- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Human Papillomavirus DNA and E6/E7 mRNA Status in Relation to Survival of Patients Treated for Cervical Squamous Cell Carcinoma
Ruth Holm*, 1, Irene Kraus2, 3, Hanne Skomedal3, Anita Langerød4, Gunnar B Kristensen5, 6, Heidi Lyng7
Abstract
The prognostic significance of human papillomavirus (HPV) DNA and E6/E7 mRNA, the presence of specific types, and the physical state of HPV DNA, were studied in 202 cervical squamous cell carcinomas. Absence or non-detectable levels of high-risk (types 16, 18, 31, 33, 35, 45, 52 and 58) E6/E7 mRNA, using the real-time nucleic acid sequence based amplification (NASBA) assay, and absence of HPV high-risk/HPV 6, 26, 66, 69, 73 (all methods collectively) were associated with poor overall survival in univariate analysis (P = 0.04 and P = 0.03, respectively) and had independent prognostic value in multivariate analysis (P = 0.01 and P = 0.03, respectively) including FIGO stage and age. Based on the individual results of type-specific PCR and in situ hybridization (ISH), the presence of HPV DNA was not found to be a prognostic factor. Likewise, concerning the presence of specific HPV types and the HPV integration status (determined by ISH), no prognostic significance was found. Mutation analyses of the TP53 gene revealed mutations in 3 of the 6 HPV negative samples (50%).
Article Information
Identifiers and Pagination:
Year: 2008Volume: 2
First Page: 74
Last Page: 81
Publisher Id: TOVJ-2-74
DOI: 10.2174/1874357900802010074
Article History:
Received Date: 1/7/2008Revision Received Date: 11/7/2008
Acceptance Date: 26/9/2008
Electronic publication date: 24/10/2008
Collection year: 2008
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Division of Pathology, The Norwegian Radium Hospital, Montebello, 0310 Oslo, Norway; Tel: (47) 22934207; Fax: (47) 22730164; E-mail: ruth.holm@radiumhospitalet.no
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 1-7-2008 |
Original Manuscript | Human Papillomavirus DNA and E6/E7 mRNA Status in Relation to Survival of Patients Treated for Cervical Squamous Cell Carcinoma | |
INTRODUCTION
Human papillomavirus (HPV) is known to be a major risk factor in the development of cervical cancer [1Franco EL. Viral etiology of cervical cancer: a critique of the evidence Rev Infect Dis 1991; 13: 1195-206.]. However, the role of HPV for the clinical outcome of cervical carcinomas is debatable. Previous studies have reported various results on whether the presence of HPV, and also whether the presence of a specific type over another, may have prognostic value for the clinical outcome of women with cervical cancer. Some studies have reported that patients with HPV negative cervical tumours have a worse prognosis than those with HPV positive tumours [2Riou G, Favre M, Jeannel D, et al. Association between poor prognosis in early-stage invasive cervical carcinomas and non-detection of HPV DNA Lancet 1990; 335: 1171-4.-6Graf AH, Cheung AL, Hauser-Kornberger C, et al. Clinical relevance of HPV 16/18 testing methods in cervical squamous cell carcinoma Appl Immunohistochem Mol Morphol 2000; 8: 300-9.]; other studies have found the presence of HPV to be unrelated to clinical outcome [7King LA, Tase T, Twiggs LB, et al. Prognostic significance of the presence of human papillomavirus DNA in patients with invasive carcinoma of the cervix Cancer 1989; 63: 897-900.-10Fule T, Csapo Z, Mathe M, et al. Prognostic significance of high-risk HPV status in advanced cervical cancers and pelvic lymph nodes Gynecol Oncol 2006; 100: 570-8.]. In most of these studies, HPV DNA was detected by PCR [2Riou G, Favre M, Jeannel D, et al. Association between poor prognosis in early-stage invasive cervical carcinomas and non-detection of HPV DNA Lancet 1990; 335: 1171-4., 4DeBritton RC, Hildesheim A, De Lao SL, et al. Human papillomaviruses and other influences on survival from cervical cancer in Panama Obstet Gynecol 1993; 81: 19-24., 5Biedermann K, Dandachi N, Trattner M, et al. Comparison of real-time PCR signal-amplified in situ hybridization and conventional PCR for detection and quantification of human papillomavirus in archival cervical cancer tissue J Clin Microbiol 2004; 42: 3758-65., 9Kristensen GB, Karlsen F, Jenkins A, et al. Human papilloma virus has no prognostic significance in cervical carcinoma Eur J Cancer 1996; 32A: 1349-53.-16Bachtiary B, Obermair A, Dreier B, et al. Impact of multiple HPV infection on response to treatment and survival in patients receiving radical radiotherapy for cervical cancer Int J Cancer 2002; 102: 237-43.] or in situ hybridization (ISH) [3Higgins GD, Davy M, Roder D, et al. Increased age and mortality associated with cervical carcinomas negative for human papillomavirus RNA Lancet 1991; 338: 910-3., 6Graf AH, Cheung AL, Hauser-Kornberger C, et al. Clinical relevance of HPV 16/18 testing methods in cervical squamous cell carcinoma Appl Immunohistochem Mol Morphol 2000; 8: 300-9., 7King LA, Tase T, Twiggs LB, et al. Prognostic significance of the presence of human papillomavirus DNA in patients with invasive carcinoma of the cervix Cancer 1989; 63: 897-900.]. When it comes to different HPV genotypes, we do know that different types have different oncogenic potential in causing cervical cancer. The question is however whether these also may exert different pressure in terms of the clinical outcome of women with cervical cancer. In previous studies, a reduced survival has in particular been correlated with HPV 18 positive tumours [11Nakagawa S, Yoshikawa H, Onda T, et al. Type of human papillomavirus is related to clinical features of cervical carcinoma Cancer 1996; 78: 1935-41.-15Huang HJ, Huang SL, Lin CY, et al. Human papillomavirus genotyping by a polymerase chain reaction-based genechip method in cervical carcinoma treated with neoadjuvant chemotherapy plus radical surgery Int J Gynecol Cancer 2004; 14: 639-49.]. Reduced survival has also been demonstrated for women with multiple HPV types [16Bachtiary B, Obermair A, Dreier B, et al. Impact of multiple HPV infection on response to treatment and survival in patients receiving radical radiotherapy for cervical cancer Int J Cancer 2002; 102: 237-43.]. Nevertheless, no definite conclusions regarding a different impact of different types on clinical outcome have been disclosed.
