- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

Current Chemical Genomics and Translational Medicine
(Discontinued)
ISSN: 2213-9885 ― Volume 12, 2018
Chemical Biology of Lysine Demethylases
Tom D Heightman*, #
Abstract
Abnormal levels of DNA methylation and/or histone modifications are observed in patients with a wide variety of chronic diseases. Methylation of lysines within histone tails is a key modification that contributes to increased gene expression or repression depending on the specific residue and degree of methylation, which is in turn controlled by the interplay of lysine methyl transferases and demethylases. Drugs that target these and other enzymes controlling chromatin modifications can modulate the expression of clusters of genes, potentially offering higher therapeutic efficacy than classical agents acting on downstream biochemical pathways that are susceptible to degeneracy. Lysine demethylases, first discovered in 2004, are the subject of increasing interest as therapeutic targets. This review provides an overview of recent findings implicating lysine demethylases in a range of therapeutic areas including oncology, immunoinflammation, metabolic disorders, neuroscience, virology and regenerative medicine, together with a summary of recent advances in structural biology and small molecule inhibitor discovery, supporting the tractability of the protein family for the development of selective druglike inhibitors.
Article Information
Identifiers and Pagination:
Year: 2011Volume: 5
Issue: Suppl 1
First Page: 62
Last Page: 71
Publisher Id: CCGTM-5-62
DOI: 10.2174/1875397301005010062
Article History:
Received Date: 27/1/2011Revision Received Date: 12/4/2011
Acceptance Date: 25/4/2011
Electronic publication date: 22/8/2011
Collection year: 2011
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at Astex Therapeutics Ltd, 436 Cambridge Science Park, Milton Road, Cambridge CB4 0QA, UK; Tel: +44 1223 226200; Fax: +44 1223 226201; E-mail: T.Heightman@astex-therapeutics.com# Current address: Astex Therapeutics Ltd, 436 Cambridge Science Park, Milton Road, Cambridge CB4 0QA, UK.
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 27-1-2011 |
Original Manuscript | Chemical Biology of Lysine Demethylases | |
INTRODUCTION
Although every cell within an organism contains the same genetic information, the pattern of genes that are activated and silenced in a given cell is highly dependent on its differentiation state and environment. Individual cellular gene expression patterns are mediated by at least four distinct but interrelated molecular processes: DNA methylation, histone post-translational modifications, non-coding RNAs, and nucleosome positioning [1Portela A, Esteller M. Epigenetic modifications and human disease Nat Biotechnol 2010; 28(10 ): 1057-68.]. Together, these four processes form a complex network, sometimes referred to as an epigenetic landscape or code [2Waddington CH. The strategy of the genes, a discussion of some aspects of theoretical biology. London: Allen & Unwin 1957.], that provides a mechanism for “potentially heritable changes in gene expression that do not involve changes in DNA sequence” [3Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals Nat Genet 2003; 33(Suppl ): 245-54.]. A large body of epidemiological and molecular evidence has accumulated to demonstrate that early life experiences strongly impact on epigenetic modifications, leading to a working hypothesis for ‘developmental plasticity’, by which cellular organisms adapt their structure and function in response to environmental cues such as diet, drugs, hormones, toxins, stress and infections [4Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on adult health and disease New Eng J Med 2008; 359(1 ): 61-73.]. Epigenetic responses are most plastic during early life and then become increasingly irreversible, thereby imposing a memory effect that can modulate an individual’s phenotype and their susceptibility to disease.
