- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

Current Chemical Genomics and Translational Medicine
(Discontinued)
ISSN: 2213-9885 ― Volume 12, 2018
Promoting Illiteracy in Epigenetics: An Emerging Therapeutic Strategy
Tim J Wigle*
Article Information
Identifiers and Pagination:
Year: 2011Volume: 5
Issue: Suppl 1
First Page: 48
Last Page: 50
Publisher Id: CCGTM-5-48
DOI: 10.2174/1875397301005010048
Article History:
Electronic publication date: 22/8/2011Collection year: 2011
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Epizyme, 840 Memorial Drive, Cambridge, MA 02139, USA; Tel: 617-500-0599; E-mail: twigle@epizyme.com
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on |
Original Manuscript | Promoting Illiteracy in Epigenetics: An Emerging Therapeutic Strategy | |
Beyond the Human Genome
On the 10th anniversary of the announcement of the draft sequence of the 3 billion DNA bases in the genome by the Human Genome Project, there is still a lack of molecular targeted therapies for previously intractable diseases. It has become clear that the genomes within and across species are too similar to explain the diversity of life and the etiology of all diseases, indicating that the underlying DNA sequence is only one component of this problem. Coinciding with the accomplishment of this impressive feat led by the NIH and Celera Genomics, has been the explosion of research defined as “epigenetic”. The term epigenetics was first coined in the 1940s by British embryologist and geneticist Conrad Waddington [1Waddington CH. The epigenotype Endeavour 1942; 18-20.], who was attempting to describe “the interactions of genes with their environment, which brings the phenotype into being”. Since then, this definition has been refined to encompass the study of heritable phenotypic traits that result from modifications to a chromosome that do not alter the underlying genetic code [2Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics Genes Dev 2009; 23: 781-3.]. An increasing awareness of the importance of the temporal and spatial control over the expression of genes has elevated the study of epigenetics to a torrid pace (Fig. 1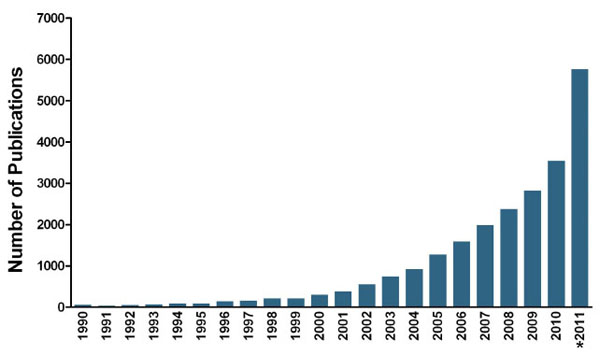 ). There is even a sequel to the Human Genome Project, the International Human Epigenome Project (IHEP), which was launched in 2010. The IHEP’s goal is to understand the patterns of DNA methylation and post-translational histone modifications that ultimately control access of specific genes to transcriptional machinery.
). There is even a sequel to the Human Genome Project, the International Human Epigenome Project (IHEP), which was launched in 2010. The IHEP’s goal is to understand the patterns of DNA methylation and post-translational histone modifications that ultimately control access of specific genes to transcriptional machinery.
Epigenetic Therapies Already in the Clinic
The potential for new classes of molecular targeted epigenetic therapies has begun to be realized with FDA-approved inhibitors of histone deacetylases (HDACs) (Vorinostast and Romidepsin) and DNA methyltransferases (DNMTs) (5-azacytidine and 5-aza-2’-deoxycytidine). However, these agents were discovered based on observation of cell phenotypes and the biochemical targets were discovered several years later. Suberoylanilide hydroxamic acid (SAHA, Vorinostat; approved 2006), the first generation clinical HDAC inhibitor brought to market, was discovered by phenotypic screens to be a potent differentiating agent whose molecular targets, the HDACs, were identified shortly thereafter [3Michaeli J, Lebedev YB, Richon VM, Chen ZX, Marks PA, Rifkind RA. Conversion of differentiation inducer resistance to differentiation inducer sensitivity in erythroleukemia cells Mol Cell Biol 1990; 10: 3535-40.-5Richon VM, Emiliani S, Verdin E, et al. A class of hybrid polar inducers of transformed cell differentiation inhibits histone deacetylases Proc Natl Acad Sci USA 1998; 95: 3003-7.]. Similarly, the azacytidines (5-azacytidine, Vidaza; approved 2004 and 5-aza-2’-deoxycytidine, Dacogen; approved 2006) were noted to have anti-proliferative effects in vitro on cancer cells in 1964, and the molecular targets, DNA methyltransferases, were determined only 15 years later [6Sorm F, Piskala A, Cihak A, Vesely J. 5-Azacytidine, a new, highly effective cancerostatic Experientia 1964; 20: 202-3., 7Friedman S. The effect of 5-azacytidine on E. coli DNA methylase Biochem Biophys Res Commun 1979; 89: 1328-33.]. These discoveries and their subsequent development into therapeutic agents indicate the potential of epigenetic therapies. As a result target-centric, bottom-up approaches to epigenetic drug discovery have rapidly intensified across both industry and academia. Epigenetic targets are now ubiquitous in drug discovery pipelines and there are now multiple biotechnology companies dedicated to pursuing epigenetic therapies, as highlighted in a recent review by Mack [8Mack GS. To selectivity and beyond Nat Biotechnol 2010; 28: 1259-66.].
