- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

Current Chemical Genomics and Translational Medicine
(Discontinued)
ISSN: 2213-9885 ― Volume 12, 2018
An Automated Approach to Efficiently Reformat a Large Collection of Compounds
Jimmy Cui#, Sergio C Chai#, Anang A Shelat, R. Kiplin Guy, Taosheng Chen*
Abstract
Large-scale screening of small organic compounds has become a standard and essential practice in the early discovery of chemical entities with potential therapeutic use. To effectively support high-throughput screening campaigns, compound collections have to be in suitable formats, which requires a process known as compound reformatting. Here we report our approach to reformat the newly-established chemical repository of a large-scale screening facility at St. Jude Children’s Research Hospital, which comprises more than half a million compounds, mostly from commercial sources. We highlight the timeline for a reformatting process, the importance of standardizing the operational procedures, and the advantages and disadvantages of using automation. The end result of our reformatting process is the concurrent generation of copies for long-term storage, screening, and “cherry-picking”; all of which facilitate compound management and high-throughput screening.
Article Information
Identifiers and Pagination:
Year: 2011Volume: 5
First Page: 42
Last Page: 47
Publisher Id: CCGTM-5-42
DOI: 10.2174/1875397301105010042
Article History:
Received Date: 28/4/2011Revision Received Date: 28/5/2011
Acceptance Date: 8/6/2011
Electronic publication date: 22/7/2011
Collection year: 2011
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at 262 Danny Thomas Place, Memphis, TN 38105 3678, USA; Tel.: +1-901-595-5937; Fax: +1-901-595- 5715; E-mail: taosheng.chen@stjude.org# Both Authors contributed equally to the completion of this manuscript.
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 28-4-2011 |
Original Manuscript | An Automated Approach to Efficiently Reformat a Large Collection of Compounds | |
INTRODUCTION
High-throughput screening has become a cornerstone in the drug discovery process, whereby active compounds are rapidly identified and used as starting points for drug development. Although robust and reliable assays are crucial for the success of a high-throughput screening campaign, the most valuable asset in any screening center is the collection of compounds. The library not only represents a large capital investment but also may consist of scarce chemicals that have been synthesized in-house, purified from natural sources, or obtained from collaborators. To efficiently support screening projects, the diverse sets of compounds should be readily available in suitable formats. Because of the advancement in technology, facilities are exploiting the benefits of miniaturization, which include reducing the use of reagents, compounds, and other consumables [1Wölcke J, Ullmann D. Miniaturized HTS technologies - uHTS Drug Discov Today 2001; 6: 637-46.].
Chemical collections are normally supplied in a 96-well format, which holds relatively large volumes. Therefore, it is necessary to compress the samples to higher density formats (eg, 384-well plates) by transferring aliquots into suitable vessels in a process known as compound reformatting [2Yasgar A, Shinn P, Jadhav A, et al. Compound management for quantitative high-throughput screening JALA 2008; 13: 79-89.]. Reformatting a library of several hundred thousand compounds can become a daunting task. Automation can expedite this process, ensure consistency, and offer reliability. Herein, we report our approach to reformat more than half a million compounds acquired from commercial sources. Our strategy enables an efficient and robust workflow that simultaneously incorporates the replating, compression, and replication processes.
MATERIALS AND METHODOLOGY
Instrumentation and Consumables
The Automated Capper and Decapper (ACD), Central Seal Press (CSP), Manual Cap Press, Automated Plate Piercer, accumulator, Mid Size Store (MSS), tube-based STBR96, and STBR384 racks were acquired from REMP AG (Switzerland, now part of NEXUS Biosystems). The Vprep Conventional Pipetting station, PlateLoc sealer, and 96LT 200 µL pipette tips were obtained from Velocity 11 (now part of Agilent Technologies, Santa Clara, CA). Polypropylene plates in a 384-well format (PP384) were purchased from Corning Incorporated (Corning, NY). LPR240 carousel-style plate hotels and STR240 automated incubators were purchased from Liconic Instruments (Woburn, MA). The laser bar code scanner was purchased from 3M (St. Paul, MN). TX40 robotic arms were built by Stäubli (Duncan, SC). The automation system and its software (Cellario version 2.4) were developed by HighRes Biosolutions (Woburn, MA).