The presence of HPV E6/E7 oncoproteins is crucial for the development and maintenance of a malignant phenotype [17Munger K, Phelps WC, Bubb V, Howley PM, Schlegel R. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes J Virol 1989; 63: 4417-21.-19Goodwin EC, DiMaio D. Repression of human papillomavirus oncogenes in HeLa cervical carcinoma cells causes the orderly reactivation of dormant tumor suppressor pathways Proc Natl Acad Sci U S A 2000; 97: 12513-8.]. Moreover, it is known that the E6 and E7 proteins of different HPV types have different oncogenic potential. Concerning HPV mRNA, one study performing HPV 16, 18, 31, and 33 mRNA detection using ISH and 125I-labelled riboprobes, reports an increased age at diagnosis and an increased mortality associated with cervical carcinomas negative for HPV mRNA [3Higgins GD, Davy M, Roder D, et al. Increased age and mortality associated with cervical carcinomas negative for human papillomavirus RNA Lancet 1991; 338: 910-3., 6Graf AH, Cheung AL, Hauser-Kornberger C, et al. Clinical relevance of HPV 16/18 testing methods in cervical squamous cell carcinoma Appl Immunohistochem Mol Morphol 2000; 8: 300-9., 7King LA, Tase T, Twiggs LB, et al. Prognostic significance of the presence of human papillomavirus DNA in patients with invasive carcinoma of the cervix Cancer 1989; 63: 897-900.].
In addition to the presence of HPV oncoproteins, integration of HPV DNA into the host genome is associated with the development of cervical cancer. As a result of HPV integration, expression of the HPV E6/E7 oncoproteins increases, abrogating cell cycle control and apoptosis mechanisms [20Munger K, Howley PM. Human papillomavirus immortalization and transformation functions Virus Res 2002; 89: 213-28.]. Alternatively to the integrated form, HPV can be found as episomes alone or in a mixed form containing both episomal and integrated virus. According to Cooper et al. [21Cooper K, Herrington CS, Stickland JE, Evans MF, McGee JO. Episomal and integrated human papillomavirus in cervical neoplasia shown by non-isotopic in situ hybridisation J Clin Pathol 1991; 44: 990-6.] and Kristiansen et al. [22Kristiansen E, Jenkins A, Holm R. Coexistence of episomal and integrated HPV16 DNA in squamous cell carcinoma of the cervix J Clin Pathol 1994; 47: 253-6.], the physical state of HPV DNA can be determined by the pattern of the ISH signals. These authors observed that ISH signals may be spread over the entire nucleus (diffuse), indicating the episomal form, concentrated in small spots (punctuate), indicating the integrated form, or in a mixture of the two, indicating both episomal and integrated forms. Previously, it has been suggested that the viral integration status may be important as a prognostic marker in cervical carcinomas, reported by two studies comparing the physical state of HPV by ISH and clinical outcome [23Unger ER, Vernon SD, Thoms WW, et al. Human papillomavirus and disease-free survival in FIGO stage Ib cervical cancer J Infect Dis 1995; 172: 1184-90., 24Vernon SD, Unger ER, Miller DL, Lee DR, Reeves WC. Association of human papillomavirus type 16 integration in the E2 gene with poor disease-free survival from cervical cancer Int J Cancer 1997; 74: 50-6.]. However, in these studies, only 47 and 50 women with cervical carcinoma were included [23Unger ER, Vernon SD, Thoms WW, et al. Human papillomavirus and disease-free survival in FIGO stage Ib cervical cancer J Infect Dis 1995; 172: 1184-90., 24Vernon SD, Unger ER, Miller DL, Lee DR, Reeves WC. Association of human papillomavirus type 16 integration in the E2 gene with poor disease-free survival from cervical cancer Int J Cancer 1997; 74: 50-6.] and thus, additional studies should be performed in order to reveal whether integration, as detected by ISH, can be used as a prognostic marker for the clinical outcome of carcinomas.
In a previous study, we have investigated the prevalence of HPV DNA and E6/E7 mRNA in a series of cervical squamous cell carcinomas (SCCs) using different PCR techniques, ISH, nucleic acid sequence based amplification (NASBA), and the NASBA based PreTect HPV-Proofer assay [25Kraus I, Molden T, Holm R, et al. Presence of E6 and E7 mRNA from human papillomavirus types 16, 18, 31, 33, and 45 in the majority of cervical carcinomas J Clin Microbiol 2006; 44: 1310-7.]. The aim of the present study was to assess whether the presence of HPV DNA and/or HPV E6/E7 mRNA, the presence of certain HPV genotypes or the physical state of HPV DNA, might have prognostic significance. To our knowledge, the present study is the first to report the association between HPV E6/E7 mRNA, detected by NASBA, and clinical outcome of women treated for SCCs.
