, and anti-inflammatory activities. This research examined the use of whole-cell biocatalyst to produce SB from soybean saponin. It was found that Aspergillus flavus, a fungus isolated from peanut pods, was capable of expressing extracellular and intracellular saponin hydrolase enzyme. However, the total enzyme activity produced using fungal whole cells (37U) in the reaction mixture was about 3 times that produced using the extracellular (12.4U) or intracellular (11.5U) enzyme. Cells with maximum hydrolytic activity for production of SB (12.2 U/g) was obtained using production medium supplemented by 2% soybean saponin, as inducer for enzyme production, adjusted at pH 9 and incubated at 30°C for 2 days. The highest yield of SB was achieved when the reaction mixture was incubated at pH 5.5 and 45°C for 48h; using 20% wet cells (corresponding to 4% cell dry weight) and soybean saponin (2%, w/v) as a substrate. Under these optimal conditions, the cells bioconversion efficiency (SB yield) increased from 5.3 to 60%. Whole cell biocatalyst has several advantages with regard to industrial applications: a consistent quality, easy to be prepared and a very low price compared with purified enzyme. Consequently, this study is significant for production of SB from soybean saponin on an industrial scale.
Article Information
Identifiers and Pagination:
Year: 2016Volume: 7
First Page: 134
Last Page: 143
Publisher Id: TOPROCJ-7-1-134
DOI: 10.2174/2210289201607010134
Article History:
Received Date: 10/08/2015Revision Received Date: 22/05/2016
Acceptance Date: 18/07/2016
Electronic publication date: 08/09/2016
Collection year: 2016
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution-Non-Commercial 4.0 International Public License (CC BY-NC 4.0) (https://creativecommons.org/licenses/by-nc/4.0/legalcode), which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Chemistry of Natural and Microbial Products Department, National Research Center, Cairo, 12622, Egypt; Tel: +202-33464472; Fax: +202-37622603; E-mail: halaamin2007@yahoo.com
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 10-08-2015 |
Original Manuscript | Whole Cell Biocatalyst for Soyasapogenol B Production from Soybean Saponin | |
INTRODUCTION
Soybean saponins are triterpenoid glycosides that possess an oleanane-type aglycone (soyasapogenols) with polysaccharide chains [1Kamo, S.; Suzuki, S.; Sato, T. The content of soyasaponin and soyasapogenol in soy foods and their estimated intake in the Japanese. Food Sci. Nutr., 2014, 2(3), 289-297.
[http://dx.doi.org/10.1002/fsn3.107] [PMID: 24936299] ]. They are reported to have many biological properties, including hyperlipidemia prevention, anticancer, immunoregulation, antioxidation, antimutagenicity and HIV infection inhibition [2Amin, H.A.; Awad, H.M.; Hanna, A.G. Comparative evaluation of in vitro cytotoxicity, antiviral and antioxidant activities of different soyasapogenols from soybean saponin. Egypt Pharm. J., 2012, 11, 73-79.]. These saponins have a common structure with glucuronic acid linked at C-3 site of soyasapogenols A or B. Soyasapogenols are generally being more bioactive compared to their glycosides. It has been reported that soyasapogenol B (SB) has various physiological activities, such as hepatoprotective, antimutagenic, antivirus, anti-inflammatory, anticomplementary, platelet aggregation suppressing effects and growth suppressing effects on cells derived from human colon cancer, human ovarian cancer, human breast cancer cells and hep G2 cells [3Rowlands, J.C.; Berhow, M.A.; Badger, T.M. Estrogenic and antiproliferative properties of soy sapogenols in human breast cancer cells in vitro. Food Chem. Toxicol., 2002, 40(12), 1767-1774.
[http://dx.doi.org/10.1016/S0278-6915(02)00181-3] [PMID: 12419690] -5Zhang, W.; Popovich, D.G. Effect of soyasapogenol A and soyasapogenol B concentrated extracts on HEP-G2 cell proliferation and apoptosis. J. Agric. Food Chem., 2008, 56(8), 2603-2608.
[http://dx.doi.org/10.1021/jf0731550] [PMID: 18361499] ].
Microbial transformation has been extensively used, to create new and useful metabolites as a substitute of chemical synthesis for preparation of pharmacologically active compounds [6Chen, G.; Chen, J. A novel cell modification method used in biotransformation of glycerol to 3-HPA by Lactobacillus reuteri. Appl. Microbiol. Biotechnol., 2013, 97(10), 4325-4332.
[http://dx.doi.org/10.1007/s00253-013-4723-2] [PMID: 23359000] , 7Omar, M.N.; Khan, N.T.; Hasali, N.H. Microbial transformations of artemisinin anti-malaria drug. Adv. Biores, 2012, 3, 27-31.]. Microbial biotransformation by the whole cell microorganisms is often advantageous as compared to isolated enzymes; it is respected economically and ecologically a competitive tool for the biotechnological professionals in search of new techniques to manufacture clean valuable chemicals, pharmaceutical, and agrochemical compounds [8Carballeira, J.D.; Quezada, M.A.; Hoyos, P.; Simeó, Y.; Hernaiz, M.J.; Alcantara, A.R.; Sinisterra, J.V. Microbial cells as catalysts for stereoselective red-ox reactions. Biotechnol. Adv., 2009, 27(6), 686-714.
[http://dx.doi.org/10.1016/j.biotechadv.2009.05.001] [PMID: 19442715] , 9Luo, J.; Liang, Q.; Shen, Y.; Chen, X.; Yin, Z.; Wang, M. Biotransformation of bavachinin by three fungal cell cultures. J. Biosci. Bioeng., 2014, 117(2), 191-196.
[http://dx.doi.org/10.1016/j.jbiosc.2013.08.001] [PMID: 24012108] ]. The enzymes are more stable in the long run as compared to free enzymes as the enzymes in cells are protected from the external environment [10Kaushik, N.; Biswas, S.; Singh, J. Biocatalysis and biotransformation processes - an insight. Scitech J., 8, 15-22.]. The search for microorganisms, which efficiently produce SB by hydrolyzing soybean saponin results in finding different filamentous fungus strains that belong to genera; Neocosmospora, Eupenicillium and Aspergillus. SB can be produced using such fungi, as they are, like a method with Aspergillus terreus [11Amin, H.A.; Mohamed, S.S. Immobilization of Aspergillus terreus on loofa sponge for soyasapogenol B production from soybean saponin. J. Mol. Catal., B Enzym., 2012, 78, 85-90.