The epigenetic code is characterized by a high degree of cross-talk between individual modifications (for an example see [5Zippo A, Serafini R, Rocchigiani M, et al. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation Cell 2009; 138(6
): 1122-0.]), and varying degrees of plasticity. While DNA methylation constitutes a binary switching mechanism (methylation at or near gene promoters correlates with gene silencing and vice versa), histone modifications are highly complex in terms of both the number of sites that can be modified, and in the variety of possible modifications [6Kouzarides T. Chromatin modifications and their function Cell 2007; 128(4
): 693-705.]. Histones H2A, H2B, H3 and H4 are comprised of DNA-binding cores, together with largely unstructured tails that project from the nucleosome particle. Both cores and tails are subject to post-translational modifications on the side chains of certain amino acid residues, including acetylation of lysine, methylation of lysine or arginine, phosphorylation of serine or threonine, SUMOylation, ubiquitination and ADP-ribosylation. These modifications occur at different sites simultaneously, leading to a bewildering array of possible combinations regulated by several hundred complementary enzymes and recognition domains that add or remove specific modifications or bind specifically to modified residues to recruit additional transcriptional regulators (Fig. 1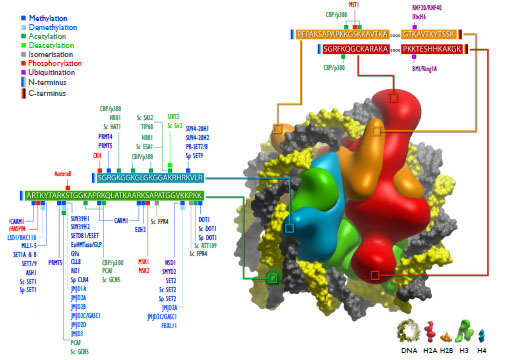 ).
).
Methylation at specific histone tail residues, in particular H3K4, is associated with actively transcribed gene loci, whereas methylation at other histone tail positions (H3K9, H3K27) generally leads to transcriptional silencing and heterochromatinisation. Dynamic regulation of lysine methylation and demethylation is effected by lysine methyltransferases (KMTs) and demethylases (KDMs) respectively, creating patterns of methylation marks that serve as binding sites for methyl binding domains (MBDs), which may form part of other histone-modifying enzymes or recruit protein complexes involved in transcriptional regulation. The known functions of individual methylation events are too complex to be described comprehensively here but have been reviewed in detail recently (see [7Lee BM, Mahadevan LC. Stability of histone modifications across mammalian genomes: Implications for 'epigenetic' marking J Cell Biochem 2009; 108(1 ): 22-34., 8Ng S, Yue W, Oppermann U, Klose R. Dynamic protein methylation in chromatin biology Cell Mol Life Sci 2009; 66(3 ): 407-22.]).
LYSINE DEMETHYLASE PROTEIN FAMILIES
Lysine demethylases fall into two major classes defined by their structure and mechanism:
- The LSD (lysine specific demethylase) family are homologues of the flavin-containing monoamine oxidases, and use the co-factor flavin adenine dinucleotide (FAD) to oxidize methylated lysines to the corresponding imine intermediate followed by hydrolysis to give the demethylated lysine and formaldehyde as byproduct (Fig. 2a
 ) [9Yang M, Culhane JC, Szewczuk LM, et al. Structural basis for the inhibition of the LSD1 histone demethylase by the antidepressant trans-2-phenylcyclopropylamine Biochem 2007; 46(27
): 8058-65.]. LSDs are incapable of demethylating trimethyllysine residues, because the quaternary ammonium group cannot form the requisite imine intermediate. To date two enzymes, LSD1 and LSD2, have been discovered in this subfamily.
) [9Yang M, Culhane JC, Szewczuk LM, et al. Structural basis for the inhibition of the LSD1 histone demethylase by the antidepressant trans-2-phenylcyclopropylamine Biochem 2007; 46(27
): 8058-65.]. LSDs are incapable of demethylating trimethyllysine residues, because the quaternary ammonium group cannot form the requisite imine intermediate. To date two enzymes, LSD1 and LSD2, have been discovered in this subfamily.

Fig. (2) Mechanisms of lysine demethylation catalysed by a) FAD-dependent LSD1; b) 2-OG dependent jumonji domain containing demethylases.
- Jumonji-domain containing demethylases belong to a relatively large family of 2-oxoglutarate (2-OG) containing oxygenases, which also includes HIF-prolyl hydroxylase. These enzymes use Fe(II) together with 2-oxoglutarate to oxygenate methyl groups on methylated lysines, generating the corresponding hydroxymethyl amine, which undergoes the same fate as in the LSD1 mechanism (Fig. 2b
 ) [10Bannister AJ, Kouzarides T. Reversing histone methylation Nature 2005; 436(7054
): 1103-6., 11Cloos PAC, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease Genes Dev 2008; 22(9
): 1115-40.]. This mechanism allows for demethylation of all three possible methylation states of lysine residues.