MATERIALS AND METHODOLOGY
Tumour material.
From the original 204 cervical SCCs, FIGO stage Ia-IVb, included in our previous study [25Kraus I, Molden T, Holm R, et al. Presence of E6 and E7 mRNA from human papillomavirus types 16, 18, 31, 33, and 45 in the majority of cervical carcinomas J Clin Microbiol 2006; 44: 1310-7.], 202 were included in the present analysis; for two women, clinical information was not available. Staging was done according to the FIGO guidelines [26Pecorelli S, Odicino F. Cervical cancer staging Cancer J 2003; 9: 390-4.]. All patients were treated at the Department of Gynaecologic Oncology, The Norwegian Radium Hospital, during the period 1995-1998. The median age was 54.1 years, ranging from 26 to 92 years. Patient information is listed in Table 1. Out of 58 patients with FIGO stage I, 13 were treated with radiation and 45 were treated by radical hysterectomy and pelvic lymphadenectomy. Postoperative adjuvant treatment was given to 17 of these patients. Of the patients with FIGO stage II-IV, 121 were treated with radiation, two with chemotheraphy and radiation, 14 with palliative radiotherapy, two with radical hysterectomy and pelvic lymphadenectomy and five had palliative treatment. After treatment, patients were followed up at The Norwegian Radium Hospital or at local hospitals. One patient was lost to follow-up after 3 months; all the others were followed until death or until August 2006. A total of 79 (39.1%) patients suffered a relapse and 75 (37.1%) died of cervical cancer. For patients still alive, median follow-up was 117 months (range from 92 to 139 months). Median follow-up for patients without relapse was 115 months (range from 76 to 138 months).
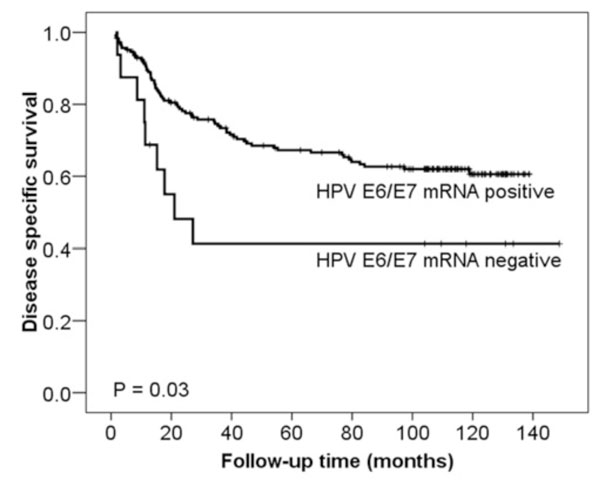 |
Fig. (1) Kaplan and Meier curves of DSS related to HPV E6/E7 mRNA expression detected by NASBA. High-risk HPV E6/E7 mRNA negative cases (n = 16) had a significantly worse DSS than those cases with detectable high-risk HPV E6/E7 mRNA (n = 186) (p = 0.03). The p-value differs slightly from that in Table 3 due to the use of the log-rank test as opposed to the Cox regression analysis in Table 3. |
Fresh frozen tissue, taken from the primary tumour at the time of diagnosis and stored at -70oC, was used for PCR and NASBA analyses. For ISH, formalin-fixed, paraffin-embedded tissue from the same tumours was used.
The study has been approved by The Regional Committee for Medical Research Ethics South of Norway (S-06381a), The Social- and Health Directorate (06/4509 and 06/4417), and The Data Inspectorate (06/01467-3).
HPV DNA analysis by PCR.
Type-specific PCR, identifying HPV 16, 18, 31, 33, 35, 45, 52 and 58 DNA, was performed as described previously [25Kraus I, Molden T, Holm R, et al. Presence of E6 and E7 mRNA from human papillomavirus types 16, 18, 31, 33, and 45 in the majority of cervical carcinomas J Clin Microbiol 2006; 44: 1310-7.]. HPV DNA detection was also performed by consensus Gp5+/6+ PCR as described previously [25Kraus I, Molden T, Holm R, et al. Presence of E6 and E7 mRNA from human papillomavirus types 16, 18, 31, 33, and 45 in the majority of cervical carcinomas J Clin Microbiol 2006; 44: 1310-7.]. Samples negative by type-specific PCR were tested by the Gp5+/6+ enzyme immunoassay (EIA) and reverse line blot (RLB) assay [27van den Brule AJ, Pol R, Fransen-Daalmeijer N, et al. GP5+/6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes J Clin Microbiol 2002; 40: 779-87.].
HPV DNA analysis by ISH.