[http://dx.doi.org/10.1016/j.molcatb.2012.03.015] ] and a method with Aspergillus parasiticus [12Amin, H.A.; Hassan, Y.M.; Yehia, S.M. Biotransformation of soybean saponin to soyasapogenol B by immobilized Aspergillus Parasiticus cells on polyurethane foam. Res. J. Pharm. Biol. Chem. Sci., 2013, 5, 332-341.]. Although, it has been found that SB is produced by these methods at high concentration in medium containing soybean saponin, but downstream processing of reaction products from the culture is difficult. This is because of the surface-active property of saponins, which increase the medium viscosity; consequently,the extraction process has to be repeated several times to improve the yield in producing the target substance from the culture [4Watanabe, M.; Mido, N.; Tamura, T. Saponin digesting enzymes, genes there of and soyasapogenol B mass production system. U.S Patent 7022508 B2, April 2006.]. Therefore, there is a need to develop a method for large-scale production of SB by an enzyme reaction using “saponin-hydrolysing enzyme”. Consequently, soybean saponin hydrolase was isolated and cloned from Neocosmospora vasinfecta var. vasinfecta PF1225 [13Watanabe, M.; Sumida, N.; Yanai, K.; Murakami, T. A novel saponin hydrolase from Neocosmospora vasinfecta var. vasinfecta. Appl. Environ. Microbiol., 2004, 70(2), 865-872.
[http://dx.doi.org/10.1128/AEM.70.2.865-872.2004] [PMID: 14766566] ] Eupenicillium brefeldianum PF122 [14Watanabe, M.; Sumida, N.; Yanai, K.; Murakami, T. Cloning and characterization of saponin hydrolases from Aspergillus oryzae and Eupenicillium brefeldianum. Biosci. Biotechnol. Biochem., 2005, 69(11), 2178-2185.
[http://dx.doi.org/10.1271/bbb.69.2178] [PMID: 16306700] ] and Aspergillus oryzae PF1224 [14Watanabe, M.; Sumida, N.; Yanai, K.; Murakami, T. Cloning and characterization of saponin hydrolases from Aspergillus oryzae and Eupenicillium brefeldianum. Biosci. Biotechnol. Biochem., 2005, 69(11), 2178-2185.
[http://dx.doi.org/10.1271/bbb.69.2178] [PMID: 16306700] ]. Moreover, the genes encoding this enzyme was successfully expressed in Trichoderma viride for preparation of the recombinant enzyme [13Watanabe, M.; Sumida, N.; Yanai, K.; Murakami, T. A novel saponin hydrolase from Neocosmospora vasinfecta var. vasinfecta. Appl. Environ. Microbiol., 2004, 70(2), 865-872.
[http://dx.doi.org/10.1128/AEM.70.2.865-872.2004] [PMID: 14766566] , 14Watanabe, M.; Sumida, N.; Yanai, K.; Murakami, T. Cloning and characterization of saponin hydrolases from Aspergillus oryzae and Eupenicillium brefeldianum. Biosci. Biotechnol. Biochem., 2005, 69(11), 2178-2185.
[http://dx.doi.org/10.1271/bbb.69.2178] [PMID: 16306700] ].
On the other hand, utilizing whole cell producing saponin hydrolase in which the purification and stabilization of the enzyme are not necessary instead of conventional immobilized enzyme for SB production is a potential way to reduce the biocatalyst cost, because immobilization can be carried out spontaneously during the process of cell cultivation [15Sun, T.; Du, W.; Liu, D. Prospective and impacts of whole cell mediated alcoholysis of renewable oils for biodiesel production. Biofuels Bioprod. Biorefin., 2009, 3, 633-639.
[http://dx.doi.org/10.1002/bbb.180] ]. For the best of our knowledge, there have been no reports of SB production using whole cells producing saponin hydrolase in the enzyme reaction mixture. However, several other biotransformation reaction trials utilizing whole cells were reported including, for example, microbial production of glycyrrhetic acid-3-O-mono-β-D-glucuronide from glycyrrhizin by Cryptococcus magnus MG-27 cells [16Kuramoto, T.; Ito, Y.; Oda, M. Microbial production of glycyrrhetic acid-3-O-mono-β-D-glucuronide from glycyrrhizin by Cryptococcus magnus MG-27. Biosci. Biotechnol. Biochem., 1994, 58, 455-458.
[http://dx.doi.org/10.1271/bbb.58.455] ] and utilizing Rhizopus oryzae (ATCC 9374) whole-cell biocatalyst for biodiesel production from soybean oil [17Hashemizadeh, S.N. A comparative study of immobilized-whole cell and commercial lipase as a biocatalyst for biodiesel production from soybean oil. Int. J. Curr. Microbiol. App. Sci., 2014, 3, 741-752.].
In this study, SB was produced by an enzyme reaction using Aspergillus flavus whole-cells producing saponin hydrolase enzyme. The conditions for cultivation of this fungus with maximum hydrolytic activity, as well as, the reaction conditions of soybean saponin with the cells for maximum yield of SB were investigated.
MATEDIALS AND METHODS
Isolation of Fungi
Samples of the peanut pods subjected to assay were obtained from Northern of El- Tahrir region during the growing season of 2014. One hundred seeds were surface disinfected by soaking in 2% sodium hypochlorite solution for 3 min; followed by three successive rinses in sterile distilled water. The sterilized seeds were platted on Potato Dextrose Agar (PDA, difco) medium and incubated for 7 days at 27±2ºC [18Foor, S.R.; Tenne, F.D.; Sinclair, J.B. Occurrence of seed borne microorganisms and germination in culture for determining seed health in soybeans. Pl. Dis. Reptre., 1976, 60, 970-977.]. Fungi growing from the seeds were isolated and purified.
Morphological Identification of Fungal Isolates
The obtained fungal colonies were cultured on PDA plates and incubated at 27±2 °C for 5 days for complete sporulation. The texture and color of the fungal colonies were observed. The color of the substrate mycelia was also recorded. The microscopic features of the fungal isolates were observed by preparing wet mounts of the fungal mycelia with Lacto-phenol Blue solution. The slides were observed in low light intensity under a 40x objective lens of a binocular light microscope. The fungal isolates were identified based on the shape of conidia and arrangement of spores on the mycelia (conidial ontogeny) according to Gilman [19Gilman, J.C. A manual of soil fungi, 2nd ed; The Iowa State University Press: Ames, Iowa, 1957. ], Barnett and Hunter [20Barnett, H.L.; Hunter, B.B. Illustrated genera of fungi, 3rd ed; Burgess Publ. Co.: Minneapolis, MN, USA, 2000. ], Domsch et al. [21Domsch, K.H.; Gams, W.; Anderson, T.H. Compendium of soil fungi, 2nd ed; APS Press, 2007. ] and Samson et al. [22Samson, R.A.; Houbraken, J.; Thrane, U. Food and indoor fungi. CBS-KNAW: Fungal Biodiversity Centre Utrecht; Institute of the Royal Netherlands Acadaemy of Arts and Science, 2010. ].
