
Current Chemical Genomics and Translational Medicine
(Discontinued)
ISSN: 2213-9885 ― Volume 12, 2018
Galectin-1 and Galectin-3 Mediate Protocadherin-24-Dependent Membrane Localization of β-catenin in Colon Cancer Cell Line HCT116
Rui Osea, b, Osamu Oharaaa, b, c, Takahiro Nagasea, *
Abstract
Protocadherin-24 (PCDH24) is linked to the suppression of tumor growth and the inhibition of cell proliferation in the colon cancer cell line HCT116. We previously observed that β-catenin is localized to the plasma membrane when PCDH24 is expressed in these cells, but the molecular mechanisms by which PCDH24 induces the membrane localization of β-catenin remain largely unknown. To clarify these mechanisms, we identified molecules that interact with ectopically expressed PCDH24 in HCT116 cells using a HaloTag® pull-down assay. We found that galectin-1 and galectin-3 physically interact with PCDH24 and are retained at the plasma membrane in association with PCDH24 expression. A luciferase-based pull-down assay using HaloTag-fused galectins revealed that an intracellular region of PCDH24 (amino acids 1186–1280) is essential for this interaction. Furthermore, the over-expression of galectin-1 or -3, or the depletion of endogenous galectins by small interfering RNA modulates β-catenin translocation. We also revealed that the retention of galectin-1 and -3 at the plasma membrane results in the inactivation of PI3K activity. From these findings, we propose a model in which the galectin-anchoring activity of PCDH24 leads to the suppression of β-catenin signaling by the localization of β-catenin at the plasma membrane in PCDH24-expressing HCT116 colon cancer cells.
Article Information
Identifiers and Pagination:
Year: 2012Volume: 6
Issue: Suppl 1
First Page: 18
Last Page: 26
Publisher Id: CCGTM-6-18
DOI: 10.2174/1875397301206010018
Article History:
Received Date: 02/3/2012Revision Received Date: 14/5/2012
Acceptance Date: 16/5/2012
Electronic publication date: 20/9/2012
Collection year: 2012
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Department of Human Genome Research, Kazusa DNA Research Institute, 2-6-7 Kazusa-Kamatari, Kisarazu, Chiba 292-0818, Japan; Tel: +81-438-52-3931; Fax: +81-438-52-3931; nagase@kazusa.or.jp
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 02-3-2012 |
Original Manuscript | Galectin-1 and Galectin-3 Mediate Protocadherin-24-Dependent Membrane Localization of β-catenin in Colon Cancer Cell Line HCT116 | |
INTRODUCTION
Protocadherin-24 (PCDH24, also known as CDHR2 and protocadherin LKC) was originally identified as a novel member of the human non-classical cadherin family. PCDH24 expression is markedly reduced in cancers derived from liver and colon tissues, while the over-expression of PCDH24 protein restores the contact inhibition of cell proliferation in the colon cancer cell line HCT116, and suppresses the growth of these cells in a nude mouse model [1Okazaki N, Takahashi N, Kojima S, Masuho Y, Koga H. Protocadherin LKC, a new candidate for a tumor suppressor of colon and liver cancers, its association with contact inhibition of cell proliferation Carcinogenesis 2002; 23: 1139-48., 2Ose R, Yanagawa T, Ikeda S, Ohara O, Koga H. PCDH24-induced contact inhibition involves downregulation of β-catenin signaling Mol Oncol 2009; 3: 54-66.]. We previously demonstrated that stably expressed PCDH24 proteins accumulate at cell-to-cell adhesion sites and regulate the localization of β-catenin to the cell membrane in the HCT116 cell line. These previous results suggested that PCDH24 is important for the contact inhibition of epithelial cells, and that the absence of PCDH24 induces tumor growth; more specifically, PCDH24 acts as a tumor suppressor protein, at least in the HCT116 cell line [1Okazaki N, Takahashi N, Kojima S, Masuho Y, Koga H. Protocadherin LKC, a new candidate for a tumor suppressor of colon and liver cancers, its association with contact inhibition of cell proliferation Carcinogenesis 2002; 23: 1139-48., 2Ose R, Yanagawa T, Ikeda S, Ohara O, Koga H. PCDH24-induced contact inhibition involves downregulation of β-catenin signaling Mol Oncol 2009; 3: 54-66.]. Although we proposed a novel model for the suppression of β-catenin signaling by PCDH24, the molecular mechanism by which PCDH24 induces the membrane localization of β-catenin has not yet been elucidated.