Reformatting Process
The compound reformatting process involves several operations that can be divided as follows: stage 1 (replating) and stage 2 (compression and replication) (Fig. 1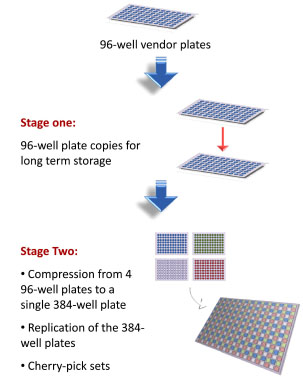 ). The first stage entails generating replicates from each vendor plate and transferring them to a long-term storage format with an identical layout. The compound solutions can then be stored in individually capped tubes arranged in bar-coded racks in a 96-well format (STBR96 tube racks). To achieve this, an ACD capped/decapped the tubes of the STBR96 racks and then aliquots (200 µL) were transferred from the vendor plates by using a Vprep liquid handling station that uses air-displacement technology and a 96LT pipettor head.
). The first stage entails generating replicates from each vendor plate and transferring them to a long-term storage format with an identical layout. The compound solutions can then be stored in individually capped tubes arranged in bar-coded racks in a 96-well format (STBR96 tube racks). To achieve this, an ACD capped/decapped the tubes of the STBR96 racks and then aliquots (200 µL) were transferred from the vendor plates by using a Vprep liquid handling station that uses air-displacement technology and a 96LT pipettor head.
To accelerate the workflow and minimize cost, the same pipette tips used in stage 1 were re-used in stage 2 to dispense their corresponding compounds (10 µL) into PP384 plates (for primary screening) and STBR384 tube racks (for “cherry-picking”). In the STBR384 tube racks, individual tubes are assembled in racks having a 384-well format. After the transfer, a CSP sealed each individual tube in the STBR384 racks with pierceable aluminum sheets. This compression stage condensed 4 vendor plates into a single 384-well plate with a checkered pattern (Fig. 1 ).
).
The automation system was assembled specifically for the compound reformatting process. Fig. (2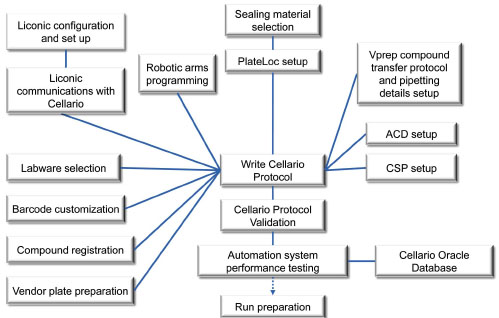 ) shows the various instrument setups, particularly how we established a communication network between the devices and the scheduler’s (ie, Cellario’s) control environment. Each individual device/instrument was tested and optimized during the validation of the automation protocol; trial-and-error approaches were often applied. The trial-and-error process involves the manual adjustment and calibration of the robots, the various automation components and instruments to ensure the proper labware movement on the deck. It is important to register each compound with a unique identifier and to plan for its tracking by customizing its bar code [3Quintero C, Kariv I. Design and implementation of an automated compound management system in support of lead optimization J Biomol Screen 2009; 14: 499-508.]. To enable this, an alphanumeric check character was included to ensure the uniqueness of the bar code so that no repeated bar code was used in the sequence.
) shows the various instrument setups, particularly how we established a communication network between the devices and the scheduler’s (ie, Cellario’s) control environment. Each individual device/instrument was tested and optimized during the validation of the automation protocol; trial-and-error approaches were often applied. The trial-and-error process involves the manual adjustment and calibration of the robots, the various automation components and instruments to ensure the proper labware movement on the deck. It is important to register each compound with a unique identifier and to plan for its tracking by customizing its bar code [3Quintero C, Kariv I. Design and implementation of an automated compound management system in support of lead optimization J Biomol Screen 2009; 14: 499-508.]. To enable this, an alphanumeric check character was included to ensure the uniqueness of the bar code so that no repeated bar code was used in the sequence.
 |
Fig. (2) Initial system setup and automation validation for a compound reformatting process. |
Preparing the system and samples before each automation run required a substantial amount of time (Fig. 3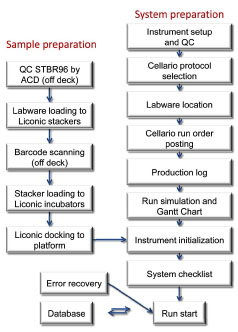 ). The STBR96 racks had to be inspected off-deck (see below), and the samples/labware were scanned offline to be registered into an alternate database, which was then compared with the log file generated during the automation process. At the beginning of the run, the scheduler generated a unique run order number based on selected protocols that were previously created. The process was then simulated by making a Gantt chart describing the steps and estimating the timing of each task.