Screening Strains
The isolated fungi were maintained on potato dextrose agar (PDA-Difco). Unless stated otherwise, soybean saponin hydrolase production was carried out in production medium consisting of (g/l) 10 soybean saponin, 40 malt extract, 20 yeast extract, 2 KH2PO4, 2 (NH4)2SO4, 0.3 MgSO4·7H2O and 0.3 CaCl2·2H2O with a pH adjusted to 7.0 [13Watanabe, M.; Sumida, N.; Yanai, K.; Murakami, T. A novel saponin hydrolase from Neocosmospora vasinfecta var. vasinfecta. Appl. Environ. Microbiol., 2004, 70(2), 865-872.
[http://dx.doi.org/10.1128/AEM.70.2.865-872.2004] [PMID: 14766566] ]. Each isolate was cultured in Erlenmeyer flasks (250 ml) containing 100 ml production medium and incubated for 72 h at 30°C on a rotary shaker at 150 rpm. Then, the enzyme activities of supernatant (extracellular enzyme) and cell extract (intracellular enzyme) were determined for each isolate in the whole fermentation process.
Analytical Methods
Thin layer chromatography (TLC) was performed on pre-coated silica gel plate (Merck, silica gel 60F-254). The plate was chromatographed for SB with a solvent system of benzene:ethyl acetate:acetic acid (12:4:0.5, v/v/v). SB having an Rf value of 0.35 was detected on TLC plates by acid charring (10% H2SO4, 120°C, 10 min).
High-pressure liquid chromatography (HPLC) was performed with an Agilent 1260 HPLC (USA) under the following conditions: column, ZORBAX Eclipse Plus C18 (4.6 by 250 mm); column temperature, 40°C; mobile phase, acetonitrile-methanol-water (50:35:15); flow rate, 0.8 ml/min; and UV detector operating 200 nm. 50 µl of ethyl acetate containing reaction products was diluted with 450 µl of the mobile phase. 10 µl of this dilution was analyzed by HPLC, and the quantity of SB in the sample was determined by comparison with authentic SB.
Aspergillus flavus Enzyme Preparations and Measurement of Enzymatic Activity
After cultivation of A. flavus, the cells were separated from the cell filtrate by filtration, and then washed with saline solution. The wet cells were weighed and used directly in the reaction mixture (whole cells), grounded with approximately twice its weight of washed cold sand in a cold mortar and extracted by acetate buffer (grinded cell extract),or re-suspended in acetate buffer (pH 5) and broken by ultra-sonicator in ice-water bath (sonicated cell extract). Saponin hydrolase activity was measured in supernatant (extracellular enzyme) and cell extract (intracellular enzyme) as follows, unless otherwise stated, to 1ml of 2% soybean saponin suspended in 0.2 M acetate (pH 5) 1 ml of enzyme solution was added, and the mixture was allowed to react at 40°C for 1 h. In case of whole cells, 1g wet weight was suspended in 5ml acetate buffer containing 1% soybean saponin and the mixture was allowed to react at 40°C for 1 h. The resulting reaction solution was extracted with an equal volume of ethyl acetate and the resulting extract was mounted on TLC plate for detection of SB. Then, it was quantitatively analyzed by HPLC. One unit of enzyme activity is defined as the amount of enzyme that produces 1 mg of aglycone per hour from the substrate. A whole cell based specific activity can be defined as mg of aglycone per hour per g dry wt cells.

Whole Cell Bioconversion in Bioreactor
The seed culture was prepared by inoculation of 100 ml of fresh production medium; its pH was adjusted initially at pH 9 and containing 2% soybean saponin, with fungal spore suspension of 5d old slant. The culture was cultivated for 48h at 30°C and 200 rpm. For preparation of the batch culture, the flask culture was inoculated into a 2.5 l New Brunswick Bioflo 3 bioreactor (UK) at 10% (v/v) inoculation volume and 1 l operating volume. Dissolved oxygen was provided by injection of filtered air at a flow rate of 1 vvm, and the agitation speed was maintained at 150 rpm. The obtained cells were harvested after 48h by filtration and washed with saline solution. Bioconversion was initiated subsequently by re-suspending cells in acetate buffer (1000 ml, pH 5.5) containing 2% soybean saponin and adjusting the temperature to 45°C for 48h with stirring.
Isolation of SB from Reaction Mixture
After the completion of reactions, whole-broth extraction with two volumes of ethyl acetate was repeated 4 times. Ethyl acetate phase was collected, dried over Na2SO4 and filtered, followed by evaporation yielding a crude product in the form of a yellow-brownish residue. Where pure compounds were needed, the dried residue (9.63 g) was dissolved in chloroform: methanol (1: 1) and mixed with 10 g silica gel. The solvent was distilled off and the dried mixture applied to a silica gel column (5 x 120 cm) and eluted first with benzene, then the polarity was increased by the gradual addition of ethyl acetate (2.5, 5, 7.5, 10, 15%). Fractions of 100 ml were collected, concentrated and monitored by TLC using benzene: ethyl acetate: acetic acid (12/4/0.5, v/v) as a developing system. Identical fractions containing SB eluted with benzene: ethyl acetate (9:1) were combined and the solvent was distilled off. The isolated compound (6.5 g) was further purified by re-crystallization from methanol to yield pure SB.
Statistical Analysis
All the experiments were performed in triplicate, and the values expressed as the means of duplicate measurements of three independent samples. Data were analyzed by one-way analysis of variance (ANOVA) followed by Duncan’s multiple range test (DMRT). The least significant differences at confidence level of 5% (LSD .05) and 1% (LSD .01) were calculated to compare the influence of different treatments on the enzyme activity and bioconversion efficiency.
RESULTS AND DISCUSSION
Morphological Identification of Fungal Isolates
Fifteen isolates of filamentous fungi associated with peanut seeds (Fig. 1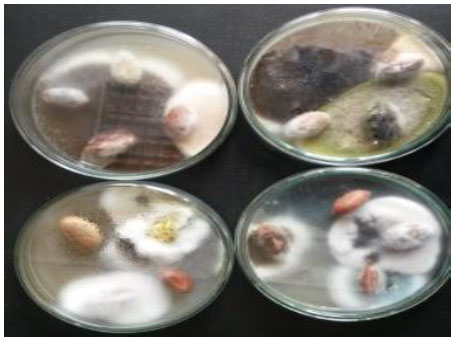 ) were obtained, among which four isolates showed saponin hydrolase activity, through detection of SB in the hydrolyzing mixture of soybean soponin by TLC. An isolate showed yellow green woolly or granular colonies on PDA. The colonies were thin, with radial grooves. Microscopic observation of a fungal isolate revealed colourless, long coarsely roughened, erect conidiophores with globose to subglubose vesicles bearing chains of conidia. Conidia appeared globose to subglobose, colorless to yellow green in colour. Sterigmata appeared either single or double with radiate or very loosely columnar heads. Reference to the modern identification proposed by Domsch et al. [21Domsch, K.H.; Gams, W.; Anderson, T.H. Compendium of soil fungi, 2nd ed; APS Press, 2007. ] and Samson et al. [22Samson, R.A.; Houbraken, J.; Thrane, U. Food and indoor fungi. CBS-KNAW: Fungal Biodiversity Centre Utrecht; Institute of the Royal Netherlands Acadaemy of Arts and Science, 2010. ] the colony morphology; pigmentation and microscopic characteristics of the isolate resembled that of A. flavus. The other fungal isolates were identified as A. terreus, A. parasiticus and A. tamari.