Galectin-1 and galectin-3 are members of the galectin family of carbohydrate-binding proteins that have a binding affinity to β-galactoside. The galectins have multiple biological functions, including effects on RNA processing, transcription regulation, cell growth, cellular adhesion, immune response, apoptosis, and metastasis [3Heilmann S, Hummel T, Margolis FL, Kasper M, Witt M. Immunohistochemical distribution of galectin-1, galectin-3, and olfactory marker protein in human olfactory epithelium Histochem Cell Biol 2000; 113: 241-5.-8Cedeno-Laurent F, Dimitroff CJ. Galectin-1 research in T cell immunity: past, present and future Clin Immunol 2012; 142: 107-6.]. The galectins are widely expressed in humans, but the expression level varies according to the origin of the tissues—specifically, the expression and subcellular localization of galectins in normal tissues differs from those in tumor cells [5Dumic J, Dabelic S, Flögel M. Galectin-3: An open-ended story Biochim Biophys Acta 2006; 1760: 616-35., 9Demydenko D, Berest I. Expression of galectin-1 in malignant tumors Exp Oncol 2009; 31: 74-9., 10Çay T. Immunhistochemical expression of galectin-3 in cancer: A review of the literature Turk Patoloji Derg 2012; 28: 1-10.]. Recent studies have suggested that galectin-3 activates Wnt signaling in a wide variety of cancer cells. Mutations in the genes involved in the Wnt signaling pathway are frequently observed in human cancers [11Morin PJ, Sparks AB, Korinek V, et al. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC Science 1997; 275: 1787-90.-13Polakis P. Wnt signaling and cancer Genes Dev 2000; 14: 1837-51.]. Activation of the Wnt signaling pathway leads to accumulation of nuclear β-catenin, resulting in activation of the transcription of specific target genes, including c-Myc and cyclin D1. Conversely, suppression of the Wnt signal results in degradation of β-catenin as a result of its phosphorylation by CK1α and glycogen synthase kinase-3β (GSK-3β) and subsequent ubiquitination [12Sparks AB, Morin PJ, Vogelstein B, Kinzler KW. Mutational analysis of the APC/β-catenin/Tcf pathway in colorectal cancer Cancer Res 1998; 58: 1130-4.-14Dihlmann S, von Knebel Doeberitz M. Wnt/β-catenin-pathway as a molecular target for future anti-cancer therapeutics Int J Cancer 2005; 113: 515-24.]. Recent studies reported that galectin-3 is an important component in the Wnt signaling, since it binds directly to the β-catenin and TCF4 complex and induces transcription of the Wnt signaling target genes [15Shimura T, Takenaka Y, Tsutsumi S, Hogan V, Kikuchi A, Raz A. Galectin-3, a novel binding partner of β-catenin Cancer Res 2004; 64: 6363-7., 16Shimura T, Takenaka Y, Fukumori T, et al. Implication of galectin-3 in Wnt signaling Cancer Res 2005; 65: 3535-7.]. These findings indicated that there is a strong relationship between galectins and β-catenin. Moreover, Elad-Sfadia et al. found that galectin-3, when bound to activated K-Ras promotes strong activation of Raf-1 and phosphoinositide 3-kinase (PI3K), and Song et al. reported that galectin-3 augments Wnt signaling by regulating GSK-3β phosphorylation through the PI3K/AKT pathway and β-catenin nuclear accumulation in colon cancer cells [17Song S, Mazurek N, Liu C, et al. Galection-3 mediates nuclear β-catenin accumulation and Wnt signaling in human colon cancer cells by regulation of glycogen synthase kinase-3? activity Cancer Res 2009; 69: 1343-9., 18Elad-Sfadia G, Haklai R, Balan E, Kloog Y. Galectin-3 augments K-ras activation and triggers a ras signal that attenuates ERK but not phosphoinositide 3-kinase activity J Biol Chem 2004; 279: 34922-0.].
In this report, we identified galectin-1 and galectin-3 as PCDH24-interacting molecules by a pull-down experiment using HaloTag® technology, and also showed that the galectin-anchoring activity of PCDH24 leads to inactivation of PI3K, resulting in suppression of the β-catenin signaling by localization of β-catenin at the plasma membrane. These results provide new insight into the tumor suppressor functions of PCDH24 in the colon cancer cell line HCT116.
MATERIALS AND METHODOLOGY
Establishment of a Stable HCT116 Cell Line Expressing PCDH24-HaloTag
The open reading frame (ORF) sequence of PCDH24 was cloned from the pCEP-PCDH24 plasmid [1Okazaki N, Takahashi N, Kojima S, Masuho Y, Koga H. Protocadherin LKC, a new candidate for a tumor suppressor of colon and liver cancers, its association with contact inhibition of cell proliferation Carcinogenesis 2002; 23: 1139-48.] by PCR and ligated with the pT-REx-DEST30 vector (Life Technologies, Carlsbad, CA). Then, we amplified the fragment containing the PCDH24 ORF and CMV-TetO2 promoter sequence by PCR and ligated the fragment into the pFC14A vector (Promega, Madison, WI); the complete fragment that included the CMV-TetO2 promoter, PCDH24 full-length ORF and HaloTag sequence was amplified by PCR. Finally, the complete fragment was ligated into the pJTI™ Fast DEST vector (Life Technologies). The nucleotide sequences were confirmed by sequencing. To establish cell lines that express the tetracyclin repressor (TetR) protein and PCDH24-HaloTag protein, the pcDNA6/TR plasmid was initially transfected into HCT116 cells by the liposome-mediated gene transfer method. The cells were switched, 48 h later, to a selective medium containing 50 µg/mL blasticidin. After 12 days of culture in the selective medium, independent blasticidin-resistant clones were isolated and expanded. The pJTI™ Fast DEST PCDH24-HaloTag plasmid was transfected into TetR-expressing HCT116 cells. Then, 48 h later, the cells were cultured in a selective medium containing 100 µg/mL hygromycin B. After 12 days of culture in the selective medium, independent hygromycin-resistant clones were isolated.
Cell Culture
The human colon cancer cell line HCT116 and cells of the derivate lines HCT116-PCDH24-EGFP and HCT116-PCDH24-HaloTag were cultured in Dulbecco's Modified Eagle's Medium supplemented with 10% Tet System Approved Fetal Bovine Serum (Takara Bio Inc., Shiga, Japan) and 100 U/mL penicillin and streptomycin (Life Technologies) in a CO2 (5%) incubator at 37°C. PCDH24-HaloTag cells were treated with 5 µg/mL blasticidin (InvivoGen, San Diego, CA) and 50 µg/mL hygromycin B (Life Technologies) to maintain the expression of TetR and the inducible PCDH24 ORF clone, respectively. HCT116-PCDH24-EGFP cells were maintained with 50 µg/mL G418 (Life Technologies). To induce the expression of PCDH24-HaloTag, HCT116-PCDH24-HaloTag cells were treated with 1 µg/mL of doxycycline (DOX) (MP Biomedicals, LLC, Solon, OH).
