
The Open Systems Biology Journal
(Discontinued)
ISSN: 1876-3928 ― Volume 5, 2014
MicroRNA Regulatory Patterns on the Human Metabolic Network
Chabane Tibiche1, Edwin Wang*, 1, 2
Abstract
MicroRNA (miRNA) is an emerging class of non-coding small RNAs, which post-transcriptionally regulate a large number of genes and become important regulators of a broad spectrum of biological processes. To understand the principles of miRNA regulation of metabolic networks, we systematically analyzed the relationships between miRNA targets and network nodes (enzymes) which have distinct network structural features through mapping the miRNA targets onto a human metabolic network. Our analysis showed that miRNAs preferentially regulate hub nodes, i.e., top 5% of the highly connected nodes in the network, and the network cut points which are the bottle-necks of metabolic flows, however, avoid regulating intermediate nodes which are the nodes between the hub nodes, cut points, upstream nodes and the output nodes. Furthermore, two or three consecutive linear metabolic reactions in the network are enriched with miRNA targets, while metabolic branches are depleted with miRNA targets. By targeting the network nodes with distinct network structural features, miRNA regulates metabolic networks regionally and locally to reduce specific metabolite production in a way of fine-tune modulating metabolic flows. Functional association analysis of miRNAs and metabolic pathways uncovered that miRNAs predominantly regulate central metabolic pathways such as amino acid biosynthesis, certain sugar and lipid metabolism.
Article Information
Identifiers and Pagination:
Year: 2008Volume: 1
First Page: 1
Last Page: 8
Publisher Id: TOSYSBJ-1-1
DOI: 10.2174/1876392800801010001
Article History:
Received Date: 9/10/2008Revision Received Date: 17/10/2008
Acceptance Date: 22/10/2008
Electronic publication date: 28/11/2008
Collection year: 2008
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http: //creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Biotechnology Research Institute, National Research Council Canada, Montreal, Quebec, H4P 2R2, Canada; E-mail: edwin.wang@cnrc-nrc.gc.ca
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 9-10-2008 |
Original Manuscript | MicroRNA Regulatory Patterns on the Human Metabolic Network | |
INTRODUCTION
According to the central dogma of molecular biology, RNAs are passive messengers and only take charge of transferring genetic information. However, this central dogma is being challenged by the recent findings that microRNA (miRNA), small noncoding RNA, is able to negatively regulate protein-coding genes. miRNAs regulate gene expression at the post-transcriptional and translational levels by base-paring with the cis-elements located on the 3-terminus of target message RNAs (mRNAs), which results in the cleavage of target mRNAs or repression of their productive translation. A growing volume of evidence has revealed that miRNAs are involved in a variety of biological processes, such as embryonic development, cell proliferation, cell differentiation, apoptosis and energy balance [1]. In terms of metabolism, miRNAs have been found to regulate amino acid catabolism, carbohydrate and lipid metabolism, although the molecular mechanisms of miRNA regulation of metabolism are not clear [2]. It is currently estimated that miRNAs account for ~ 1% of predicted genes in higher eukaryotic genomes and that up to one-third of human genes might be regulated by miRNAs. Thus miRNAs play important roles as negative regulators and add a new level of regulation and fine-tuning for gene expression. Because miRNAs regulate a substantial fraction of genes in animal genomes, miRNAs may form a complex network that in turn intertwines with other cellular networks such as cellular signaling and metabolic networks.
Metabolites are critical in biological systems. Some metabolites such as amino acids and fatty acids take part in the cellular processes for growth, development and reproduction, while some others are involved in defending against parasites and cell signaling. Many metabolites are shared by different metabolic pathways and further intertwined to form complex networks. Thus, various cellular activities are accompanied with the changes of metabolism. It is essential to control the rates of metabolic processes in response to the changes in an internal or external environment for living cells. Mechanisms that control metabolic networks are complex and involve transcriptional, post-transcriptional and translational regulations. Traditionally we think that the enzymes of metabolic networks are tightly controlled by transcription factors. Moreover, the principles of transcriptional regulation of metabolic networks by transcription factors have been illustrated through an integrative analysis of gene expression profiles and the yeast metabolic network [3, 4]. Since miRNAs have been emerged as an abundant class of negative regulators, it is reasonable to think that miRNAs might regulate cellular networks extensively. Several recent computational analyses have been attempted to understand the miRNA regulations in a network context. We and others have studied the principles of miRNA regulation of the networks of human cellular signaling, gene regulation and protein interactions [5-8]. However, so far it is not clear to what extent miRNAs regulate metabolic networks, nor how miRNAs mechanistically regulate metabolic networks.