) were obtained, among which four isolates showed saponin hydrolase activity, through detection of SB in the hydrolyzing mixture of soybean soponin by TLC. An isolate showed yellow green woolly or granular colonies on PDA. The colonies were thin, with radial grooves. Microscopic observation of a fungal isolate revealed colourless, long coarsely roughened, erect conidiophores with globose to subglubose vesicles bearing chains of conidia. Conidia appeared globose to subglobose, colorless to yellow green in colour. Sterigmata appeared either single or double with radiate or very loosely columnar heads. Reference to the modern identification proposed by Domsch et al. [21Domsch, K.H.; Gams, W.; Anderson, T.H. Compendium of soil fungi, 2nd ed; APS Press, 2007. ] and Samson et al. [22Samson, R.A.; Houbraken, J.; Thrane, U. Food and indoor fungi. CBS-KNAW: Fungal Biodiversity Centre Utrecht; Institute of the Royal Netherlands Acadaemy of Arts and Science, 2010. ] the colony morphology; pigmentation and microscopic characteristics of the isolate resembled that of A. flavus. The other fungal isolates were identified as A. terreus, A. parasiticus and A. tamari.
 |
Fig. (1) Different fungi associated with peanut seeds after incubation on PDA plates for 5 days at 27±2 °C. |
Screening of Isolates for Soybean Saponin Hydrolase Production
Four strains of genus Aspergillus were screened for their ability to produce saponin hydrolase that can degrade soybean saponin and generate SB. Enzyme activities were monitored by measuring the amount of SB released in the enzyme reaction. In this context, Kudou et al. [23Kudou, S.; Tsuizaki, I.; Shimoyamada, M. Screening for microorganisms producing soybean saponin hydrolase. Agric. Biol. Chem., 1990, 54, 3035-3037.] cultured 158 strains of the genus Aspergillus in a medium containing soybean saponin and reported that 26 of them had marked soybean saponin hydrolase activity. It could be noticed from data presented in Table 1 that these strains manifested large variations in capacity to produce saponin hydrolase, which proved to be a strain dependent property. Among tested strains, A. flavus showed the highest extracellular and intracellular enzyme activities (10.55 and 9.39 U,respectively). Additionally, A. terreus and A. parasiticus were not significantly different in terms of extracellular enzyme activities, while A. parasiticus exhibited significantly higher intracellular enzyme activity. On the other hand, significantly lower extracellular and intracellular enzyme activities were associated with A. tamari.
Effect of Different A. flavus Enzyme Preparations on Saponin Hydrolase Activity
The enzyme activity of extracellular (supernatant), intracellular enzyme (sonicated or grinded cell extract) preparations and whole cells from A. flavus fermentation were investigated. Results illustrated in Fig. (2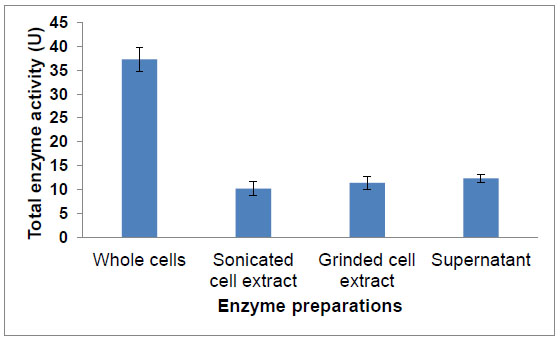 ) show that there were no significant difference between the extracellular and intracellular enzyme preparations. While, whole cells showed significantly higher total enzyme activity among different enzyme preparations, which represented about 3 times that produced by extracellular, intracellular enzyme preparations (Fig. 2
) show that there were no significant difference between the extracellular and intracellular enzyme preparations. While, whole cells showed significantly higher total enzyme activity among different enzyme preparations, which represented about 3 times that produced by extracellular, intracellular enzyme preparations (Fig. 2 ). This could be attributed to the possibility of being this enzyme an associated or integral membrane protein. It was reported that membrane proteins are difficult to be purified because they are present in low levels and they require detergents to become soluble in an aqueous solution [24Lin, S.H.; Guidotti, G. Methods in Enzymology, 192-Guide to Protein Purification, 2nd ed; Richard, R.B.; Murray, P.D., Eds.; Academic Press, Elsevier Inc., 2009, pp. 619-629.
). This could be attributed to the possibility of being this enzyme an associated or integral membrane protein. It was reported that membrane proteins are difficult to be purified because they are present in low levels and they require detergents to become soluble in an aqueous solution [24Lin, S.H.; Guidotti, G. Methods in Enzymology, 192-Guide to Protein Purification, 2nd ed; Richard, R.B.; Murray, P.D., Eds.; Academic Press, Elsevier Inc., 2009, pp. 619-629.
[http://dx.doi.org/10.1016/S0076-6879(09)63035-4] ]. Accordingly, the direct use of whole-cell biocatalysts in the enzyme reaction mixture is suspected to improve the cost effectiveness of the bioconversion processes [25Narita, J.; Okano, K.; Tateno, T.; Tanino, T.; Sewaki, T.; Sung, M.H.; Fukuda, H.; Kondo, A. Display of active enzymes on the cell surface of Escherichia coli using PgsA anchor protein and their application to bioconversion. Appl. Microbiol. Biotechnol., 2006, 70(5), 564-572.
[http://dx.doi.org/10.1007/s00253-005-0111-x] [PMID: 16133338] ]. In contrary to the present study, different saponin hydrolases were isolated from the cultural filtrate of Neocosmospora vasinfecta var. vasinfecta PF1225 [13Watanabe, M.; Sumida, N.; Yanai, K.; Murakami, T. A novel saponin hydrolase from Neocosmospora vasinfecta var. vasinfecta. Appl. Environ. Microbiol., 2004, 70(2), 865-872.
[http://dx.doi.org/10.1128/AEM.70.2.865-872.2004] [PMID: 14766566] ] Eupenicillium brefeldianum PF122 [14Watanabe, M.; Sumida, N.; Yanai, K.; Murakami, T. Cloning and characterization of saponin hydrolases from Aspergillus oryzae and Eupenicillium brefeldianum. Biosci. Biotechnol. Biochem., 2005, 69(11), 2178-2185.
[http://dx.doi.org/10.1271/bbb.69.2178] [PMID: 16306700] ] and Aspergillus oryzae PF1224. Another advantage of whole cell systems is that they do not need extra cofactors, as they possess all the necessary enzymes for cofactor regeneration [26Stewart, J.D.; Rodriquez, S.; Kayser, M.M. Enzyme technology for pharmaceutical and biotechnological applications; Marcel Dekker: New York, 2001. , 27Bommarius, A.S.; Riebel, B.R. Biocatalysis: fundamentals and applications; Wiley-VCH Verlag GmbH&Co. : KGaA, Weinheim, 2004.