Histone Methylation: The Next Epigenetic Therapy?
An extensive literature search reveals at least 232 enzymes that are proven or postulated to add or remove histone post-translational modifications (Fig. 2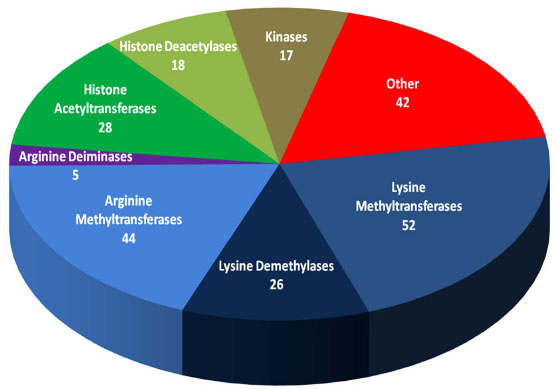 ). Given that histone acetylation and DNA methylation have already been exploited with approved therapeutics, this issue of Current Chemical Genomics focuses on histone lysine methylation, which is emerging as an attractive target for drug discovery. Amongst histone-modifying enzymes, the lysine and arginine methyltransferases (KMTs and RMTs), and lysine demethylases (KDMs) comprise more than half of the total number, yet there are no inhibitors of these enzymes in clinical studies. Furthermore, a growing body of evidence suggests that genetically driven alteration of their enzymatic activities and specificities drives disease progression. For example, point mutations to the lysine methyltransferase EZH2 that change the product specificity of the PRC2 complex from monomethyl- to trimethyl-lysine have been characterized in a subset of lymphoma patients. As a result, heterozygosity leads to the coordinated activities of mutant and wild-type enzymes in the trimethylation of lysine 27 on histone H3, a notorious mark that is ubiquitous in many cancers [9Morin RD, Johnson NA, Severson TM, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin Nat Genet 2010; 42: 181-5., 10Sneeringer CJ, Scott MP, Kuntz KW, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas Proc Natl Acad Sci USA 2010; 107: 20980-5.]. Translocations creating fusion proteins with aberrant activity have also been observed in disease, such as the MLL-partner fusions that result in the mistargeting of H3K79 methylation by DOT1L in childhood leukemias [11Okada Y, Feng Q, Lin Y, et al. hDOT1L links histone methylation to leukemogenesis Cell 2005; 121: 167-78.] or NSD1-nucleoporin 98 fusions that are associated with acute myeloid leukemia [12Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis Nat Cell Biol 2007; 9: 804-12.]. Recently, overexpression of the lysine methyltransferase SETDB1 was found to accelerate the progression of melanoma in an elegant zebrafish model [13Ceol CJ, Houvras Y, Jane-Valbuena J, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset Nature 2011; 471: 513-7.], and similarly, overexpression of the lysine methyltransferases SMYD3 [14Hamamoto R, Silva FP, Tsuge M, et al. Enhanced SMYD3 expression is essential for the growth of breast cancer cells Cancer Sci 2006; 97: 113-8.] and G9a [15Chen MW, Hua KT, Kao HJ, et al. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM Cancer Res 2010; 70: 7830-40.] have been observed in a variety of cancers. In the case of the latter, the recent development of selective, sub-nanomolar inhibitors [16Liu F, Chen X, Allali-Hassani A, et al. Protein lysine methyltransferase G9a inhibitors: design, synthesis, and structure activity relationships of 2,4-diamino-7-aminoalkoxy-quinazolines J Med Chem 2010; 53: 5844-7.] will enable its investigation as a target for cancer therapy. In addition, the lysine demethylases LSD1 and JARID1B are found to be overexpressed in prostate cancer [17Kahl P, Gullotti L, Heukamp LC, et al. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence Cancer Res 2006; 66: 11341-7.], and inhibitors of LSD1 have shown promise in controlling the proliferation of cancer cells using xenograft models [18Huang Y, Stewart TM, Wu Y, et al. Novel oligoamine analogues inhibit lysine-specific demethylase 1 and induce reexpression of epigenetically silenced genes Clin Cancer Res 2009; 15: 7217-28.]. Ultimately, these examples represent the intersection of genetics and epigenetics, and define a targeted patient population that will enhance the probability of clinical success.