Proliferation Assay
HCT116, PCDH24-HaloTag, and PCDH24-EGFP cells were cultured in their respective cell culture conditions and 1.0 × 106 cells were seeded on 35-mm dishes (Thermo Fisher Scientific, Waltham, MA). The cells were harvested by trypsinization and the number of cells was counted using a hemocytometer every 2 days for 12 days. For the wound-healing assay, cell movement was monitored for 24 hours using a time-lapse application of Biozero BZ-8000 (KEYENCE, Osaka, Japan).
Fluorescent Immunostaining and Microscopic Observation
To observe endogenous proteins, fluorescent immunostaining was applied. The cells were cultured in a 5% CO2 incubator at 37°C. Cultured cells were fixed in 3.7% formaldehyde/phosphate buffer saline (PBS), permeabilized by 0.1% Triton X-100/PBS, blocked by 1% casein/PBS blocking solution, and incubated with diluted primary antibodies in 1% casein/PBS. The cells were then incubated with the secondary antibodies, and mounted with Vectashield® Mounting Medium (Vector Laboratories Inc., Burlingame, CA). Anti-β-catenin monoclonal (BD Biosciences), anti-galectin-1 monoclonal (M01) (Abnova), anti-β-catenin (H-102) polyclonal (sc-7199) and anti-galectin-3 (H-160) polyclonal (sc-20157) (Santa Cruz Biotechnology, Santa Cruz, CA) were used as primary antibodies. Alexa Fluor® 546-conjugated goat anti-mouse, Alexa Fluor® 555-conjugated goat anti-rabbit Alexa Fluor® 633-conjugated goat anti-rabbit, and Alexa Fluor® 633-conjugated goat anti-mouse (Molecular Probes, Eugene, OR) were used as secondary antibodies. Axiovert S100 (Carl Zeiss, Jena, Germany) and Biozero BZ-8000 microscopes were used for phase-contrast and fluorescence images. Confocal microscopic analysis using an LSM510 microscope (Carl Zeiss) was performed according to a previous report [1Okazaki N, Takahashi N, Kojima S, Masuho Y, Koga H. Protocadherin LKC, a new candidate for a tumor suppressor of colon and liver cancers, its association with contact inhibition of cell proliferation Carcinogenesis 2002; 23: 1139-48.]. To visualize the PCDH24-HaloTag protein, HaloTag was labeled with HaloTag® TMR ligand, R110Direct™ ligand or coumarin ligand according to the manufacturer’s instructions (Promega; “No-Wash” protocol).
Pull-down assay Followed by Mass Spectrometry Analysis
We performed a pull-down assay using HaloLinkTM resin (Promega). Whole cell lysate including HaloTag-fusion proteins from 4.0 × 107 cells was extracted with 8 mL lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100 and 0.1% sodium deoxycholate). The cell extracts were mixed with 250 µL of HaloLinkTM resin (pre-equilibrated, 25% slurry) overnight at 4°C. After washing the resin five times with PBS, the endogenous proteins bound to the HaloTag-fusion proteins were recovered in 100 µL of sodium dodecyl sulfate (SDS) elution buffer (50 mM Tris-HCl, pH 7.5, 1% SDS). The eluates were concentrated using a Microcon centrifugal filter device YM-10 (Millipore Corp., Billerica, MA). The concentrated samples were resolved by 5–20% SDS-polyacrylamide gel electrophoresis (PAGE) and then subjected to imidazole-zinc reverse staining. Several bands were digested with 10 µg/mL trypsin (Promega). After dilution with 1% trifluoroacetic acid, the resulting peptide mixture was subjected to liquid chromatography (LC)-mass spectrometry (MS)/MS analysis as previously described [19Koga H, Yuasa S, Nagase T, et al. A comprehensive approach for establishment of the platform to analyze functions of KIAA proteins II: public release of inaugural version of InGaP database containing gene/protein expression profiles for 127 mouse KIAA genes/proteins DNA Res 2004; 11: 293-304.]. The resulting MS/MS data were analyzed using the Mascot search engine (Matrix Science, London, UK).
Luciferase-based Pull-down Assay
We seeded 2.0 × 106 HCT116 cells on a tissue culture plastic plate (diameter, 100 mm) at 24 h before transfection. Expression clones for luciferase-fused full-length PCDH24 and the C-terminal deletion mutants were co-transfected with the HaloTag or HaloTag-fused galectin expression clones using FuGENE HD (Roche Diagnostics, Mannheim, Germany). At 48 h after the transfection, the cells were lysed with CelLyticTM M (Sigma, St. Louis, MO) and an aliquot of the lysate was used for a pull-down assay using HaloLinkTM resin. The lysate was incubated with HaloLinkTM resin for 30 min at room temperature. Luciferase-fusion proteins bound to the HaloTag-fusion proteins were recovered by washing five times with 0.1% Tween-20/PBS and used in the luciferase assay. As an input control, an aliquot of the whole cell lysate was also used in the assay. The luciferase assay was performed using a Dual-Glo™ Luciferase Assay System (Promega) and a GloMAX™ 96 Microplate Luminometer (Promega) according to the manufacturer’s instructions.