[http://dx.doi.org/10.1002/3527602364] ]. This could explain their higher enzyme activity compared to the extra- and intra-enzyme preparations.
 |
Fig. (2) Saponin hydrolase total activity of different A. flavus enzyme preparations. |
Optimization of A. flavus Cultivation Conditions for Soybean Saponin Hydrolase Production
As the optimization of the growth environment is important to achieving maximal enzyme production, the evaluation of the effects of soybean saponin concentration, medium pH and the incubation period was investigated. Kudou et al. [28Kudou, S.; Tsuizaki, I.; Uchida, T.; Okubo, K. Purification and some properties of soybean saponin hydrolase from Aspergillus oryzae KO-2. Agric. Biol. Chem., 1991, 55(1), 31-36.
[PMID: 1368672] ] reported that saponin hydrolase was an enzyme induced by the existence of soybean saponin, as it has high substrate specificity for the glucuronide bonds of glycosides. As shown in Fig. (3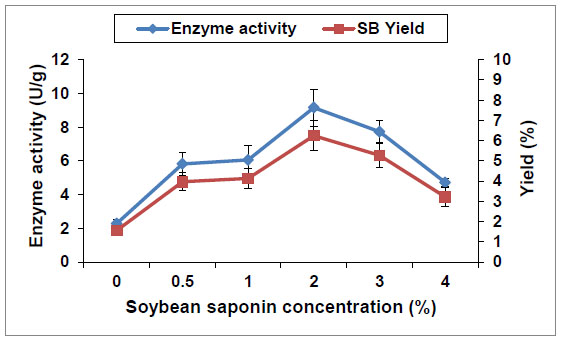 ), saponin hydrolase activity is significantly affected by presence of soybean saponin in the production medium. Increasing of soybean saponin concentation from 0 to 2% was accompanied by about 4 folds increase in the enzyme activity (from 2.3 to 9.2 U/g, respectively). Simultaneously, 2% soybean saponin concentration resulted in the highest SB yield of 6.2%. Thereafter, higher soybean saponin concentrations (3 and 4%) resulted in lower enzyme activities. This can be attributed to increase of the medium viscosity because of the surface active property of saponins. Moreover, the course of A. flavus growth and the hydrolysis activities is shown in Fig. (4
), saponin hydrolase activity is significantly affected by presence of soybean saponin in the production medium. Increasing of soybean saponin concentation from 0 to 2% was accompanied by about 4 folds increase in the enzyme activity (from 2.3 to 9.2 U/g, respectively). Simultaneously, 2% soybean saponin concentration resulted in the highest SB yield of 6.2%. Thereafter, higher soybean saponin concentrations (3 and 4%) resulted in lower enzyme activities. This can be attributed to increase of the medium viscosity because of the surface active property of saponins. Moreover, the course of A. flavus growth and the hydrolysis activities is shown in Fig. (4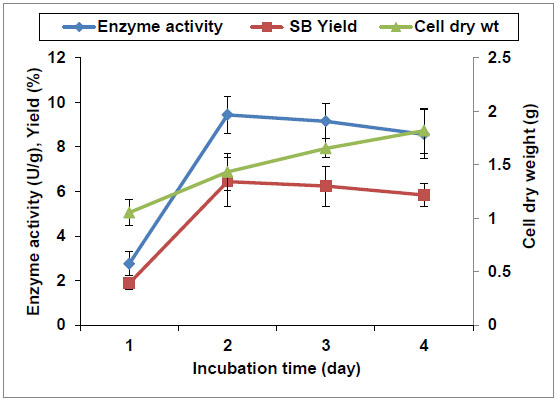 ). The cells with the maximum enzyme activity (9.4 U/g) were obtained after cultivation for 48 h. However, more biomass was produced with increasing the incubation period up to 96 h. Consequently, the growth of A. flavus was partial coupled with enzyme production and the highest enzyme activity was recorded in the middle of exponential phase. Additionally, to determine the optimal H+ concentration for saponin hydrolase production, A. flavus was cultivated over the range of pH 5-10 (data not shown). The saponin hydrolase enzyme was highly active in the range of pH 8-9, and the maximal activity of 12.4 U/g was observed at pH 9. This in accordance with Guçlu-Ustundag and Mazza [29Güçlü-Ustündağ, O.; Mazza, G. Saponins: properties, applications and processing. Crit. Rev. Food Sci. Nutr., 2007, 47(3), 231-258.
). The cells with the maximum enzyme activity (9.4 U/g) were obtained after cultivation for 48 h. However, more biomass was produced with increasing the incubation period up to 96 h. Consequently, the growth of A. flavus was partial coupled with enzyme production and the highest enzyme activity was recorded in the middle of exponential phase. Additionally, to determine the optimal H+ concentration for saponin hydrolase production, A. flavus was cultivated over the range of pH 5-10 (data not shown). The saponin hydrolase enzyme was highly active in the range of pH 8-9, and the maximal activity of 12.4 U/g was observed at pH 9. This in accordance with Guçlu-Ustundag and Mazza [29Güçlü-Ustündağ, O.; Mazza, G. Saponins: properties, applications and processing. Crit. Rev. Food Sci. Nutr., 2007, 47(3), 231-258.
[http://dx.doi.org/10.1080/10408390600698197] [PMID: 17453922] ] who reported that a sharp increase in the solubility of soyasaponin Bb was observed in the pH range 6.5-7.3, which was very low in the acidic region. Accordingly, shifting pH to the alkaline region increase the solubility of soybean saponin in the medium allowing more induction for enzyme production.
 |
Fig. (3) Effect of soybean saponin concentration in the production medium on A. flavus cells saponin hydrolase activity. |
 |
Fig. (4) Time course of soybean saponin hydrolase production by A. flavus cells. |
Optimization of Soybean Saponin Hydrolase Reaction Conditions
The enzymatic activity of A. flavus cells, produced under the optimized cultivation conditions, was assayed at varying temperature (25-60°C), pH (4-9), wet cell weight (2.5- 20%), substrate concentration (0.5-3%) and at different time intervals (1- 96 h) to achieve high product concentrations. Results (data not shown) showed that the reaction rate was significantly increased by increasing reaction temperature up to 45°C, at which the highest yield of SB (12.3%) was attained. On the other hand, relatively high SB yields were obtained at pH range of 5-7 (data not shown).Maximum SB output of 16.42% was obtained at pH 5.5. These results is closely related to that obtained by Aspergillus oryzae KO-2 saponin hydrolase, where its optimum pH was pH 4.5 to 5.0 and its optimum temperature was 50°C [23Kudou, S.; Tsuizaki, I.; Shimoyamada, M. Screening for microorganisms producing soybean saponin hydrolase. Agric. Biol. Chem., 1990, 54, 3035-3037.].