). The STBR96 racks had to be inspected off-deck (see below), and the samples/labware were scanned offline to be registered into an alternate database, which was then compared with the log file generated during the automation process. At the beginning of the run, the scheduler generated a unique run order number based on selected protocols that were previously created. The process was then simulated by making a Gantt chart describing the steps and estimating the timing of each task.
 |
Fig. (3) System and sample preparation before beginning a routine automation run. |
The workflow for a routine reformatting run is depicted in Fig. (4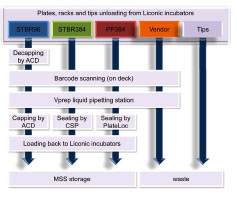 ). The vendor plates, the STBR96 racks, and the tip boxes were placed in 3 separate Liconic incubators. A fourth Liconic incubator was used exclusively to store the STBR384 racks and PP384 plates. Incubators 3 and 4 performed the additional step of loading back the STBR96, STBR384, and PP384 copies. Because Liconic incubators can hold 11 stackers each, different densities of Liconic stackers were used to hold different consumables: 22-level stackers for STBR384 tube racks and PP384 plates, 17-level stackers for STBR96 racks, and 8-level stackers for deep-well plates and pipette tip boxes.
). The vendor plates, the STBR96 racks, and the tip boxes were placed in 3 separate Liconic incubators. A fourth Liconic incubator was used exclusively to store the STBR384 racks and PP384 plates. Incubators 3 and 4 performed the additional step of loading back the STBR96, STBR384, and PP384 copies. Because Liconic incubators can hold 11 stackers each, different densities of Liconic stackers were used to hold different consumables: 22-level stackers for STBR384 tube racks and PP384 plates, 17-level stackers for STBR96 racks, and 8-level stackers for deep-well plates and pipette tip boxes.
 |
Fig. (4) The workflow of a routine compound reformatting run. |
To initiate the reformatting process, the empty STBR96 racks were first moved to the ACD where the tubes were decapped and scanned to detect bar codes before being transferred to the Vprep pipetting station. The other 3 types of labware were similarly scanned and transferred. After the liquid-pipetting cycle, the empty vendor plates and used tips were disposed. Meanwhile, the STBR96 racks were capped by the ACD. The STBR384 racks and the PP384 were sealed by the CSP and the PlateLoc, respectively. Finally, the vessels containing the reformatted compounds were sent back to their respective Liconic incubators. The reformatting process was performed on a HigRes Biosolution automated robotic deck featuring 2 Stäubli robotic arms, each with custom, end-effector, collision-sensing tools and a pair of pneumatic grippers that shuffles the labware components between stations.
The entire automation deck is fully enclosed, establishing a controlled environment. A dehumidifier kept the humidity less than 20%, minimizing water absorption of the DMSO-dissolved compounds [4Cheng X, Hochlowski J, Tang H, et al. Studies on repository compound stability in DMSO under various conditions J Biomol Screen 2003; 8: 292-304.]. A dry nitrogen line also controlled humidity and oxygen levels in the chamber, preventing degradation of the chemicals by oxidation. Throughout the reformatting process, the chamber’s temperature was between 23°C and 28°C [5Kozikowski BA, Burt TM, Tirey DA, et al. The effect of freeze/thaw cycles on the stability of compounds in DMSO J Biomol Screen 2003; 8: 210-5.].
Cellario software was used as a dynamic time scheduler, which assigns operational steps, controls timing during the continuous batched-reformatting process, and uses barcodes to track each labware’s location. Protocols created using Cellario optimize the trafficking on the deck on the basis of particular timings, delay and priority logics, instrument availability, and input event sequences. Cellario includes a customized Oracle database and is accessible through an SQL interface. The automation log data and compound-mapping files of vendor plates and those of newly reformatted plates can be subsequently extracted. Compound tracking throughout the process was accomplished by tracking well numbers, custom bar codes on each plate, and the unique identifiers of each compound in the collection.