To address these questions, we systematically analyzed the relationships between miRNA targets and network nodes (enzymes) which have distinct network structural features through mapping the miRNA targets onto a human metabolic network. Our analysis showed that miRNAs preferentially regulate hub nodes, network cut points and avoid regulating intermediate nodes. By targeting the network nodes with distinct network structural features, miRNA regulates metabolic networks regionally and locally to reduce specific metabolite production in a way of fine-tune modulating metabolic flows.
MATERIALS AND METHODOLOGY
Metabolic Network Construction and microRNA Target Gene Mapping
We used the human metabolic pathways from KEGG database (ftp://ftp.genome.jp/pub/kegg/xml/organisms/hsa) to construct the metabolic network. We identified all the reactions and their associated enzymes and genes. We used a reaction-centric model to represent the metabolic network. The network is presented as a directed graph in which nodes represent enzymes or reactions. For two nodes (reactions) A and B, we linked A → B if a product metabolite of A is a substrate metabolite of B.
The most commonly used molecules such as ATP, NAD, CO2 in biochemical reactions were excluded in the network. For each node in the metabolic network, we identified all the genes associated with it. Human metabolic network contains 1,549 nodes and 4,740 directed links. In the network, over 70% of the nodes are linked to form the largest network component. The largest component of the human metabolic network contains 1,099 nodes and 4,196 links. In this study, we used only the largest contiguous component of the network to perform analyses.
We used genome-wide computationally predicted miRNA target genes in human genome from a recent study [9]. Updated miRNA targets from the prediction were downloaded from authors’ website.
Classifying Network Node Types with Distinct Structural Features
In a reaction-centric network, nodes represent enzymes or reactions. We classified the nodes into five categories based on their structural features in the network. The nodes that have no incoming links are called upstream nodes (UPNs, Fig. 1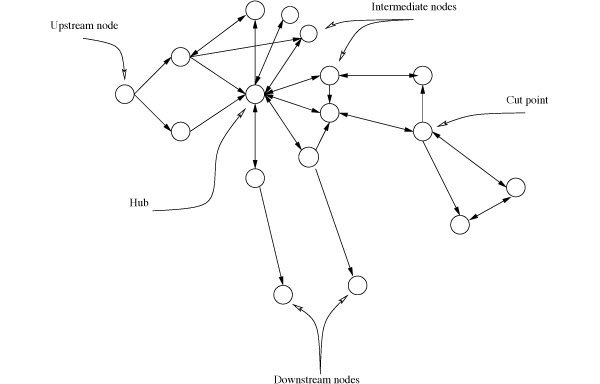 , Supplementary Data
1), where the metabolites are often up-taken from the extracellular space, are used to produce downstream metabolites of the network. Therefore, the UPNs are the most upstream reactions that many other downstream reactions and metabolites rely on. On the other hand, the nodes that have no outgoing links are called output nodes or downstream nodes (DSNs, Fig. 1
, Supplementary Data
1), where the metabolites are often up-taken from the extracellular space, are used to produce downstream metabolites of the network. Therefore, the UPNs are the most upstream reactions that many other downstream reactions and metabolites rely on. On the other hand, the nodes that have no outgoing links are called output nodes or downstream nodes (DSNs, Fig. 1 , Supplementary Data
1), where many pathway output metabolites such as amino acids of the network are produced. These metabolites are then used for various cellular activities such as acting as cell membrane components or regulating signaling pathways. A cut vertex or a cut point (CP, Fig. 1
, Supplementary Data
1), where many pathway output metabolites such as amino acids of the network are produced. These metabolites are then used for various cellular activities such as acting as cell membrane components or regulating signaling pathways. A cut vertex or a cut point (CP, Fig. 1 , and Supplementary Data
1) is such a bottleneck node that its deletion will disconnect at least one component from the network. CPs are in crucial network positions and become bottlenecks of the network, and therefore control metabolic flows from a part to another in the network. The removal of one node from a network results in more network components than in the original network, we then regarded that node as a CP. To identify CPs, we examined each node in the network to check whether its removal results in more network components. In the network, highly connected nodes (hubs, Fig. 1
, and Supplementary Data