ISH was carried out with biotinylated probes against HPV 16 and 18 (gifts from profs. Harald zur Hausen and Ethel-Michele de Villiers, Germany) [28Durst M, Gissmann L, Ikenberg H, zur HH. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions Proc Natl Acad Sci U S A 1983; 80: 3812-5., 29Boshart M, Gissmann L, Ikenberg H, et al. A new type of papillomavirus DNA, its presence in genital cancer biopsies and in cell lines derived from cervical cancer EMBO J 1984; 3: 1151-7.] and HPV 31 and 33 (Eurodiagnostics BV) and an alkaline phosphatase anti-alkaline phosphatase detection system, as described previously [25Kraus I, Molden T, Holm R, et al. Presence of E6 and E7 mRNA from human papillomavirus types 16, 18, 31, 33, and 45 in the majority of cervical carcinomas J Clin Microbiol 2006; 44: 1310-7.]. The physical state of HPV DNA was determined by the pattern of the ISH signal as described in Cooper et al. [21Cooper K, Herrington CS, Stickland JE, Evans MF, McGee JO. Episomal and integrated human papillomavirus in cervical neoplasia shown by non-isotopic in situ hybridisation J Clin Pathol 1991; 44: 990-6.] and Kristiansen et al. [22Kristiansen E, Jenkins A, Holm R. Coexistence of episomal and integrated HPV16 DNA in squamous cell carcinoma of the cervix J Clin Pathol 1994; 47: 253-6.]: A diffuse signal indicates episomal DNA, a punctuate signal indicates integrated DNA and a mixture of these two patterns indicates the presence of both episomal and integrated DNA.
HPV E6/E7 mRNA analysis by NASBA.
The real-time multiplex NASBA assay, including PreTectTM HPV-Proofer, identifying HPV 16, 18, 31, 33, 35, 45, 52 and 58 E6/E7 mRNA, was performed as suggested by the manufacturer (NorChip AS, Klokkarstua, Norway) and as published previously [25Kraus I, Molden T, Holm R, et al. Presence of E6 and E7 mRNA from human papillomavirus types 16, 18, 31, 33, and 45 in the majority of cervical carcinomas J Clin Microbiol 2006; 44: 1310-7., 30Molden T, Kraus I, Skomedal H, Nordstrom T, Karlsen F. PreTect HPV-Proofer: real-time detection and typing of E6/E7 mRNA from carcinogenic human papillomaviruses J Virol Methods 2007; 142: 204-12.].
TP53 mutation analysis.
TP53 mutation analysis was performed in tumour DNA by resequencing exon 2-11 using the 3730 DNA Analyzer (Applied Biosystems, Applied Biosystems, Foster City, California, USA). All fragments were sequenced in both directions and the primers used were linked to universal M13 sequences in the 5’ position to simplify the sequencing reaction (Table 2). 25 ng DNA was amplified in a 10 μl PCR reaction using HotStarTaq DNA Polymerase (Qiagen) and touchdown PCR with annealing temperatures moving from 68°C to 56°C. SeqScape® Software v2.5 (Applied Biosystems) was used for alignment to reference sequence and the scoring were carried out by two experienced operators independently. GeneBank accession number NM_000546 was used as reference sequence.
Statistical analysis.
For statistical evaluation and based on previous studies [2Riou G, Favre M, Jeannel D, et al. Association between poor prognosis in early-stage invasive cervical carcinomas and non-detection of HPV DNA Lancet 1990; 335: 1171-4., 7King LA, Tase T, Twiggs LB, et al. Prognostic significance of the presence of human papillomavirus DNA in patients with invasive carcinoma of the cervix Cancer 1989; 63: 897-900., 9Kristensen GB, Karlsen F, Jenkins A, et al. Human papilloma virus has no prognostic significance in cervical carcinoma Eur J Cancer 1996; 32A: 1349-53., 13Schwartz SM, Daling JR, Shera KA, et al. Human papillomavirus and prognosis of invasive cervical cancer: a population-based study J Clin Oncol 2001; 19: 1906-15., 15Huang HJ, Huang SL, Lin CY, et al. Human papillomavirus genotyping by a polymerase chain reaction-based genechip method in cervical carcinoma treated with neoadjuvant chemotherapy plus radical surgery Int J Gynecol Cancer 2004; 14: 639-49., 23Unger ER, Vernon SD, Thoms WW, et al. Human papillomavirus and disease-free survival in FIGO stage Ib cervical cancer J Infect Dis 1995; 172: 1184-90.], the cases were divided into different groups. Furthermore, the different HPV detection methods were evaluated either individually and/or collectively (Table 3).
The concordance between the different methods was evaluated by McNemar test. Differences in proportions were evaluated by the Person χ2 test. Survival was calculated by the method described by Kaplan and Meier and comparisons performed by the log-rank test. Progression free survival (PFS) was defined as the interval between diagnosis and relapse of cervical cancer and disease specific survival (DSS) as the interval between diagnosis and cervical cancer related death. A Cox proportional hazards regression model was used for both univariate and multivariate evaluation of survival rates. In the multivariate analysis, forward and backward stepwise regression was used. All calculations were performed using the SPSS 15.0 statistical software package (SPSS, Chicago, IL). A p < 0.05 was considered statistical significant.