The replating and compression stages from a single run of a vendor plate generated 1 REMP96 copy for long-term storage, 2 PP384 copies for use directly in primary screens, and 3 REMP384 copies for cherry-picking and follow-up experiments. All resulting copies were maintained in a fully automated and humidity-controlled −20°C storage facility (REMP MSS). The facility houses a one-aisle robot equipped with rack grippers, a handling platform, and a punching device, allowing plate delivery and retrieval. This robot can also hand in selected tubes from STBR96 and STBR384 racks.
To minimize compound degradation, the reformatted plates are stored at -20 ºC under low humidity in the MSS store, which is designed for long-term storage. In addition, the STBR96 and STBR384 rack systems allow for individual compound cherry-picking, minimizing the chances of compound degradation due to multiple freeze-thaw cycles. Even though we utilize one of the most favorable conditions for long-term storage of compounds, degradation is still possible due to the labile nature of certain compounds. Compounds that are known to be very reactive were filtered out and not included in our collection.
Quality Control and Performance
The automation protocols created using Cellario were first validated by performing dry runs (i.e. runs without compounds or reagents). Because the software has real-time monitoring capabilities, the labware location indicated by Cellario was confirmed by visually tracking the location of the physical items on the deck. The event records generated for each operational step were subsequently analyzed for consistency. Additionally, bar code and time records were extracted from the database to corroborate the location of the compounds within a given plate.
The liquid-handling performance was regularly verified to ensure pipetting accuracy and precision. Fluorescence measurements using fluorescein isothiocyanate (FITC) were used to calculate liquid-dispensing variations among pipette tips and volume deviations among wells. Additionally, the total volume of liquid delivered was monitored by measuring the weight of the transferred solutions [6Rhode H, Schulze M, Renard S, et al. An improved method for checking HTS/uHTS liquid-handling systems J Biomol Screen 2004; 9: 726-33.].
Problems in the ACD’s capping and decapping steps could lead to system failure, resulting in cessation of all operations. To ensure that the caps were smoothly decapped and correctly recapped, each STBR96 and STBR384 plate was visually inspected for damage, and a dedicated stand-alone ACD was used to manually perform the capping/decapping operations before loading the tube racks into the automation system.
Many of the devices on the automation system require an air supply with steady pressure to ensure consistency during operations. We used dry air generated by an in-house air compressor and stored in reserve tanks to ensure that proper air pressure was supplied to each device. All devices were equipped with gages and valves for pressure control. To ensure continuous power during a reformatting process, an uninterruptible power supply battery for the automation system was wired in place to handle unexpected short-term power loss and sudden voltage fluctuations.
RESULTS
In a span of 42 weeks, we were able to reformat approximately 5.2 × 105 compounds (Fig. 5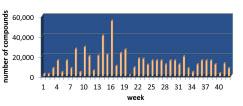 ) at an average rate of 3000 compounds per week. The first few weeks were allocated to performing pilot reformatting before increasing the throughput, reaching a maximal output of 55 040 compounds per week by week 16. For a reformatting run involving 88 vendor plates, the entire reformatting process took 9 h. Cellario’s dynamic scheduling capabilities reduced the process by 6 h compared to a static method, which would have taken 15 h for completion (Fig. 6
) at an average rate of 3000 compounds per week. The first few weeks were allocated to performing pilot reformatting before increasing the throughput, reaching a maximal output of 55 040 compounds per week by week 16. For a reformatting run involving 88 vendor plates, the entire reformatting process took 9 h. Cellario’s dynamic scheduling capabilities reduced the process by 6 h compared to a static method, which would have taken 15 h for completion (Fig. 6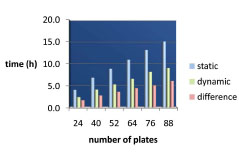 ). In a static mode, the specific operations follow a linear succession, and the system is on hold until a specific task is concluded. In contrast, in a dynamic environment, the workflow progresses based on resource allocation (eg, when labware is being handled by a specific device, the robotic arms shuffle other items to optimize time). The time saved by using a dynamic approach instead of a static one increases as the number of plates increases from 24 to 88 (Fig. 6
). In a static mode, the specific operations follow a linear succession, and the system is on hold until a specific task is concluded. In contrast, in a dynamic environment, the workflow progresses based on resource allocation (eg, when labware is being handled by a specific device, the robotic arms shuffle other items to optimize time). The time saved by using a dynamic approach instead of a static one increases as the number of plates increases from 24 to 88 (Fig. 6 ). It is therefore apparent that keeping the production at maximal capacity would increase efficiency. However, knowing that a system failure could lead to loss of a significant number of compounds, it is advisable to have somebody monitor the entire process to enable prompt troubleshooting and error recovery. Therefore, the throughput is limited to some extent by the availability of such a designated monitor. Out of a total of 8251 vendor plates, 23 plates were lost or discarded due to automation error, resulting in an overall failure rate of 0.28%.