Western Blotting
Western blotting was performed as described previously [20Koga H, Shimada K, Hara Y, et al. A comprehensive approach for establishment of the platform to analyze functions of KIAA proteins: Generation and evaluation of anti-mKIAA antibodies Proteomics 2004; 4: 1412-6.]. Rabbit anti-PC-LKC (PCDH24) (209-318) polyclonal (Abnova, Taipei, Taiwan), anti-alpha-tubulin (DM1A; Calbiochem, Darmstadt, Germany), anti-β-catenin monoclonal, anti-galectin-1 monoclonal (M01) (Abnova), anti-galectin-3 (H-160) polyclonal (sc-20157), anti-Akt (40D4) (Cell Signaling Technology, Danvers, MA) and anti-phospho Akt (Ser473) (D9E) (Cell Signaling Technology) were used for the primary antibodies. After incubation with the primary antibody, membranes were incubated with the appropriate horseradish peroxidase-conjugated secondary antibodies and visualized using a chemiluminescence (ECL plus) detection system (GE Healthcare, Buckinghamshire, UK) with a Luminescent Image Analyzer LAS-3000 or LAS-4000 (Fujifilm, Tokyo, Japan). Whole cell lysate was extracted using CelLyticTM M unless otherwise indicated. For enrichment of integral membrane proteins from cultured mammalian cells, a Mem-PER® Eukaryotic Membrane Protein Extraction Reagent Kit (Thermo Fisher Scientific) was used.
PI3K Activity Assay
The PI3K activity assay was performed using a PI3K Assay Kit (BPS Bioscience, San Diego, CA) according to the manufacturer’s instruction. Whole cell lysate extracted using CelLyticTM M was used. Fluorescent signals were measured at 540 nm excitation and 580 nm emission using a SpectraMax M2 (Molecular Devices, LLC, Sunnyvale, CA). CelLyticTM M buffer was used as a mock sample.
siRNA Experiment
HCT116 cells were seeded at 24 h before small interfering RNA (siRNA) transfection. Synthetic galectin-1 or -3 siRNA (Japan Bio Services Co., LTD., Saitama, JAPAN) was transfected using the Lipofectamine™ 2000 transfection reagent (Life Technologies) according to the manufacturer’s instructions. Anti-β-catenin, anti-galectin-1 and anti-galectin-3 antibodies were used for fluorescent immunostaining analysis at 48 h post-transfection. The suppression levels of the galectins by the respective siRNAs were analyzed using Western blot analysis with anti-galectin-1 and anti-galectin-3 antibodies. The amount of β-catenin localized to the cell membrane was also analyzed with anti-β-catenin monoclonal antibody.
RESULTS
Characterization of HCT116 Cells Expressing PCDH24-HaloTag
We previously showed that the constitutive expression of C-terminal EGFP-fused PCDH24 decreases the saturation density of monolayer cell cultures and suppresses the piling up of HCT116 colon cancer cells [1Okazaki N, Takahashi N, Kojima S, Masuho Y, Koga H. Protocadherin LKC, a new candidate for a tumor suppressor of colon and liver cancers, its association with contact inhibition of cell proliferation Carcinogenesis 2002; 23: 1139-48.]. To identify the molecules related to this alteration of cell behavior, we established an HCT116 colon cancer cell line with DOX-inducible C-terminal HaloTag-fused PCDH24 (PCDH24-HaloTag). This stable transformant allowed us not only to monitor the translocation of target proteins in the absence or presence of PCDH24 in a cell line but also to efficiently pull-down PCDH24-interacting proteins using HaloTag® technology. First, we obtained a stable transformant of HCT116 that expresses the TetR protein, a transcriptional repressor for the tetracycline repressor/operator system. Then, we obtained TetR-expressing HCT116 stable transformants containing CMV/TetO2-driven PCDH24-HaloTag-expression plasmids (Fig. 1a ). In fact, high levels of PCDH24-HaloTag expression were observed when the cells were exposed to DOX at a final concentration of 1 µg/mL for 48 h. The result of Western blot analysis showed that HCT116 and PCDH24-HaloTag cells cultured without DOX did not express PCDH24, but only PCDH24-HaloTag cells maintained with DOX expressed PCDH24 (Fig. 1b
). In fact, high levels of PCDH24-HaloTag expression were observed when the cells were exposed to DOX at a final concentration of 1 µg/mL for 48 h. The result of Western blot analysis showed that HCT116 and PCDH24-HaloTag cells cultured without DOX did not express PCDH24, but only PCDH24-HaloTag cells maintained with DOX expressed PCDH24 (Fig. 1b ). PCDH24-HaloTag was specifically visualized in the cell lysate on an SDS-PAGE gel by HaloTag® TMR ligand-labeling using a fluoroimager at high resolution as well as by HaloTag® R110Direct ligand-labeling in living cells with a fluorescent microscope (Fig. 1b
). PCDH24-HaloTag was specifically visualized in the cell lysate on an SDS-PAGE gel by HaloTag® TMR ligand-labeling using a fluoroimager at high resolution as well as by HaloTag® R110Direct ligand-labeling in living cells with a fluorescent microscope (Fig. 1b and 1c
and 1c , respectively). The saturation density of HCT116 cells expressing PCDH24-HaloTag fusion proteins was reduced compared with that of the HCT116 cells without DOX in their culture medium, even though no significant differences in the growth rates were observed during the exponential phase (Fig. 1d
, respectively). The saturation density of HCT116 cells expressing PCDH24-HaloTag fusion proteins was reduced compared with that of the HCT116 cells without DOX in their culture medium, even though no significant differences in the growth rates were observed during the exponential phase (Fig. 1d ). To elucidate whether the change in cellar phenotype leads to altered cell motility as observed in HCT116 cells expressing PCDH24-EGFP, we performed a migration assay following wound injury. As expected, we observed that PCDH24-HaloTag-expressing cells also migrated at a faster rate and closed the wound in a shorter time than the DOX-untreated cells (Fig. 1e
). To elucidate whether the change in cellar phenotype leads to altered cell motility as observed in HCT116 cells expressing PCDH24-EGFP, we performed a migration assay following wound injury. As expected, we observed that PCDH24-HaloTag-expressing cells also migrated at a faster rate and closed the wound in a shorter time than the DOX-untreated cells (Fig. 1e ). These data indicated that the behavior of PCDH24-HaloTag-expressing HCT116 cells is similar to that of HCT116 cells expressing EGFP-fused PCDH24.