RESULTS
In total, 196 of the 202 (97%) samples have previously been found to harbour an HPV infection [25Kraus I, Molden T, Holm R, et al. Presence of E6 and E7 mRNA from human papillomavirus types 16, 18, 31, 33, and 45 in the majority of cervical carcinomas J Clin Microbiol 2006; 44: 1310-7.]. By type-specific PCR, high-risk HPV (types 16, 18, 31, 33, 35, 45, 52 and 58) DNA has previously been detected in 186 of the 202 (92.1%) samples. By NASBA, high-risk HPV (types 16, 18, 31, 33, 35, 45, 52 and 58) E6/E7 mRNA was detected in 186 of the 202 (92.1%) samples. The results for these two methods were similar (p = 1.00) (Table 4), with divergence in only 4 cases, as two of the cases positive by type-specific PCR (HPV 16 and 33) were negative for HPV E6/E7 mRNA (case 3307 and 3959), and two cases positive for HPV E6/E7 mRNA (HPV 31 and 45) were negative by type-specific PCR [25Kraus I, Molden T, Holm R, et al. Presence of E6 and E7 mRNA from human papillomavirus types 16, 18, 31, 33, and 45 in the majority of cervical carcinomas J Clin Microbiol 2006; 44: 1310-7.]. It is important to notice, that sample 3307 and 3959 originally were negative by all methods and became positive by type-specific PCR only when tested on 10 times more concentrated material (nevertheless remaining RNA negative). The quality of the RNA within the sample was not investigated at the time of HPV detection as NASBA in general was performed successfully as shown by detectable mRNA expressed from the housekeeping gene U1A. The positivity rate for NASBA and type-specific PCR is presented in Table 1. Out of 16 cases testing negative by type-specific PCR and NASBA, seven cases were previously found to be positive for another HPV type (HPV 6, 26, 66, 69, and 73) by consensus Gp5+/6+ PCR and RLB [25Kraus I, Molden T, Holm R, et al. Presence of E6 and E7 mRNA from human papillomavirus types 16, 18, 31, 33, and 45 in the majority of cervical carcinomas J Clin Microbiol 2006; 44: 1310-7.]. Moreover, one additional HPV 18 infection was found by RLB and ISH.
ISH detected HPV 16, 18, 31 and 33 DNA in 158 of the 202 (78.2%) samples (Table 1). Detailed results have been presented previously [25Kraus I, Molden T, Holm R, et al. Presence of E6 and E7 mRNA from human papillomavirus types 16, 18, 31, 33, and 45 in the majority of cervical carcinomas J Clin Microbiol 2006; 44: 1310-7.]. The physical state of HPV DNA was determined by the pattern of the ISH signal. In 84 (53.2%) of samples a punctuate signal, indicating integrated virus DNA was seen, in 74 (46.8%) samples both diffuse and punctuate signals indicating both integrated and episomal viral DNA were seen, while none of the samples had only diffuse signal indicative of episomal viral DNA only.
Mutation analyses of the TP53 gene revealed mutations in 3 of the 6 HPV negative samples (50%) (Table 5).
HPV of any type tended to be detected less frequently by increasing age of the patients. This trend obtained statistical significance for high-risk HPV (group no. 1, Table 3) when based on results obtained with NASBA (p = 0.04) and all methods collectively (p = 0.03). In line with this, absence of HPV 16/18 with all methods (group no. 5, Table 3) was significantly correlated with higher age (p = 0.03). No correlation was found to FIGO stage.
FIGO stage was a strong indicator of survival in both univariate and multivariate analysis as shown in Table 6. Age was of statistical significance in univariate but not in the multivariate analysis.
Based on the HPV E6/E7 mRNA (NASBA) results, patients testing negative had a statistically significant poorer DSS in univariate analysis than those testing positive, as shown in Fig. (1 ) (p = 0.03), while the difference in PFS did not obtain statistical significance (p = 0.10). Comparing high-risk HPV vs HPV 6, 26, 66, 69 and 73 vs HPV negative (group no. 8, Table 3, all methods collectively) HPV negative cases had a statistically significant poorer DSS in univariate analysis that the two other groups (p = 0.03) (Fig. 2
) (p = 0.03), while the difference in PFS did not obtain statistical significance (p = 0.10). Comparing high-risk HPV vs HPV 6, 26, 66, 69 and 73 vs HPV negative (group no. 8, Table 3, all methods collectively) HPV negative cases had a statistically significant poorer DSS in univariate analysis that the two other groups (p = 0.03) (Fig. 2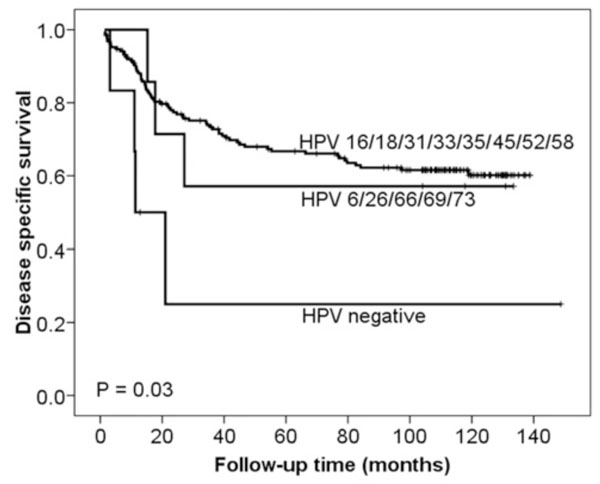 ), whereas no statistical significance was seen for PFS (p = 0.18). For the multivariate analysis, we combined the HPV high-risk group with the HPV 6, 26, 66, 69, 73 group because women in these categories were found to be of similar risk. In the multivariate analysis of DSS (Table 6), the high-risk HPV E6/E7 mRNA (NASBA) and HPV high-risk/HPV 6, 26, 66, 69, 73 (all methods collectively) negative status obtained independent significance. The differences in both PFS and DSS became smaller when based on the results from the other groups and methods for HPV detection and was not of statistical significance.
), whereas no statistical significance was seen for PFS (p = 0.18). For the multivariate analysis, we combined the HPV high-risk group with the HPV 6, 26, 66, 69, 73 group because women in these categories were found to be of similar risk. In the multivariate analysis of DSS (Table 6), the high-risk HPV E6/E7 mRNA (NASBA) and HPV high-risk/HPV 6, 26, 66, 69, 73 (all methods collectively) negative status obtained independent significance. The differences in both PFS and DSS became smaller when based on the results from the other groups and methods for HPV detection and was not of statistical significance.