1) is such a bottleneck node that its deletion will disconnect at least one component from the network. CPs are in crucial network positions and become bottlenecks of the network, and therefore control metabolic flows from a part to another in the network. The removal of one node from a network results in more network components than in the original network, we then regarded that node as a CP. To identify CPs, we examined each node in the network to check whether its removal results in more network components. In the network, highly connected nodes (hubs, Fig. 1 , Supplementary Data
1) are the reactions that share metabolites with many other reactions. The metabolites of these highly connected nodes are also major suppliers for the peripheral region of the network where many output metabolites are produced. In this study, we defined the top 5% of the highly connected network nodes, which have higher degrees (in- and out-degrees together) as hubs. Besides these four types of nodes described above, all other nodes in the network are called intermediate nodes (ITNs, Fig. 1
, Supplementary Data
1) are the reactions that share metabolites with many other reactions. The metabolites of these highly connected nodes are also major suppliers for the peripheral region of the network where many output metabolites are produced. In this study, we defined the top 5% of the highly connected network nodes, which have higher degrees (in- and out-degrees together) as hubs. Besides these four types of nodes described above, all other nodes in the network are called intermediate nodes (ITNs, Fig. 1 , and Supplementary Data
1).
, and Supplementary Data
1).
 |
Fig. (1) Node types with distinct network structural features. |
Randomization Tests
To test the statistical significance of observations, we performed randomization tests. A more detailed explanation of randomization tests was previously described by Wang and Purisima [10]. Briefly description of the randomization tests was the followings:
To test the statistical significance of miRNA regulation of a multiple-gene-node, we calculated the fraction of miRNA targets of the genes which are associated with the multiple-gene-node. We then randomly shuffled the miRNA targets (genes) 5,000 times among the metabolic genes and calculated the P values. We further corrected the P values by applying False Recovery Rate (FDR). FDR corrected P values (<0.25) were considered as significant.
To test the statistical significance of the node types which are either enriched or less-enriched with miRNA targets, we calculated the fraction of miRNA targets of each node type, and then randomly shuffled the miRNA targets (nodes) 5,000 times among the network nodes to calculate the P values.
To test the statistical significance of the linear network motifs which are enriched with miRNA targets, we calculated the fraction of the linear motifs in which all the nodes are miRNA targets, and then randomly shuffled the miRNA targets (nodes) 10,000 times among the network nodes to determine the P values.
Metabolic flows are arranged as linear or branching reactions. To test the statistical significance of the miRNA regulatory patterns on metabolic branches, we enumerated all the possible miRNA targeting patterns for each branch type (see Table 1) then calculated the fraction of each pattern. Finally, we randomly shuffled the miRNA targets (nodes) 10,000 times among the network nodes to determine the P values.
Functional Association of microRNA Regulation
We obtained human metabolic pathways from the KEGG database. We conducted the analysis by considering two situations: (a) miRNA-target enrichment in each individual pathway; (b) in a network context, a pathway that contains miRNA-targeted downstream nodes (output nodes, DSNs) or miRNA-targeted CPs who are the parents of the pathway’s DSNs. To determine the statistical significance of the enrichment of miRNA targets of a pathway, for each pathway we first calculated the number of the reactions, which are miRNA-targeted nodes that have been determined in this study, and then randomly swapped the miRNA targets among the metabolic reactions (nodes). In each swap, we recalculated the fraction of the miRNA targets in each pathway and compared to that of each pathway. The P values were corrected by FDR. FDR corrected P values (<0.25) were considered as significant. To examine the pathways that are regulated by miRNA in a network context, we considered: (a) if a DSN of a pathway is the target of miRNA, we classified that pathway as miRNA-regulated; (b) when a miRNA targets a CP which is a parent of a DSN of a pathway, we also classified that pathway as miRNA-regulated.