). These data indicated that the behavior of PCDH24-HaloTag-expressing HCT116 cells is similar to that of HCT116 cells expressing EGFP-fused PCDH24.
Identification of Proteins Interacting with PCDH24
To identify endogenous molecules that interact with PCDH24, we performed a pull-down assay using HaloTag® technology. PCDH24-HaloTag-expressing HCT116 cells (DOX+), PCDH24-HaloTag-non-expressing HCT116 cells (DOX-), and HCT116 cells transiently expressing HaloTag as a control were used for the assay and the protein coprecipitates were separated by SDS-PAGE. Some discrete bands were commonly observed in all lanes, suggesting these bands were probably the result of non-specific interaction. Several specific bands were observed in the DOX+ sample lane on the gel images and subjected to LC-MS/MS analysis (Fig. 2a ). As shown in Supplementary Table 1, some proteins were specifically enriched in the PCDH24 precipitate. Two major bands at ~220 kDa were derived from PCDH24, and the smaller band might be a degradation product of PCDH24. As major precipitates, we detected galectin-1 and -3 (Fig. 2a
). As shown in Supplementary Table 1, some proteins were specifically enriched in the PCDH24 precipitate. Two major bands at ~220 kDa were derived from PCDH24, and the smaller band might be a degradation product of PCDH24. As major precipitates, we detected galectin-1 and -3 (Fig. 2a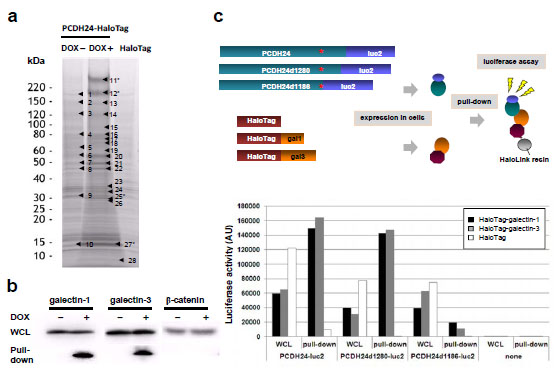 , No. 27 and No. 25, respectively), which are well known as β-galactose-binding lectin proteins.
, No. 27 and No. 25, respectively), which are well known as β-galactose-binding lectin proteins.
Western blot analysis revealed that there was no significant difference in the protein levels of galectin-1 or -3 between the parental cells and PCDH24-expressing cells (Fig. 2b ). On the other hand, HaloTag® pull-down assay followed by Western blot analysis using anti-galectin antibodies showed that galectin-1 and -3 were specifically enriched in the DOX+ lane, suggesting a physiological interaction between PCDH24 and galectin-1 and -3 (Fig. 2b
). On the other hand, HaloTag® pull-down assay followed by Western blot analysis using anti-galectin antibodies showed that galectin-1 and -3 were specifically enriched in the DOX+ lane, suggesting a physiological interaction between PCDH24 and galectin-1 and -3 (Fig. 2b ). We previously demonstrated that PCDH24 regulates the localization of β-catenin in the plasma membrane and we could not rule out the possibility that PCDH24 directly interacts with β-catenin; however, we did not detect an interaction between PCDH24 and β-catenin using the HaloTag® pull-down system (Fig. 2b
). We previously demonstrated that PCDH24 regulates the localization of β-catenin in the plasma membrane and we could not rule out the possibility that PCDH24 directly interacts with β-catenin; however, we did not detect an interaction between PCDH24 and β-catenin using the HaloTag® pull-down system (Fig. 2b ). To delineate the interaction domains of PCDH24 with galectins, luciferase-fused PCDH24 and two intracellular domain deletion mutants were constructed. After co-transfection of HaloTag-fused galectins and luciferase-fused PCDH24 derivatives into HCT116 cells, luciferase-based pull-down assays were performed (Fig. 2c
). To delineate the interaction domains of PCDH24 with galectins, luciferase-fused PCDH24 and two intracellular domain deletion mutants were constructed. After co-transfection of HaloTag-fused galectins and luciferase-fused PCDH24 derivatives into HCT116 cells, luciferase-based pull-down assays were performed (Fig. 2c ). Although luciferase activity was observed in the whole cell lysates of PCDH24-luc2 derivatives, activity for the co-purified proteins was only detected for PCDH24-luc2 and PCDH24d1280-luc2 (amino acids Δ1281–1310 aa), while interactions between PCDH24 and galectines were not observed when most of the intracellular domain of PCDH24 was deleted (PCDH24d1280-luc2, amino acids Δ1187–1310 aa), indicating that a part of PCDH24 (i.e., amino acids 1186–1280) is essential for the interactions.