DISCUSSION
In this study, we have investigated the relation of the presence of HPV DNA and/or HPV E6/E7 mRNA, as well as the physical state of HPV DNA, with the clinical outcome of 202 women treated for cervical SCC. We were particularly interested in comparing the physical state of HPV DNA detected by ISH, and HPV E6/E7 mRNA expression, detected by NASBA, with clinical outcome. Only two studies have previously compared the physical state of HPV DNA, detected by ISH, and clinical outcome [23Unger ER, Vernon SD, Thoms WW, et al. Human papillomavirus and disease-free survival in FIGO stage Ib cervical cancer J Infect Dis 1995; 172: 1184-90., 24Vernon SD, Unger ER, Miller DL, Lee DR, Reeves WC. Association of human papillomavirus type 16 integration in the E2 gene with poor disease-free survival from cervical cancer Int J Cancer 1997; 74: 50-6.] and no study has this far investigated the association between HPV E6/E7 mRNA detected by NASBA and prognosis.
Regarding the relation between HPV status and age, it has previously been reported that women with HPV DNA negative tumours were older than women with HPV DNA positive tumours [3Higgins GD, Davy M, Roder D, et al. Increased age and mortality associated with cervical carcinomas negative for human papillomavirus RNA Lancet 1991; 338: 910-3., 4DeBritton RC, Hildesheim A, De Lao SL, et al. Human papillomaviruses and other influences on survival from cervical cancer in Panama Obstet Gynecol 1993; 81: 19-24., 9Kristensen GB, Karlsen F, Jenkins A, et al. Human papilloma virus has no prognostic significance in cervical carcinoma Eur J Cancer 1996; 32A: 1349-53.]. In a study of Schwartz and colleagues [13Schwartz SM, Daling JR, Shera KA, et al. Human papillomavirus and prognosis of invasive cervical cancer: a population-based study J Clin Oncol 2001; 19: 1906-15.], however, no association between HPV DNA data and age was demonstrated. Also, in some previous studies, infections with HPV 16 [10Fule T, Csapo Z, Mathe M, et al. Prognostic significance of high-risk HPV status in advanced cervical cancers and pelvic lymph nodes Gynecol Oncol 2006; 100: 570-8., 11Nakagawa S, Yoshikawa H, Onda T, et al. Type of human papillomavirus is related to clinical features of cervical carcinoma Cancer 1996; 78: 1935-41.], HPV 33/35 [2Riou G, Favre M, Jeannel D, et al. Association between poor prognosis in early-stage invasive cervical carcinomas and non-detection of HPV DNA Lancet 1990; 335: 1171-4.] and HPV types other than HPV 16/18 [11Nakagawa S, Yoshikawa H, Onda T, et al. Type of human papillomavirus is related to clinical features of cervical carcinoma Cancer 1996; 78: 1935-41.] have been found to increase with age, whereas in other studies, HPV 16 [2Riou G, Favre M, Jeannel D, et al. Association between poor prognosis in early-stage invasive cervical carcinomas and non-detection of HPV DNA Lancet 1990; 335: 1171-4.] and HPV 18 [11Nakagawa S, Yoshikawa H, Onda T, et al. Type of human papillomavirus is related to clinical features of cervical carcinoma Cancer 1996; 78: 1935-41.] infections were found to be frequent also in lower age groups. In the present study, high-risk HPV negatives by NASBA, as well as HPV negatives by all methods, was significantly correlated with higher age. This was also the case for HPV 16/18 negatives based on all methods collectively. In summary, such conflicting results may be explained by variation in terms of HPV detection methods, number and type of HPV types detected by the assay used, patient selection, or number of patients included in the different studies.
The differences in HPV status between the results obtained by NASBA and type-specific PCR were small with discordance in only 4 of 202 samples (2%) and therefore, the clinical relevance is questionable. It is possible that the minor differences between type-specific PCR and NASBA may reflect accidental circumstances or poor sample quality, rather than the actual absence of HPV DNA or mRNA in the respective samples. This is supported by the fact that the samples 3307 and 3959 became positive by type-specific PCR only when tested on 10 times more concentrated material. Furthermore, we cannot exclude the possibility that these differences may be explained by a possible degradation of HPV mRNA, for example in case of suboptimal freezing procedures for the tissue directly following the operation of the patient. On the other hand, we cannot either exclude that the differences actually reflect a higher amount of DNA present as compared to E6/E7 mRNA, and vice versa, as may be seen for example in relation to the integrated form of the virus: Several viral copies may be integrated as concatemers in the human genome; however, expression of the viral oncogenes may be silenced by host regulatory mechanisms. Yet, since the E6 and E7 proteins are shown to be necessary for malignancy, this situation is questionable. Alternatively, only one or two viral copies may be integrated, exerting a high level of E6/E7 expression, explaining the DNA negative/mRNA positive result; the negative DNA result may therefore be explained by low analytical sensitivity of the PCR method. Another explanation for the different results by the two methods might be mutations or deletions in the HPV target sequences. Deletions of larger regions of the HPV genome can be seen for example upon chromosomal integration, with the interruption within the E1 or E2 gene and the possible deletion of downstream sequences as the E5, L2, and L1 genes.