Regarding the effect of cell-wet weight in the reaction mixture on the enzyme activity and SB yield, Fig. (5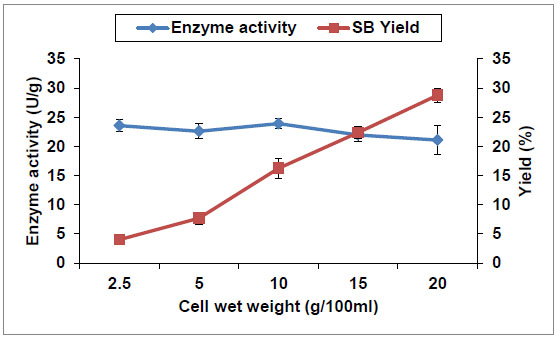 ) shows that SB yield is significantly increased with increasing cell-wet weight up to 20% compared with control (10%). Simultaneously, there was no significant change in the enzyme activity per gram dry weight (U/g). This means that cell weight had no effect on cells enzyme activity, but it had a significant effect on SB yield. It was increased from 4 to 28.8% by increasing the cell wet weight from 2.5 to 20%, respectively. This could be attributed to increase of the saponin hydrolase enzyme concentration produced by the used cells by increasing their weight in the reaction mixture. Therefore, A. flavus cells wet weight of 20% (corresponding to 4% cell dry weight) was selected as a suitable catalyst concentration based on the final yield of SB (28.8%).
) shows that SB yield is significantly increased with increasing cell-wet weight up to 20% compared with control (10%). Simultaneously, there was no significant change in the enzyme activity per gram dry weight (U/g). This means that cell weight had no effect on cells enzyme activity, but it had a significant effect on SB yield. It was increased from 4 to 28.8% by increasing the cell wet weight from 2.5 to 20%, respectively. This could be attributed to increase of the saponin hydrolase enzyme concentration produced by the used cells by increasing their weight in the reaction mixture. Therefore, A. flavus cells wet weight of 20% (corresponding to 4% cell dry weight) was selected as a suitable catalyst concentration based on the final yield of SB (28.8%).
Substrate concentration is another important parameter in the catalytic process, and it requires careful investigation because high substrate concentrations may lead to substrate inhibition. Results in Fig. (6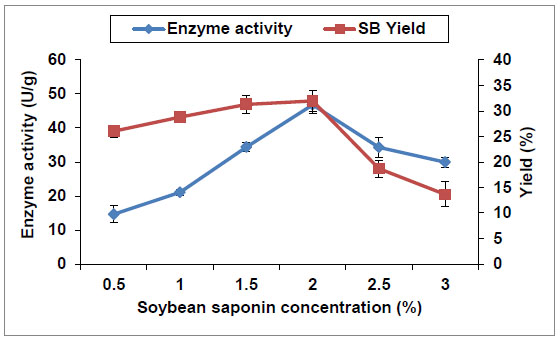 ) show that the substrate concentration in the reaction mixture had a significant effect on saponin hydrolase activity. When the substrate concentration increased from 0.5 to 2%, the enzyme activity increased rapidly (from 14.6 to 46.8, respectively), while SB yield increased slowly (from 26 to 32%, respectively). This reveals the enhancement effect of the substrate on cells enzyme activity. Thereafter, both of them decreased substantially with the increase of substrate concentration beyond 2%. Similar behavior has been described for production of Chiral Chemicals from by E. coli whole cells recombinant strain producing (2R,3R)-2,3-butanediol dehydrogenase [30Xiao, Z.; Lv, C.; Gao, C.; Qin, J.; Ma, C.; Liu, Z.; Liu, P.; Li, L.; Xu, P. A novel whole-cell biocatalyst with NAD+ regeneration for production of chiral chemicals. PLoS One, 2010, 5(1), e8860.
) show that the substrate concentration in the reaction mixture had a significant effect on saponin hydrolase activity. When the substrate concentration increased from 0.5 to 2%, the enzyme activity increased rapidly (from 14.6 to 46.8, respectively), while SB yield increased slowly (from 26 to 32%, respectively). This reveals the enhancement effect of the substrate on cells enzyme activity. Thereafter, both of them decreased substantially with the increase of substrate concentration beyond 2%. Similar behavior has been described for production of Chiral Chemicals from by E. coli whole cells recombinant strain producing (2R,3R)-2,3-butanediol dehydrogenase [30Xiao, Z.; Lv, C.; Gao, C.; Qin, J.; Ma, C.; Liu, Z.; Liu, P.; Li, L.; Xu, P. A novel whole-cell biocatalyst with NAD+ regeneration for production of chiral chemicals. PLoS One, 2010, 5(1), e8860.
[http://dx.doi.org/10.1371/journal.pone.0008860] [PMID: 20126645] ].
As shown in Fig. (7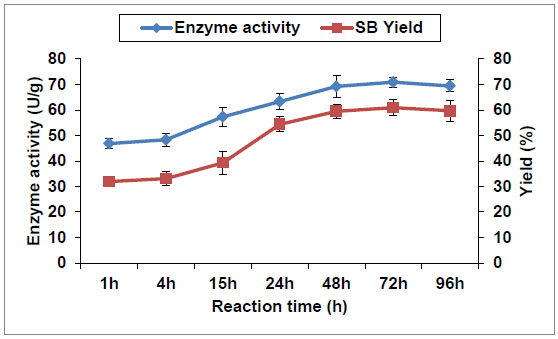 ) cells enzyme activity was significantly increased by increasing time to reach a maximum of 69.2U/g after 48 h accompanied by a SB yield of 59.5%, which represented approximately twice that obtained after 1h (46.83U/g and 31.9%, respectively). A previous study [13Watanabe, M.; Sumida, N.; Yanai, K.; Murakami, T. A novel saponin hydrolase from Neocosmospora vasinfecta var. vasinfecta. Appl. Environ. Microbiol., 2004, 70(2), 865-872.
) cells enzyme activity was significantly increased by increasing time to reach a maximum of 69.2U/g after 48 h accompanied by a SB yield of 59.5%, which represented approximately twice that obtained after 1h (46.83U/g and 31.9%, respectively). A previous study [13Watanabe, M.; Sumida, N.; Yanai, K.; Murakami, T. A novel saponin hydrolase from Neocosmospora vasinfecta var. vasinfecta. Appl. Environ. Microbiol., 2004, 70(2), 865-872.