RESULTS AND DISCUSSIONS
In this study, we systematically analyzed the human metabolic network by integrating miRNA target genes onto the network. In general, miRNAs could have two ways to regulate cellular metabolism: miRNAs could regulate transcription factors or signaling proteins, which in turn regulate metabolic enzymes. For example, Krutzfeldt and colleagues showed that miR-122 in mouse liver negatively regulate some transcriptional repressors which control plasma cholesterol concentration through modulating a cluster of cholesterol-biosynthesis genes, including HMG-CoA reductase, the rate-limiting step of cholesterol synthesis [11]. Alternatively, miRNAs could regulate the production of certain metabolites by directly regulating the genes that encode metabolic enzymes. In this study, we mainly focused on this latter type of regulation.
In general, biochemical characterizations of enzymes and reactions of metabolic pathways have been documented extensively. Comparing to gene regulatory and cellular signaling networks, metabolic networks are relatively more comprehensive and well characterized. Furthermore, analyses of various cellular networks including metabolic networks through integrating high-throughput datasets have resulted in a series of interesting discoveries [12-16]. Therefore, a global analysis of metabolic networks through integrating miRNA target genes would result in a comprehensive understanding how miRNA regulates metabolic networks. Since miRNAs are natural negative regulators that could reduce the enzyme concentration and then regulate certain metabolite production, we expect that the principles of miRNA regulation of the networks would help us learning nature’s tricks for metabolic controlling, which in turn helps designing drugs to perturb metabolic networks and control production of particular metabolites in living organisms.
To understand the principles of miRNA regulation of metabolic networks from a systems-wide perspective, we systematically analyzed the relationships between miRNA targets and the network nodes having different network structural features. To do so, we first constructed a human metabolic network using KEGG pathway database [17]. Using the database, we identified all chemical reactions and their associated enzymes, genes and metabolites within human metabolism and then constructed a reaction-centric network, in which we joined the reactions that share a common metabolite as either a product or reactant. In the network one node represents one or a set of genes that encode the enzymes that catalyze one reaction. The outgoing or incoming connections from a node indicate that a metabolic reaction is producing metabolites for other reactions or consuming metabolites produced by other reactions. In the network, a giant, connected component (subgraph) was found. The largest subgraph of human metabolic network contains 1,099 nodes, 4,169 links and represents over 70% of the nodes, nearly 90% of the links of the human network and most metabolic pathways including all the central pathways (Supplementary Data 2). Therefore, we took this largest subgraph as the network in the study.
To analyze miRNA regulation of the networks, we mapped the computationally predicted miRNA target genes onto the network. The current miRNA target prediction methods are mainly based on the principle of miRNA-target interactions [9, 18]. Although several miRNA-target prediction algorithms have been developed, TargetScan seems to be the best among these algorithms based on a recent proteomic survey of miRNA targets [19]. Among the 246 miRNAs which have been predicted for targets by TargetScan, 234 (95%) of miRNAs have at least one metabolic gene as targets. In the network, each node is associated with one or more genes, therefore, we grouped the network nodes into two groups: one group (single-gene-node group) contains the nodes such that each node is encoded by only one gene, while the other group (multiple-gene-node group) contains the nodes such that each node is encoded by multiple genes. Finally, 558 single-gene-nodes and 541 multiple-gene-nodes of the network were mapped with miRNA targets. For the single-gene-nodes, we considered a node as a miRNA target as long as the gene that is associated with that node is a miRNA target. Because each multiple-gene-node is associated with more than one genes, it could have higher chance to be associated with miRNA targets by random. Therefore, we performed randomization tests to determine whether a multiple-gene-node is significantly regulated by miRNAs (see Methodology). By doing so, we defined 79 multiple-gene-nodes as miRNA targets. We merged the miRNA targets of single-gene-nodes with the multiple-gene-nodes, and found that 238 (22%) nodes are miRNA targets (Supplementary Data 3).