). Given that histone acetylation and DNA methylation have already been exploited with approved therapeutics, this issue of Current Chemical Genomics focuses on histone lysine methylation, which is emerging as an attractive target for drug discovery. Amongst histone-modifying enzymes, the lysine and arginine methyltransferases (KMTs and RMTs), and lysine demethylases (KDMs) comprise more than half of the total number, yet there are no inhibitors of these enzymes in clinical studies. Furthermore, a growing body of evidence suggests that genetically driven alteration of their enzymatic activities and specificities drives disease progression. For example, point mutations to the lysine methyltransferase EZH2 that change the product specificity of the PRC2 complex from monomethyl- to trimethyl-lysine have been characterized in a subset of lymphoma patients. As a result, heterozygosity leads to the coordinated activities of mutant and wild-type enzymes in the trimethylation of lysine 27 on histone H3, a notorious mark that is ubiquitous in many cancers [9Morin RD, Johnson NA, Severson TM, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin Nat Genet 2010; 42: 181-5., 10Sneeringer CJ, Scott MP, Kuntz KW, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas Proc Natl Acad Sci USA 2010; 107: 20980-5.]. Translocations creating fusion proteins with aberrant activity have also been observed in disease, such as the MLL-partner fusions that result in the mistargeting of H3K79 methylation by DOT1L in childhood leukemias [11Okada Y, Feng Q, Lin Y, et al. hDOT1L links histone methylation to leukemogenesis Cell 2005; 121: 167-78.] or NSD1-nucleoporin 98 fusions that are associated with acute myeloid leukemia [12Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis Nat Cell Biol 2007; 9: 804-12.]. Recently, overexpression of the lysine methyltransferase SETDB1 was found to accelerate the progression of melanoma in an elegant zebrafish model [13Ceol CJ, Houvras Y, Jane-Valbuena J, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset Nature 2011; 471: 513-7.], and similarly, overexpression of the lysine methyltransferases SMYD3 [14Hamamoto R, Silva FP, Tsuge M, et al. Enhanced SMYD3 expression is essential for the growth of breast cancer cells Cancer Sci 2006; 97: 113-8.] and G9a [15Chen MW, Hua KT, Kao HJ, et al. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM Cancer Res 2010; 70: 7830-40.] have been observed in a variety of cancers. In the case of the latter, the recent development of selective, sub-nanomolar inhibitors [16Liu F, Chen X, Allali-Hassani A, et al. Protein lysine methyltransferase G9a inhibitors: design, synthesis, and structure activity relationships of 2,4-diamino-7-aminoalkoxy-quinazolines J Med Chem 2010; 53: 5844-7.] will enable its investigation as a target for cancer therapy. In addition, the lysine demethylases LSD1 and JARID1B are found to be overexpressed in prostate cancer [17Kahl P, Gullotti L, Heukamp LC, et al. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence Cancer Res 2006; 66: 11341-7.], and inhibitors of LSD1 have shown promise in controlling the proliferation of cancer cells using xenograft models [18Huang Y, Stewart TM, Wu Y, et al. Novel oligoamine analogues inhibit lysine-specific demethylase 1 and induce reexpression of epigenetically silenced genes Clin Cancer Res 2009; 15: 7217-28.]. Ultimately, these examples represent the intersection of genetics and epigenetics, and define a targeted patient population that will enhance the probability of clinical success.
The discovery of potent chemical probes of KMTs and KDMs are a critical first step in the dissection of the biological pathways they regulate and in understanding the consequences of genetically-driven misregulation of their activities. These enzymes appear primed for drug discovery, with a wealth of structural information now available to guide medicinal chemistry efforts, and this is reviewed in this issue by articles from Heightman and Shapira. The readers of lysine methylation, including PHD fingers, MBT domains, Tudor domains, chromodomains, PWWP repeats and WD40 repeats, which may also make interesting drug targets, are reviewed by Herold et al. The assay technologies that will enable chemical exploration of these targets are quickly evolving and are highlighted by Quinn and Simeonov and Zee et al. Finally, pioneering efforts in HTS and medicinal chemistry have indicated that indeed, these enzymes appear chemically tractable, and the current state of chemical matter targeting histone methylation is reviewed by Yost et al. and Heightman in this issue.
While it is likely that modulators of histone lysine methylation will yield clinical candidates in the future, the study of histone lysine methylation and its effect on biological processes is still in its relative infancy. It remains to be seen how many enzymes or proteins will make for good therapeutic targets. The integration of current efforts in genomics, chemical biology and drug discovery efforts should begin to deliver greater understanding of the potential of epigenetic targets. The research undertakings and accomplishments reviewed in this issue will be critical in the validation of small molecule modulators of methyl-lysine writers, readers and erasers as first-in-class molecular targeted therapies. These targeted agents should be an improvement over current treatments, and will have a profound impact on patients with unmet medical needs.
ACKNOWLEDGEMENT
I thank Dr. Margaret Porter Scott, Dr. Mikel Moyer and Dr. Robert Copeland for their support and helpful discussions in the preparation of this editorial.