The survival analysis of our data suggests that patients negative for high-risk HPV (types 16, 18, 31, 33, 35, 45, 52 and 58) E6/E7 mRNA (NASBA) or negative for HPV high-risk/HPV 6, 26, 66, 69 and 73 (all methods collectively) had a poorer DSS than patients positive for these HPV groups. This is in accordance with the data of Higgins et al. [3Higgins GD, Davy M, Roder D, et al. Increased age and mortality associated with cervical carcinomas negative for human papillomavirus RNA Lancet 1991; 338: 910-3.] investigating HPV 16, 18, 31 and 33 mRNA expression using ISH and 125I-labelled riboprobes and the work of DeBritton et al. [4DeBritton RC, Hildesheim A, De Lao SL, et al. Human papillomaviruses and other influences on survival from cervical cancer in Panama Obstet Gynecol 1993; 81: 19-24.] identifying HPV 16, 18 and 33 using PCR, Southern blot and slot blot techniques. However, possible underlying biological mechanisms of this phenomenon remain to be elucidated. One possible explanation may be related to HPV unrelated genetic aberrations. We have previously found that in HPV negative cervical carcinomas, compared to HPV positive cases, TP53 mutations are frequently identified [31Helland A, Karlsen F, Due EU, et al. Mutations in the TP53 gene and protein expression of p53, MDM 2 and p21/WAF-1 in primary cervical carcinomas with no or low human papillomavirus load Br J Cancer 1998; 78: 69-72.], which results in lack of functional p53 protein and a possibly more aggressive tumour. This is supported by this study, where we show that 3 of 6 (50%) of the HPV negative cases had TP53 mutations. In general, in cervical carcinomas lacking HPV E6/E7 mRNA expression (or in which the oncogenic expression is kept low), also additional molecular aberrations may exist that increase the carcinogenic potential and aggressiveness of the tumour or which may decrease the sensitivity to radiation therapy. In other words, other progression factors than the action of the E6/E7 oncoproteins may in a small number of cases play a more important role at a later stage in the progression of cancer. It should also be mentioned that the number of cases negative for HPV was low and the results have to be confirmed in larger studies.
Based on the results of PCR and ISH, the presence of HPV DNA was not found to be a prognostic factor in cervical SCCs. Similar findings have been reported by others [7King LA, Tase T, Twiggs LB, et al. Prognostic significance of the presence of human papillomavirus DNA in patients with invasive carcinoma of the cervix Cancer 1989; 63: 897-900.-10Fule T, Csapo Z, Mathe M, et al. Prognostic significance of high-risk HPV status in advanced cervical cancers and pelvic lymph nodes Gynecol Oncol 2006; 100: 570-8.]. However, these observations in turn contradict other investigations; using multivariate analysis, Riou et al. [2Riou G, Favre M, Jeannel D, et al. Association between poor prognosis in early-stage invasive cervical carcinomas and non-detection of HPV DNA Lancet 1990; 335: 1171-4.] and deBritton et al. [4DeBritton RC, Hildesheim A, De Lao SL, et al. Human papillomaviruses and other influences on survival from cervical cancer in Panama Obstet Gynecol 1993; 81: 19-24.] reported that patients negative for HPV DNA had a poorer prognosis than patients positive for HPV DNA. These conflicting results may be due to the small number of negative cases included and additional studies are needed in order to reveal whether the detection of HPV by a specific method may be of prognostic value in assessing survival.
When it comes to the individual HPV types, reduced survival has in particular been correlated with HPV 18 positive tumours [11Nakagawa S, Yoshikawa H, Onda T, et al. Type of human papillomavirus is related to clinical features of cervical carcinoma Cancer 1996; 78: 1935-41.-15Huang HJ, Huang SL, Lin CY, et al. Human papillomavirus genotyping by a polymerase chain reaction-based genechip method in cervical carcinoma treated with neoadjuvant chemotherapy plus radical surgery Int J Gynecol Cancer 2004; 14: 639-49.]. Our results, however, show that neither HPV 18 DNA nor HPV 18 E6/E7 mRNA had any correlation to clinical outcome. Also, it is known that adenocarcinomas have a higher prevalence of HPV 18 than SCCs [12Burger RA, Monk BJ, Kurosaki T, et al. Human papillomavirus type 18: association with poor prognosis in early stage cervical cancer J Natl Cancer Inst 1996; 88: 1361-8.-15Huang HJ, Huang SL, Lin CY, et al. Human papillomavirus genotyping by a polymerase chain reaction-based genechip method in cervical carcinoma treated with neoadjuvant chemotherapy plus radical surgery Int J Gynecol Cancer 2004; 14: 639-49.] and for patients with advanced adenocarcinoma, a significantly worse prognosis has been reported [32Smith HO, Tiffany MF, Qualls CR, Key CR. The rising incidence of adenocarcinoma relative to squamous cell carcinoma of the uterine cervix in the United States--a 24-year population-based study Gynecol Oncol 2000; 78: 97-105.]. The differences may therefore be explained by the fact that our series consisted of SCCs only, whereas in the other studies both SCCs, adenocarcinomas and adenosquamous carcinomas were included [11Nakagawa S, Yoshikawa H, Onda T, et al. Type of human papillomavirus is related to clinical features of cervical carcinoma Cancer 1996; 78: 1935-41.-15Huang HJ, Huang SL, Lin CY, et al. Human papillomavirus genotyping by a polymerase chain reaction-based genechip method in cervical carcinoma treated with neoadjuvant chemotherapy plus radical surgery Int J Gynecol Cancer 2004; 14: 639-49.]. Reduced survival has also been demonstrated for women with multiple HPV types. Our results show no correlation of multiple types with clinical outcome.