). Although luciferase activity was observed in the whole cell lysates of PCDH24-luc2 derivatives, activity for the co-purified proteins was only detected for PCDH24-luc2 and PCDH24d1280-luc2 (amino acids Δ1281–1310 aa), while interactions between PCDH24 and galectines were not observed when most of the intracellular domain of PCDH24 was deleted (PCDH24d1280-luc2, amino acids Δ1187–1310 aa), indicating that a part of PCDH24 (i.e., amino acids 1186–1280) is essential for the interactions.
PCDH24 Co-localizes with Galectin-1 and -3 in the Cell Membrane
From the results of the pull-down assay followed by Western blot analysis or by luciferase assay, we hypothesized that galectin-1 and -3 are anchored to the intracellular domain of PCDH24, resulting in the modulation of their activity. To test this hypothesis, we determined the subcellular localization of galectin-1 and -3 in a fluorescent immunostaining assay. We observed that both galectin-1 and -3 are localized to the cytoplasm and nucleus in PCDH24-HaloTag (DOX-) cells (Fig. 3a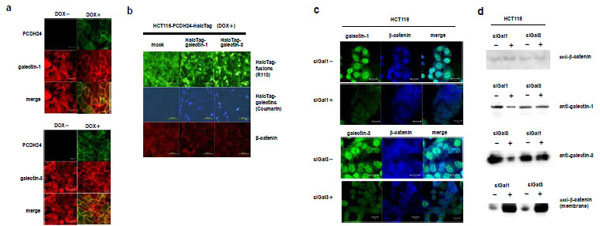 ). In contrast, both are localized to the cell membrane in PCDH24-HaloTag (DOX+) cells. These data support the hypothesis that galectin-1 and -3 interact with the PCDH24 protein at the cell membrane.
). In contrast, both are localized to the cell membrane in PCDH24-HaloTag (DOX+) cells. These data support the hypothesis that galectin-1 and -3 interact with the PCDH24 protein at the cell membrane.
Effects of Galectin-1 and -3 on the Localization of β-catenin
A previous study indicated that the expression of PCDH24 induces the localization of β-catenin at the cell membrane. We examined whether or not the localization of galectin-1 and galectin-3 is associated with the localization of β-catenin by fluorescent immunostaining. If the galectins physiologically interact with β-catenin, the over-expression of galectins would affect the localization of β-catenin, resulting in the disruption of the cell-membrane localization of β-catenin. We transfected N-terminal HaloTag-fused galectin-1 and -3 expression clones into HCT116 cells expressing PCDH24-HaloTag. The over-expression of the galectins disrupted the membrane-localization of β-catenin even though PCDH24 was ectopically expressed (Fig. 3b ). To clarify whether membrane-localization of the galectins with PCDH24 directly localizes β-catenin to the cell membrane, the effects of galectin-1 and -3 on the localization of β-catenin were analyzed in the presence of siRNA for galectin-1 and -3. Fluorescent immunostaining and Western blot analyses revealed that the amount of endogenous galectin-1 and -3 proteins in cells was specifically reduced in the presence of their respective siRNA (Fig. 3c
). To clarify whether membrane-localization of the galectins with PCDH24 directly localizes β-catenin to the cell membrane, the effects of galectin-1 and -3 on the localization of β-catenin were analyzed in the presence of siRNA for galectin-1 and -3. Fluorescent immunostaining and Western blot analyses revealed that the amount of endogenous galectin-1 and -3 proteins in cells was specifically reduced in the presence of their respective siRNA (Fig. 3c and 3d
and 3d , respectively). Interestingly, β-catenin accumulated at cell-to-cell adhesion sites in association with the reduction of the galectins. An increase in the amount of β-catenin at the cell membrane was shown by Western blot analysis of the membrane fraction of the cells in the presence and absence of siRNA for galectins. These results indicate that the amount of galectin-1 and -3 in the cytoplasm affects the nuclear localization of β-catenin and the presence of both galectins is necessary for the enhancement of β-catenin localization to the cell membrane. Moreover, β-catenin seems not to directly bind to the galectins at the cell membrane, because the amount of β-catenin at the cell membrane increased despite the reduction of the galectins.
, respectively). Interestingly, β-catenin accumulated at cell-to-cell adhesion sites in association with the reduction of the galectins. An increase in the amount of β-catenin at the cell membrane was shown by Western blot analysis of the membrane fraction of the cells in the presence and absence of siRNA for galectins. These results indicate that the amount of galectin-1 and -3 in the cytoplasm affects the nuclear localization of β-catenin and the presence of both galectins is necessary for the enhancement of β-catenin localization to the cell membrane. Moreover, β-catenin seems not to directly bind to the galectins at the cell membrane, because the amount of β-catenin at the cell membrane increased despite the reduction of the galectins.