miRNAs Preferentially Regulate Hubs and Cut Point Nodes But Avoid Regulating Intermediate Nodes
To further explore miRNA regulatory principles, we asked if miRNA targets are enriched in certain node types, i.e. hubs, UPNs, DSNs CPs and ITNs. To address this question, we calculated the fractions of miRNA targets for each node type and performed randomization tests. To perform the randomization tests, we randomly assigned 238 targets onto the network nodes and examined the statistical significance of the miRNA targets in each node type (see Methodology). As shown in Table 2, miRNA targets are less enriched in ITNs in the network (P=0.032, Table 2, Supplementary Data 4), suggesting that miRNAs avoid regulating ITNs. On the other hand, miRNA targets are significantly enriched in the hubs and CPs of the network (P=0.015 and P=0.006, respectively, Table 2, Supplementary Data 4). These results suggest that miRNAs could regulate metabolism globally by preferentially regulating hubs, and control the downstream metabolic flux of the CPs with a local regulation strategy by preferentially regulating CPs. Hubs are the reactions that are shared by many pathways. miRNA regulation of hubs would affect many pathways and therefore such a regulation has a global effect on metabolism. For example, citrate synthase gene, encoding a major enzyme in citrate cycle (TCA cycle), could be regulated by a set of miRNAs: miR-152, miR-148a, miR-148b, miR-299-5p, miR-19b, miR-122a, miR-421, miR-494 and miR-19a. When these miRNAs regulate the citrate synthase gene, 78 pathways such as purine metabolism, pentose phosphate pathway, fatty acid biosynthesis, which represent lipid, carbon, nucleotide and amino acid metabolism, could be affected (Supplementary Data 5). This example illustrates that miRNA could regulate metabolism globally. On the other hand, GANAB, encoding glucosidase, a cut point in the network, could be regulated by miR-133a, miR-133b, miR-199a and miR-199b. Once GANAB is regulated by these miRNAs, the ouputs of three pathways, biosynthesis of steroids, pyrimidine metabolism and glycine, serine and threonine metabolism could be affected at the same time. In this way, a miRNA could regulate a few pathways in a coordinated manner.
Taken together, miRNAs preferentially regulate network hubs and cut point nodes but avoid regulating the intermediate nodes. These results suggest that miRNAs have strategies to regulate metabolism by preferentially regulating certain types of nodes. In cellular signaling networks, we found that miRNAs avoid regulating the upstream nodes, i.e., ligands and receptors [5], while in metabolic networks, miRNAs avoid regulating intermediate nodes. In this study, we found that miRNAs preferentially regulate metabolic network hubs, which is in agreement with the previous studies that miRNAs preferentially regulate hubs in protein interaction networks and gene regulatory networks [6, 7]. It becomes a general principle that except in signaling networks, miRNAs preferentially regulate hubs of the cellular networks. In addition, miRNA preferentially regulates CPs for targeted tuning metabolic flows, while a similar local regulatory strategy of miRNA has been found in signaling networks, i.e., miRNA preferentially regulating the downstream components of adaptors in signaling networks [5].
Patterns of miRNA Regulating Metabolic Flows
Metabolic flows are arranged as linear or branching reactions. We first examined whether 2 and more consecutive linear reactions are enriched with miRNA targets. We found that 307 and 218 cases of the 2 and 3 consecutive linear reactions are all miRNA-targeted, respectively. Randomization tests showed that these cases are statistically significant (P<0.0002 and P<0.01, respectively, see Methodology). When performing the same analysis using the targets of individual miRNAs, no such statistically significant results were obtained. These results suggest that certain reaction regions (i.e., two or more consecutive linear reactions) are preferentially regulated by miRNA. For example, in N-Glycan biosynthetic pathway, fucosyltransferase 8 (FUT8), mannosyl (alpha-1,6-)-glyco-protein beta-1,2-N-acetylglucosaminyltransferase (MGAT2) and mannosidase (MAN2A1) are three enzymes that catalyze 3 consecutive reactions for N-Glycan biosynthesis. Their genes could be regulated by (miR-34c, miR-342, miR-449, miR-449b, miR-122a, miR-34a, miR-494 and miR-377), (miR-200b, miR-200c, miR-429, miR-181b, miR-181a, miR-495, miR-181c and miR-181d) and (miR-128b, miR-92b, miR-128a, miR-27a, miR-490, miR-32, miR-363, miR-500, miR-26b, miR-367, miR-25, miR-218, miR-92, miR-26a, miR-135b, miR-27b, miR-135a), respectively.