In addition to specific HPV types, HPV DNA integration status has also been suggested to play a role for the clinical outcome of cervical SCCs. In the present study, integrated HPV was detected by ISH; however did not correlate with either DSS or PFS free survival. Previously, only one research group has used ISH to compare the physical state of HPV and clinical outcome for patients with cervical carcinoma [23Unger ER, Vernon SD, Thoms WW, et al. Human papillomavirus and disease-free survival in FIGO stage Ib cervical cancer J Infect Dis 1995; 172: 1184-90., 24Vernon SD, Unger ER, Miller DL, Lee DR, Reeves WC. Association of human papillomavirus type 16 integration in the E2 gene with poor disease-free survival from cervical cancer Int J Cancer 1997; 74: 50-6.]. In these studies, including 47 and 50 patients respectively, PFS was shorter for the women with tumours containing integrated HPV DNA than for the women with tumours containing episomal or episomal/integrated HPV DNA [23Unger ER, Vernon SD, Thoms WW, et al. Human papillomavirus and disease-free survival in FIGO stage Ib cervical cancer J Infect Dis 1995; 172: 1184-90., 24Vernon SD, Unger ER, Miller DL, Lee DR, Reeves WC. Association of human papillomavirus type 16 integration in the E2 gene with poor disease-free survival from cervical cancer Int J Cancer 1997; 74: 50-6.]. In two studies using the E2/E6 ratio PCR assay for determining the integration status (based on the fact that E2 is generally disrupted upon integration [33Schwarz E, Freese UK, Gissmann L, et al. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells Nature 1985; 314: 111-4.]), including 50 and 158 patients respectively, it was reported that viral integration was significantly associated with decreased PFS [24Vernon SD, Unger ER, Miller DL, Lee DR, Reeves WC. Association of human papillomavirus type 16 integration in the E2 gene with poor disease-free survival from cervical cancer Int J Cancer 1997; 74: 50-6., 34Kalantari M, Karlsen F, Kristensen G, et al. Disruption of the E1 and E2 reading frames of HPV 16 in cervical carcinoma is associated with poor prognosis Int J Gynecol Pathol 1998; 17: 146-53.]. Another study including 40 patients, reported that HPV 16 positive tumours with a disrupted E2 gene had a tendency towards a shorter progression free survival [35Lindel K, Burri P, Studer HU, et al. Human papillomavirus status in advanced cervical cancer: predictive and prognostic significance for curative radiation treatment Int J Gynecol Cancer 2005; 15: 278-84.]. In contrast to these studies, but in agreement with our data, Szarka et al. [36Szarka K, Veress G, Juhasz A, et al. Integration status of virus DNA and p53 codon 72 polymorphism in human papillomavirus type 16 positive cervical cancers Anticancer Res 2000; 20: 2161-7.] used southern blot hybridization and E1/E2 vs E6 specific PCR on a series of 41 cervical cancers and found that HPV 16 integration did not correlate with progression free survival. One explanation for the discrepancy of previous results [23Unger ER, Vernon SD, Thoms WW, et al. Human papillomavirus and disease-free survival in FIGO stage Ib cervical cancer J Infect Dis 1995; 172: 1184-90., 24Vernon SD, Unger ER, Miller DL, Lee DR, Reeves WC. Association of human papillomavirus type 16 integration in the E2 gene with poor disease-free survival from cervical cancer Int J Cancer 1997; 74: 50-6., 35Lindel K, Burri P, Studer HU, et al. Human papillomavirus status in advanced cervical cancer: predictive and prognostic significance for curative radiation treatment Int J Gynecol Cancer 2005; 15: 278-84., 36Szarka K, Veress G, Juhasz A, et al. Integration status of virus DNA and p53 codon 72 polymorphism in human papillomavirus type 16 positive cervical cancers Anticancer Res 2000; 20: 2161-7.] may be the small number of patients investigated, or may possibly also be related to differences in terms of the detection assays. In general, the clinical outcome and values of prognostic significance are obviously strongly associated with the method used and also with the number of and specific types included in the assay.
CONCLUSION
The presence of high-risk HPV DNA (types 16, 18, 31, 33, 35, 45, 52 and 58), as well as the physical state of the virus, was not significantly associated with clinical outcome. Moreover, no differences in survival of women harbouring specific HPV types were seen. Concerning high-risk HPV E6/E7 mRNA (detected by NASBA) and HPV high-risk/HPV 6, 26, 66, 69 and 73 (all methods collectively), absence may appear to be associated with poor DSS; however, due to the low number of cases negative for HPV, the results should be confirmed in larger studies. To our point of view, in order to better consider the prognosis of women with cervical carcinoma, a better delineation of the total genetic changes (including among others TP53 mutations), not only the HPV status, would be necessary.
ACKNOWLEDGEMENTS
This work was supported by grant 14316/213 from the Research Council of Norway and from the Norwegian Cancer Society. We thank Karin Skogsfjord, Vivi Bassøe, Eldri Undlien Due, Phuong Vu, Mai Thi Phuong Nguyen, Petter Grønn, Inger Marie Falang and Marit Øder Øye for technical assistance. We are also grateful to Profs. Harald zur Hausen and Ethel-Michele de Villiers, Germany for HPV 16 and 18 plasmids.
POTENTIAL CONFLICT OF INTEREST
Hanne Skomedal is a shareholder in NorChip AS. The NASBA based assay PreTect HPV-Proofer used in the study has been developed and manufactured by NorChip AS. Hanne Skomedal and Irene Kraus have been involved in the NASBA and PCR analysis, but they have no influence on the end results of this paper since Gunnar B. Kristensen was responsible for collection of clinical data and Heidi Lyng and Ruth Holm performed the statistical analysis.