). It is therefore apparent that keeping the production at maximal capacity would increase efficiency. However, knowing that a system failure could lead to loss of a significant number of compounds, it is advisable to have somebody monitor the entire process to enable prompt troubleshooting and error recovery. Therefore, the throughput is limited to some extent by the availability of such a designated monitor. Out of a total of 8251 vendor plates, 23 plates were lost or discarded due to automation error, resulting in an overall failure rate of 0.28%.
 |
Fig. (5) Number of compounds reformatted from vendor plates to STBR96, STBR384, and 384-well screening plates in a given week. |
The movement of the Stäubli robotic arms accounted for 46 % of the time needed to complete the entire protocol (Fig. 7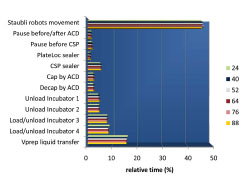 ), and the Vprep liquid transfer accounted for 16 %. Time in incubators 1 and 2 accounted for only 4% of the total time, and time in incubators 3 and 4 accounted for 8%. Notably, capping/decapping of the STBR96 by the ACD, sealing of STBR384 by the CSP, and sealing of the PP384 screening plates by the PlateLoc occupied only a small fraction of the total time. The relative time for each task remained virtually constant when the number of vendor plates was increased from 24 to 88 because there is a proportional increase in every task as the number of plates increases (Fig. 7
), and the Vprep liquid transfer accounted for 16 %. Time in incubators 1 and 2 accounted for only 4% of the total time, and time in incubators 3 and 4 accounted for 8%. Notably, capping/decapping of the STBR96 by the ACD, sealing of STBR384 by the CSP, and sealing of the PP384 screening plates by the PlateLoc occupied only a small fraction of the total time. The relative time for each task remained virtually constant when the number of vendor plates was increased from 24 to 88 because there is a proportional increase in every task as the number of plates increases (Fig. 7 ).
).
One STBR96 copy, 2 PP384 copies, and 3 STBR384 copies were generated from each vendor plate. Although only 1 copy of STBR96 was created, this step took 27 % of the total process time (Fig. 8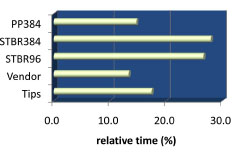 ). Because four 96-well vendor plates were compressed into a single 384-well plate or rack, 96-well labware were shuffled by the robotic arms to the Vprep station 4 times for every 384-well vessel, which remained on designated Vprep shelves until the 4-cycle process was completed. In our 384-well, screening-ready formats, columns 1, 2, 13, and 14 correspond with empty wells of the original vendor plates and are reserved for control or reference samples needed for subsequent biologic assays.
). Because four 96-well vendor plates were compressed into a single 384-well plate or rack, 96-well labware were shuffled by the robotic arms to the Vprep station 4 times for every 384-well vessel, which remained on designated Vprep shelves until the 4-cycle process was completed. In our 384-well, screening-ready formats, columns 1, 2, 13, and 14 correspond with empty wells of the original vendor plates and are reserved for control or reference samples needed for subsequent biologic assays.