Branching metabolic flows could be convergent or divergent. To understand how miRNA regulate the branching metabolic flows, we extracted all the convergent or divergent units which are associated with three distinct reactions from the network. Convergent units could integrate metabolic flows while divergent units could allow a metabolic flow to diverge in two directions. We enumerated and extracted all the possible miRNA targeting patterns for the convergent or divergent units from the network and examined the statistical significance for each pattern. We found that both the convergent and divergent units without any miRNA targets are significantly enriched (Patterns 6 and 12 in Table 1, P<1.0 x 10-4, randomization tests, see Methodology). In agreement with these observations, both the convergent and divergent units containing at least 2 miRNA-targeted nodes are significantly less-enriched (Patterns 1, 2, 5, 7, 8 and 11 in Table 1, randomization tests, see Methodology). Collectively, these results suggest that miRNAs preferentially regulate two or more consecutive linear metabolic reactions but avoid regulating metabolic branches.
Functional Associations of miRNA Regulation
To discover which metabolic pathways are regulated by miRNAs, we queried the KEGG human metabolic pathways using the reactions (nodes in the network), which are the miRNA targets, and conducted two types of analyses for determining the pathways that are significantly regulated by miRNA. We first performed an enrichment analysis for miRNA-targeted pathways using randomization tests. Using this method, we found that six pathways such as N-Glycan biosynthesis are enriched with miRNA targets (Supplementary Data 6, see Methodology). In the second type of analysis, we considered a pathway as miRNA-regulated pathway, if the pathway contains a miRNA-targeted reaction, which is a DSN or a CP that is the parent of the DSN in the network. Using this approach, we found that thirty pathways such as amino acid synthesis or degradation and other central metabolisms are extensively regulated by miRNA through targeting DSNs or CPs. Theses results suggest that miRNAs prefer to regulate DSNs or their parent-CPs. We merged the results from the two approaches, and found that in total 32 pathways are regulated by miRNAs (Table 3).
As shown in Table 3, many biosynthetic pathways such as amino acid synthesis are extensively regulated by miRNAs. Glycan biosynthesis, pantothenate and CoA biosynthesis are also regulated by miRNAs. miRNAs also regulate certain lipid metabolism such as sphingolipid metabolism and glycerophospholipid metabolism. Similarly, miRNA regulation of central metabolic enzymes has been reported in Drosophila, i.e., miR-277 seems to be a regulatory switch for amino acid metabolism [20]. Amino acids are the building elements of proteins. Lipids are components of the plasma membrane or involved in cell signaling, while glycan and CoA are the major metabolic substrates for cell energy. These examples showed that miRNAs are predominantly involved in central metabolic pathways and thus play an important role in cell metabolism.
CONCLUSIONS
Our analysis provides a systems-level understanding of miRNA post-transcriptional regulation of metabolic networks by exploring the relationships between the miRNA targets and the network nodes with distinct network structural properties. We found that miRNAs avoid regulating ITNs but preferentially regulate highly linked nodes and cut points. Furthermore, miRNAs preferentially regulate two or three consecutive linear metabolic reactions but avoid regulating metabolic branches. By regulating network hubs, miRNA could regulate the metabolism globally, while by regulating CPs and certain consecutive linear reactions, miRNA could control the production of certain metabolites via a local regulatory strategy. Functional association analysis of miRNAs and metabolic pathways showed that miRNAs predominantly regulate basic metabolic pathways such as amino acid biosynthesis/degradation and certain lipid metabolisms, suggesting that miRNAs exert their functions in central metabolic activities of cells. These results imply that miRNA is extensively involved in many biological processes and modulating metabolic flux alterations. As we mentioned earlier, miRNA could regulate transcription factors and signaling proteins, which in turn regulate metabolic enzymes. Furthermore, miRNAs could regulate mRNAs through chromatin remodeling [21]. At this stage, it is unclear how to define the miRNA targets when miRNA acts on chromatin remodeling. In this study, our analysis is focusing on the direct regulation of enzymes by miRNA. Nevertheless, this analysis enhances our understanding of how miRNA post-transcriptionally regulates metabolic networks, and reveals molecular strategies for controlling metabolic flux by miRNAs in living organisms.
ACKNOWLEDGEMENTS
We thank Dr. Tyler MacKenzie for his comments on the manuscript. This work is partially supported by Genome and Health Initiatives.
SUPPLEMENTARY MATERIAL
This article also contain supplementary data and it can be viewed at www.bentham.org/open/tosysbj