Downstream Signaling of Galectin-1 and -3
One of the critical questions that arose is how galectin-1 and -3 affect the subcellular localization of β-catenin. Several reports have demonstrated that they regulate PI3K activity. We thought that inhibition of PI3K activity could lead to the localization of β-catenin at the cell membrane, because a recent study demonstrated that increasing PI3K activity disrupts the cell membrane localization of β-catenin [21Li J, Mizukami Y, Zhang X, Jo WS, Chung DC. Oncogenic K-ras stimulates Wnt signaling in colon cancer through inhibition of GSK-3 Gastroenterology 2005; 128: 1907-8.]. We hypothesized that PI3K activity is inhibited by the translocation of galectin-1 and -3 to the cell membrane. To verify this hypothesis, we analyzed PI3K activity by using the PI3K Kinase Assay Kit with fluorescently labeled phosphatidylinositol as a substrate. We observed that PI3K activity was decreased in PCDH24-HaloTag (DOX+) cells, but not in PCDH24-HaloTag (DOX-) cells (Fig. 4a ). These data indicate that PCDH24 expression suppresses PI3K activity. Then we performed the PI3K activity assay in the presence of an excess amount of ectopically expressed galectin-1 or -3. PI3K activity was not suppressed regardless of PCDH24 expression when either galectin was over-expressed, indicating that the over-expression of galectin-1 or -3 disrupts the suppression of PI3K kinase activity by PCDH24. To confirm this, we also monitored AKT/PKB activity, which is induced by phosphorylation, downstream of PI3K signaling modified by the PCDH24 and/or the galectins by Western blot analysis using the antibodies against Akt and phosphorylated Akt. In the same way as PI3K activity, we observed a certain level of phosphorylated Akt in PCDH24-HaloTag (DOX-) cells, but not in PCDH24-HaloTag (DOX+) cells and over-expression of galectin-1 or -3 overcomes the decrease of phosphorylated AKT/PKB by PCDH24. Conversely, this phenomenon was not observed in HCT116 cells which expresses no PCDH24. As a control, we showed that the phosphorylation of AKT/PKB was prevented by the PI3K-specific inhibitor LY294002 in HCT116 cells (Fig. 4b
). These data indicate that PCDH24 expression suppresses PI3K activity. Then we performed the PI3K activity assay in the presence of an excess amount of ectopically expressed galectin-1 or -3. PI3K activity was not suppressed regardless of PCDH24 expression when either galectin was over-expressed, indicating that the over-expression of galectin-1 or -3 disrupts the suppression of PI3K kinase activity by PCDH24. To confirm this, we also monitored AKT/PKB activity, which is induced by phosphorylation, downstream of PI3K signaling modified by the PCDH24 and/or the galectins by Western blot analysis using the antibodies against Akt and phosphorylated Akt. In the same way as PI3K activity, we observed a certain level of phosphorylated Akt in PCDH24-HaloTag (DOX-) cells, but not in PCDH24-HaloTag (DOX+) cells and over-expression of galectin-1 or -3 overcomes the decrease of phosphorylated AKT/PKB by PCDH24. Conversely, this phenomenon was not observed in HCT116 cells which expresses no PCDH24. As a control, we showed that the phosphorylation of AKT/PKB was prevented by the PI3K-specific inhibitor LY294002 in HCT116 cells (Fig. 4b ). Recent study indicated that β-catenin localization was regulated through PI3K/AKT pathway. We also demonstrated that β-catenin was localized to the cell membrane in the presence of LY294002 in a fluorescent staining assay (Fig. 4c
). Recent study indicated that β-catenin localization was regulated through PI3K/AKT pathway. We also demonstrated that β-catenin was localized to the cell membrane in the presence of LY294002 in a fluorescent staining assay (Fig. 4c ), indicating that the PI3K activity affects localization of β-catenin.
), indicating that the PI3K activity affects localization of β-catenin.
DISCUSSION
Contact inhibition suppresses cell proliferation via cell-to-cell contact and is an essential phenomenon to maintain normal tissue, and the disruption of this system induces tumorigenesis. The purpose of this study was to identify molecules that interact with PCDH24 and to examine their role in PCDH24 signaling, because the over-expression of PCDH24 leads to the suppression of tumor growth and the inhibition of cell proliferation in HCT116 colon cancer cells. We previously found that PCDH24 modulates β-catenin signaling by the translocation of β-catenin to the cell membrane, but a direct interaction between PCDH24 and β-catenin has not been proved. Moreover, we could not identify molecular candidates that act as intermediates between PCDH24 and β-catenin with a conventional pull-down assay using a specific antibody [2Ose R, Yanagawa T, Ikeda S, Ohara O, Koga H. PCDH24-induced contact inhibition involves downregulation of β-catenin signaling Mol Oncol 2009; 3: 54-66.]. E-cadherin, which forms an adhesion complex with β-catenin [22Aberle H, Butz S, Stappert J, Weissig H, Kemler R, Hoschuetzky H. Assembly of the cadherin-catenin complex in vitro with recombinant proteins J Cell Sci 1994; 107: 3655-63.-24Stockinger A, Eger A, Wolf J, Beug H, Foisner R. E-cadherin regulates cell growth by modulating proliferation-dependent beta-catenin transcriptional activity J Cell Biol 2001; 154: 1185-96.], does not appear to interact with β-catenin, because the expression of E-cadherin is suppressed in PCDH24-expressing HCT116 cells [2Ose R, Yanagawa T, Ikeda S, Ohara O, Koga H. PCDH24-induced contact inhibition involves downregulation of β-catenin signaling Mol Oncol 2009; 3: 54-66.]. In this study, we applied a novel pull-down assay using HaloTag® technology [25Los GV, Encell LP, McDougall MG, et al. HaloTag: A novel protein labeling technology for cell imaging and protein analysis ACS Chem Biol 2008; 3: 373-82.-27Nagase T, Yamakawa H, Tadokoro S, et al. Exploration of human ORFeome: High-throughput preparation of ORF clones and efficient characterization of their protein products DNA Res 2008; 15: 137-49.] that can efficiently recover HaloTag-fusion proteins from cell lysates [28Ohana RF, Hurst R, Vidugiriene J, Slater MR, Wood KV, Urh M. HaloTag-based purification of functional human kinases from mammalian cells Protein Expr Purif 2010; 76: 154-64., 29Itokawa Y, Yanagawa T, Yamakawa H, Watanabe N, Koga H, Nagase T. KAP1-independent transcriptional repression of SCAN-KRAB-containing zinc finger proteins Biochem Biophys Res Commun 2009; 388: 689-94.], and identified galectin-1 and -3 as PCDH24-interacting molecules. Although Shimura and colleagues revealed that galectin-3 binds to the β-catenin/TCF complex and activates Wnt signaling in human breast cancer cells [15Shimura T, Takenaka Y, Tsutsumi S, Hogan V, Kikuchi A, Raz A. Galectin-3, a novel binding partner of β-catenin Cancer Res 2004; 64: 6363-7., 16Shimura T, Takenaka Y, Fukumori T, et al. Implication of galectin-3 in Wnt signaling Cancer Res 2005; 65: 3535-7.], we could find no involvement of β-catenin in complex formation of PCDH24 and galectins in our pull-down assay. Moreover, we found that the over-expression of the galectins disturbs the membrane localization of β-catenin, while reduced levels of galectins augment the membrane localization of β-catenin in spite of the nuclear localization of most of the galectins. From these data, we speculate that PCDH24-anchored galectin-1 and -3 indirectly localize β-catenin to the cell membrane at least in colon cancer cells.