 |
Fig. (8) Relative time allocated for copying PP384, STBR384, STBR96, and for the transfer of tips and vendor plates. |
DISCUSSION
Preserving compound integrity is vital for the quality and reliability of screening campaigns and follow-up confirmations [5Kozikowski BA, Burt TM, Tirey DA, et al. The effect of freeze/thaw cycles on the stability of compounds in DMSO J Biomol Screen 2003; 8: 210-5., 7Kozikowski BA, Burt TM, Tirey DA, et al. The effect of room-temperature storage on the stability of compounds in DMSO J Biomol Screen 2003; 8: 205-9.]. Therefore, our approach was to store each sample in individual tubes by using REMP Tube Technology consumables: only those compounds of interest are taken out of the −20°C store facility. In comparison to the use of traditional microtiter plates, the tube system reduces multiple freeze-thaw cycles of irrelevant compounds and decreases the chance of cross-contamination [5Kozikowski BA, Burt TM, Tirey DA, et al. The effect of freeze/thaw cycles on the stability of compounds in DMSO J Biomol Screen 2003; 8: 210-5.]. Therefore, compounds for long-term storage and cherry-picking are stocked in STBR96 and STBR384, respectively. This standard reformatting protocol was also developed to generate compound copies to be used directly for screening in a 384-well format (PP384). The screening copies can be ordered from the MSS and returned when the screening is complete, with freeze-and-thaw cycles automatically recorded by the system.
Condensing compounds from 96-well plates to higher density vessels conserves reagents, compounds, and labware while reducing screening time. Therefore, the effort invested in compressing compounds can be advantageous in the long term, particularly in the case of a major project involving millions of compounds. Concomitantly executing the replating, compression, and replication stages also leads to cost savings. First, the reformatting time is condensed. Second, operating costs can be reduced by reusing the pipette tips when the same compounds are transferred during different stages. Additionally, having all the compounds readily available in a consistent layout facilitates the screening process, data analysis, and interpretation.
In general, our process could be expedited by increasing the speed of the robotic arms. However, this would be at the expense of increasing the chances of dropping plates or racks from the grippers, which would squander precious chemicals. Additionally, the Vprep pipetting station had a limited number of shelves, constraining the amount of labware that it could handle. Therefore, integrating a second pipetting station could reduce the burden of the robotic arms and increase the throughput. However, this approach would introduce the risk of triggering system deadlock or a traffic jam. The 2 robots that were programmed to move 5 different types of labware among 8 locations consumed nearly half of the total process time. The routes between these different devices, as well as robot acceleration and deceleration can be further optimized. In addition, changes in the layout of the stations on the deck can increase process speed. To increase the throughput, we used a dynamic software environment and concurrently executed the replating, compression, and replication processes. System maintenance, compound registration into the database, and labware restocking were some of the factors that prevented a constant throughput.
Most academic institutions perform the reformatting process in a semi-automation and static mode, generating only 384PP plates for long-term and immediate usage. This approach is more economically feasible, but it increases the likelihood for human error, cross-contamination, compound degradation, while increasing process time. Some of the screening centers in the private sector employ strategies that focus on the long-term stability of the compounds by storing them solely in individual tubes as solutions or dry powder, which result in significant costs and the lack of screening copies for immediate use. Our strategy incorporates elements from the above-mentioned methods to generate concomitantly multiple copies for short and long-term use, while minimizing cost and process time. The compound repository of St. Jude Children’s Research Hospital is composed of over half a million compounds, with a growing number from both external and internal sources. Compound reformatting has become a routine practice in our facility. Reformatted compound plates resulting from the procedure described in this report have been used in 9 full-deck (ie, entire library) and 12 partial-deck screening campaigns. This method can be applied in institutions with limited resources if the dynamic process used simulates a priority to optimize the timing of events; therefore, our experience completing this major task is of use to emerging screening facilities, particularly those in academic institutions.
CONFLICT OF INTEREST
None Declared.
ACKNOWLEDGEMENTS
We thank Nick Mills, David Smithson, and Fu-Yue Zeng for their input in the design of the reformatting process; David Bouck, Fu-Yue Zeng, and Wenwei Lin for their assistance in the reformatting process; other members of the Department of Chemical Biology and Therapeutics for valuable discussions; and Dr. Cherise Guess of the Department of Scientific Editing for editing the manuscript. This work was supported by the American Lebanese Syrian Associated Charities (ALSAC), St. Jude Children’s Research Hospital, and National Cancer Institute grant P30CA027165.
ABBREVIATIONS
| ACD | =Automated Capper and Decapper |
| CSP | =Central Seal Press |
| MSS | =Mid Size Store |
| PP384 | =384-well polypropylene plates |