[http://dx.doi.org/10.1128/AEM.70.2.865-872.2004] [PMID: 14766566] ] found that 1,000 ml culture broth of N. vasinfecta var. vasinfecta PF1225 showed a total soybean saponin hydrolase enzyme activity of 16,380 U (1U=1 nmole of SB/min) after 1h incubation. The analysis of the data obtained as well as of the previous finding after 1h suggests that the latter activity is representing about half that obtained by 18 g dry A. flavus cells, comes from 1,000ml culture, after 1h incubation with soybean saponin, which calculated to be 842.9 U (1U= 1mg of SB/h) equivalent to 30,674 U (1U=1nmole of SB/min). Thereafter, there was no significant difference in the obtained product yields with further incubation up to 96h. This observed stability after 48h in the enzyme activity and SB yield could be attributed to the presence of high concentration of SB in the reaction mixture, which inhibits (feedback inhibition) further conversion of soybean saponin. The enzyme showing such behavior is called allosteric enzyme, which is controlled by a feedback or end product inhibition at an allosteric site different from the active site [31Gupta, P.K. Genetics Classical To Modern; Rastogi Publications: New Dehli, 2007. ]. The interaction of the end product at an allosteric site changes the structure of the enzyme so that the active site is also changed. Meitian et al. [32Meitian, X.; Yayan, H.; Chun, M.; Yanghao, G. Kinetics of asymmetric reduction of phenylglyoxylic acid to R-(-)-mandelic acid by Saccharomyces cerevisiae FD11b. Chin. J. Chem. Eng., 2006, 14, 73-80.
[http://dx.doi.org/10.1016/S1004-9541(06)60040-2] ] reported that accumulation of R-(-)-mandelic acid caused a severe feed-back inhibition for its production from phenylglyoxylic acid by Saccharomyces cerevisiae FD11b when the product concentration was above 60mmol·l-1.
Production and Isolation of SB
Based on the previously mentioned results, the production of SB from soybean saponin by A. flavus whole cells was done under the optimized saponin hydrolase production and reaction conditions in 2.5 l continuously stirred bioreactor. A. flavus was grown in the enzyme production medium containing 2% soybean saponin at pH 9 and 30°C for 48h. Subsequent to cell harvesting and resuspension, whole cell bioconversion of soybean saponin to SB was performed in 0.2M acetate (pH 5.5) buffer containing 2% soybean saponin for 48h at 45°C. The mixture was extracted two times with double its volume of ethyl acetate (2000 ml). The organic layer was distilled off and the dried mixture applied to a silica gel column (5 x 120 cm). The column was eluted with 10% ethyl acetate (7L), which sufficient to desorb SB completely. The 10% ethyl acetate elute was distilled off to give SB (6.5 g, yield: 67%). Recrystallization of the product from chloroform/methanol gave completely pure SB for identification. Identification of SB was established by comparison of its mp., optical rotation and EI-MS, NMR spectra, with those of our previously published data [33Amin, H.A.; Hanna, A.G.; Mohamed, S.S. Comparative studies of acidic and enzymatic hydrolysis for production of soyasapogenols from soybean saponin. Biocatal. Biotrans., 2011, 29, 311-319.
[http://dx.doi.org/10.3109/10242422.2011.632479] ]. This procedure not only allows the pH of the reaction to be controlled to some extent, but also results in cleaner transformation, the organism having been removed from the complex mixture of chemicals that accumulates during growth, and thus can simplify downstream processing of reaction products [34Grogan, G. Practical biotransformations: A Beginner's Guide; John Wiley & Sons: New York, 2009. ].
CONCLUSION
This study is considered as the first record investigating SB production using whole cells producing saponin hydrolase in the enzyme reaction mixture. Among tested isolated fungi, Aspergillus flavus was the best saponin hydrolase producer. Whole cells showed 3times higher enzyme activity compared to the extra- and intra-enzyme preparations. Under optimized cultivation and reaction conditions, the cells bioconversion efficiency increased from 5.3 to 60%. Based on the results, A. flavus was cultivated in production medium containing 2% soybean saponin at pH 9 and 30°C for 48h. The reaction of the resulting wet cells (20%) and soybean saponin (2%) in 1l acetate buffer at pH 5.5and 45°C for 48h in stirred bioreactor afforded SB in a yield of 67%. Subsequently, the reaction mixture was extracted, purified on silica gel column and recrystallized to give completely pure SB. Merits of the suggested method include; simplicity, relatively high SB yield, cost effective and easy downstream processing of products. The current results could be the groundwork for production of SB from soybean saponin on an industrial scale.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
ACKNOWLEDGEMENTS
The authors would like to thank for financial support by the National Research Center Foundation of Egypt via tenth research plan grant (2013-2016) project (No.10070301).
REFERENCES
| [1] | Kamo, S.; Suzuki, S.; Sato, T. The content of soyasaponin and soyasapogenol in soy foods and their estimated intake in the Japanese. Food Sci. Nutr., 2014, 2(3), 289-297. [http://dx.doi.org/10.1002/fsn3.107] [PMID: 24936299] |
| [2] | Amin, H.A.; Awad, H.M.; Hanna, A.G. Comparative evaluation of in vitro cytotoxicity, antiviral and antioxidant activities of different soyasapogenols from soybean saponin. Egypt Pharm. J., 2012, 11, 73-79. |
| [3] | Rowlands, J.C.; Berhow, M.A.; Badger, T.M. Estrogenic and antiproliferative properties of soy sapogenols in human breast cancer cells in vitro. Food Chem. Toxicol., 2002, 40(12), 1767-1774. [http://dx.doi.org/10.1016/S0278-6915(02)00181-3] [PMID: 12419690] |
| [4] | Watanabe, M.; Mido, N.; Tamura, T. Saponin digesting enzymes, genes there of and soyasapogenol B mass production system. U.S Patent 7022508 B2, April 2006. |