Wnt signaling plays a key role in colon carcinogenesis, and Song et al. suggested that galectin-3 enhances Wnt signaling by regulating GSK-3β phosphorylation through the PI3K/AKT pathway and the nuclear accumulation of β-catenin [17Song S, Mazurek N, Liu C, et al. Galection-3 mediates nuclear β-catenin accumulation and Wnt signaling in human colon cancer cells by regulation of glycogen synthase kinase-3? activity Cancer Res 2009; 69: 1343-9.]. Fang et al. demonstrated that AKT phosphorylatesβ-catenin at Ser552, which leads to its transcriptional activity [30Fang D, Hawke D, Zheng Y, et al. Phosphorylation of β-catenin by AKT promotes β-catenin transcriptional activity J Biol Chem 2007; 282: 11221-9.]. Oka et al. found that the over-expression of galectin-3 activates AKT in human bladder cancer cells [31Oka N, Nakahara S, Takenaka Y, et al. Galectin-3 inhibits tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by activating Akt in human bladder carcinoma cells Cancer Res 2005; 65: 7546-53.], and Elda-Sfadia et al. also found that galectin-3 binds to K-Ras and induces the activation of RAF1 and PI3K [18Elad-Sfadia G, Haklai R, Balan E, Kloog Y. Galectin-3 augments K-ras activation and triggers a ras signal that attenuates ERK but not phosphoinositide 3-kinase activity J Biol Chem 2004; 279: 34922-0.]. Taking these results into consideration, we focused on the PI3K pathway in this study, although galectin-1 and -3 have been implicated in a variety of biological pathways. However, we could not rule out the possibility that PCDH24-anchored galectin-1 and/or -3 are implicated in another pathway for β-catenin translocation. We also found that PI3K activity was decreased in PCDH24-HaloTag (DOX+) cells, in which galectin-1 and -3 were anchored at the cell membrane by PCDH24, and that the over-expression of the galectins restored PI3K activity in spite of the presence of PCDH24, indicating that the galectins activate PI3K and their activity is suppressed by the galectin-anchoring activity of PCDH24. As a result, we propose the following model for the tumor suppression of HCT116 cells by PCDH24. PCDH24 anchors galectin-1 and -3 at the cell membrane, and the depletion of galectin-1 and -3 from the cytosol prevents the activation of PI3K kinase. The inactivated PI3K could not activate AKT, and this failure led to the activation of GSK-3βand the reduction of β-catenin accumulation in the nucleus (Fig. 4d ). In fact, β-catenin was translocated from the nucleus to the cell membrane following treatment with the PI3K inhibitor, LY294002, in HCT116 cells.
). In fact, β-catenin was translocated from the nucleus to the cell membrane following treatment with the PI3K inhibitor, LY294002, in HCT116 cells.
PCDH24 is known to be specifically expressed in the liver, kidney and colon; conversely, PCDH24 expression is markedly reduced in cancers derived from these tissues, indicating that PCDH24 plays important roles in these tissues by regulating cell growth and contact inhibition to maintain the normal cell conditions [1Okazaki N, Takahashi N, Kojima S, Masuho Y, Koga H. Protocadherin LKC, a new candidate for a tumor suppressor of colon and liver cancers, its association with contact inhibition of cell proliferation Carcinogenesis 2002; 23: 1139-48., 2Ose R, Yanagawa T, Ikeda S, Ohara O, Koga H. PCDH24-induced contact inhibition involves downregulation of β-catenin signaling Mol Oncol 2009; 3: 54-66.]. In this report, we revealed that one of the mechanisms by which PCDH24 regulates the localization of β-catenin to the cell membrane is via the inhibition of galectin-1 and -3 activity in HCT116 human colon cancer cells. These results provide a new insight into the tumor suppressor functions of PCDH24 in cancers arising from epithelial cells.
FUNDING
This project was supported by a special grant for the acceleration of the practical applications of biotechnology from Chiba Prefectural Government.
CONFLICTS OF INTEREST
The authors confirm that this article content has no conflicts of interest.
ACKNOWLEDGEMENTS
We would like to thank K. Sato, T. Watanabe, K. Ozawa, and T. Kimura for technical assistance. We also thank N. Suzuki for the LC MS/MS analysis. We are grateful to M. Nakayama, H. Koga and N. Okazaki for their valuable comments.