| [5] | Zhang, W.; Popovich, D.G. Effect of soyasapogenol A and soyasapogenol B concentrated extracts on HEP-G2 cell proliferation and apoptosis. J. Agric. Food Chem., 2008, 56(8), 2603-2608. [http://dx.doi.org/10.1021/jf0731550] [PMID: 18361499] |
| [6] | Chen, G.; Chen, J. A novel cell modification method used in biotransformation of glycerol to 3-HPA by Lactobacillus reuteri. Appl. Microbiol. Biotechnol., 2013, 97(10), 4325-4332. [http://dx.doi.org/10.1007/s00253-013-4723-2] [PMID: 23359000] |
| [7] | Omar, M.N.; Khan, N.T.; Hasali, N.H. Microbial transformations of artemisinin anti-malaria drug. Adv. Biores, 2012, 3, 27-31. |
| [8] | Carballeira, J.D.; Quezada, M.A.; Hoyos, P.; Simeó, Y.; Hernaiz, M.J.; Alcantara, A.R.; Sinisterra, J.V. Microbial cells as catalysts for stereoselective red-ox reactions. Biotechnol. Adv., 2009, 27(6), 686-714. [http://dx.doi.org/10.1016/j.biotechadv.2009.05.001] [PMID: 19442715] |
| [9] | Luo, J.; Liang, Q.; Shen, Y.; Chen, X.; Yin, Z.; Wang, M. Biotransformation of bavachinin by three fungal cell cultures. J. Biosci. Bioeng., 2014, 117(2), 191-196. [http://dx.doi.org/10.1016/j.jbiosc.2013.08.001] [PMID: 24012108] |
| [10] | Kaushik, N.; Biswas, S.; Singh, J. Biocatalysis and biotransformation processes - an insight. Scitech J., 8, 15-22. |
| [11] | Amin, H.A.; Mohamed, S.S. Immobilization of Aspergillus terreus on loofa sponge for soyasapogenol B production from soybean saponin. J. Mol. Catal., B Enzym., 2012, 78, 85-90. [http://dx.doi.org/10.1016/j.molcatb.2012.03.015] |
| [12] | Amin, H.A.; Hassan, Y.M.; Yehia, S.M. Biotransformation of soybean saponin to soyasapogenol B by immobilized Aspergillus Parasiticus cells on polyurethane foam. Res. J. Pharm. Biol. Chem. Sci., 2013, 5, 332-341. |
| [13] | Watanabe, M.; Sumida, N.; Yanai, K.; Murakami, T. A novel saponin hydrolase from Neocosmospora vasinfecta var. vasinfecta. Appl. Environ. Microbiol., 2004, 70(2), 865-872. [http://dx.doi.org/10.1128/AEM.70.2.865-872.2004] [PMID: 14766566] |
| [14] | Watanabe, M.; Sumida, N.; Yanai, K.; Murakami, T. Cloning and characterization of saponin hydrolases from Aspergillus oryzae and Eupenicillium brefeldianum. Biosci. Biotechnol. Biochem., 2005, 69(11), 2178-2185. [http://dx.doi.org/10.1271/bbb.69.2178] [PMID: 16306700] |
| [15] | Sun, T.; Du, W.; Liu, D. Prospective and impacts of whole cell mediated alcoholysis of renewable oils for biodiesel production. Biofuels Bioprod. Biorefin., 2009, 3, 633-639. [http://dx.doi.org/10.1002/bbb.180] |
| [16] | Kuramoto, T.; Ito, Y.; Oda, M. Microbial production of glycyrrhetic acid-3-O-mono-β-D-glucuronide from glycyrrhizin by Cryptococcus magnus MG-27. Biosci. Biotechnol. Biochem., 1994, 58, 455-458. [http://dx.doi.org/10.1271/bbb.58.455] |
| [17] | Hashemizadeh, S.N. A comparative study of immobilized-whole cell and commercial lipase as a biocatalyst for biodiesel production from soybean oil. Int. J. Curr. Microbiol. App. Sci., 2014, 3, 741-752. |
| [18] | Foor, S.R.; Tenne, F.D.; Sinclair, J.B. Occurrence of seed borne microorganisms and germination in culture for determining seed health in soybeans. Pl. Dis. Reptre., 1976, 60, 970-977. |
| [19] | Gilman, J.C. A manual of soil fungi, 2nd ed; The Iowa State University Press: Ames, Iowa, 1957. |
| [20] | Barnett, H.L.; Hunter, B.B. Illustrated genera of fungi, 3rd ed; Burgess Publ. Co.: Minneapolis, MN, USA, 2000. |
| [21] | Domsch, K.H.; Gams, W.; Anderson, T.H. Compendium of soil fungi, 2nd ed; APS Press, 2007. |
| [22] | Samson, R.A.; Houbraken, J.; Thrane, U. Food and indoor fungi. CBS-KNAW: Fungal Biodiversity Centre Utrecht; Institute of the Royal Netherlands Acadaemy of Arts and Science, 2010. |
| [23] | Kudou, S.; Tsuizaki, I.; Shimoyamada, M. Screening for microorganisms producing soybean saponin hydrolase. Agric. Biol. Chem., 1990, 54, 3035-3037. |
| [24] | Lin, S.H.; Guidotti, G. Methods in Enzymology, 192-Guide to Protein Purification, 2nd ed; Richard, R.B.; Murray, P.D., Eds.; Academic Press, Elsevier Inc., 2009, pp. 619-629. [http://dx.doi.org/10.1016/S0076-6879(09)63035-4] |
| [25] | Narita, J.; Okano, K.; Tateno, T.; Tanino, T.; Sewaki, T.; Sung, M.H.; Fukuda, H.; Kondo, A. Display of active enzymes on the cell surface of Escherichia coli using PgsA anchor protein and their application to bioconversion. Appl. Microbiol. Biotechnol., 2006, 70(5), 564-572. [http://dx.doi.org/10.1007/s00253-005-0111-x] [PMID: 16133338] |
| [26] | Stewart, J.D.; Rodriquez, S.; Kayser, M.M. Enzyme technology for pharmaceutical and biotechnological applications; Marcel Dekker: New York, 2001. |
| [27] | Bommarius, A.S.; Riebel, B.R. Biocatalysis: fundamentals and applications; Wiley-VCH Verlag GmbH&Co. : KGaA, Weinheim, 2004. [http://dx.doi.org/10.1002/3527602364] |
| [28] | Kudou, S.; Tsuizaki, I.; Uchida, T.; Okubo, K. Purification and some properties of soybean saponin hydrolase from Aspergillus oryzae KO-2. Agric. Biol. Chem., 1991, 55(1), 31-36. [PMID: 1368672] |
| [29] | Güçlü-Ustündağ, O.; Mazza, G. Saponins: properties, applications and processing. Crit. Rev. Food Sci. Nutr., 2007, 47(3), 231-258. [http://dx.doi.org/10.1080/10408390600698197] [PMID: 17453922] |
| [30] | Xiao, Z.; Lv, C.; Gao, C.; Qin, J.; Ma, C.; Liu, Z.; Liu, P.; Li, L.; Xu, P. A novel whole-cell biocatalyst with NAD+ regeneration for production of chiral chemicals. PLoS One, 2010, 5(1), e8860. [http://dx.doi.org/10.1371/journal.pone.0008860] [PMID: 20126645] |
| [31] | Gupta, P.K. Genetics Classical To Modern; Rastogi Publications: New Dehli, 2007. |
| [32] | Meitian, X.; Yayan, H.; Chun, M.; Yanghao, G. Kinetics of asymmetric reduction of phenylglyoxylic acid to R-(-)-mandelic acid by Saccharomyces cerevisiae FD11b. Chin. J. Chem. Eng., 2006, 14, 73-80. [http://dx.doi.org/10.1016/S1004-9541(06)60040-2] |
| [33] | Amin, H.A.; Hanna, A.G.; Mohamed, S.S. Comparative studies of acidic and enzymatic hydrolysis for production of soyasapogenols from soybean saponin. Biocatal. Biotrans., 2011, 29, 311-319. [http://dx.doi.org/10.3109/10242422.2011.632479] |
| [34] | Grogan, G. Practical biotransformations: A Beginner's Guide; John Wiley & Sons: New York, 2009. |