) [10Bannister AJ, Kouzarides T. Reversing histone methylation Nature 2005; 436(7054
): 1103-6., 11Cloos PAC, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease Genes Dev 2008; 22(9
): 1115-40.]. This mechanism allows for demethylation of all three possible methylation states of lysine residues.
The known FAD and 2-OG containing demethylases have been classified into several subfamilies, and a systematic KDM nomenclature system has been proposed: LSD1 (KDM1); JARID1 (JMJD5); JMJD1 (KDM3); JMJD2A-E (KDM4); UTX/JMJD3 (KDM6A/B); FBXL11 (KDM2), JMJD5 (KDM8) [12Allis CD, Berger SL, Cote J, et al. New nomenclature for chromatin-modifying enzymes Cell 2007; 131(4 ): 633-.].
SUBSTRATE SPECIFICITY
The sequence selectivity of demethylation within histones has been established for many of the demethylases (Fig. 3 ) [13Marmorstein R, Trievel RC. Histone modifying enzymes: Structures, mechanisms, and specificities Biochim Biophys Acta - Gene Regul Mechan 2009; 1789(1
): 58-68.]. Demethylase catalytic domains have an intrinsic sequence selectivity, but this can be modulated by complex formation. Hence, LSD1 has been shown to repress gene expression through the demethylation of H3K4Me1/2, while its association with the androgen receptor leads to enhanced transcription by demethylation of H3K9Me1/2 [14Metzger E, Wissmann M, Yin N, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription Nature 2005; 437(7057
): 436-9.].
) [13Marmorstein R, Trievel RC. Histone modifying enzymes: Structures, mechanisms, and specificities Biochim Biophys Acta - Gene Regul Mechan 2009; 1789(1
): 58-68.]. Demethylase catalytic domains have an intrinsic sequence selectivity, but this can be modulated by complex formation. Hence, LSD1 has been shown to repress gene expression through the demethylation of H3K4Me1/2, while its association with the androgen receptor leads to enhanced transcription by demethylation of H3K9Me1/2 [14Metzger E, Wissmann M, Yin N, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription Nature 2005; 437(7057
): 436-9.].
 |
Fig. (3) Human protein lysine demethylases and their target sites on histones [13Marmorstein R, Trievel RC. Histone modifying enzymes: Structures, mechanisms, and specificities Biochim Biophys Acta - Gene Regul Mechan 2009; 1789(1 ): 58-68.]. |
Among the 2-OG dependent demethylases, individual enzymes show methylation state selectivity apparently driven by steric accommodation: trimethyl demethylases (e.g. JMJD2A) having larger methyllysine binding pockets than dimethyl demethylases (e.g. FBXL11, PHF8) [15Ng SS, Kavanagh KL, McDonough MA, et al. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity Nature 2007; 448(7149 ): 87., 16Horton JR, Upadhyay AK, Qi HH, et al. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases Nat Struct Mol Biol 2010; 17(1 ): 38-43.].
In some cases, the sequence selectivity of demethylation is partly controlled by other domains within the enzymes, as recently described for PHF8 and KIAA1718 [16Horton JR, Upadhyay AK, Qi HH, et al. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases Nat Struct Mol Biol 2010; 17(1 ): 38-43.]. PHF8 contains a PHD finger which binds to H3K4Me3, directing the catalytic domain towards H3K9Me2 and thereby increasing its activity and selectivity by ~100-fold; whilst for KIAA1718, PHD finger binding to H3K4Me3 directs the catalytic domain to preferentially demethylate H3K27Me2. The extent to which similar binding domain control occurs in the substrate selectivity of other demethylase subfamilies (for example JMJD2A-C enzymes which contain methyllysine-binding tudor domains) remains to be explored.
As described in the introduction, a variety of modifications can co-exist on individual histone tails, creating a complex system of cross-talk between the individual marks. Indeed, the aptitude of methylated histone lysines to be demethylated is influenced by post-translational modifications at neighbouring residues: for example, phosphorylation of H3T11 by the kinase PRK1 has been shown to accelerate demethylation of H3K9Me3 by the demethylase JMJD2C [17Metzger E, Yin N, Wissmann M, et al. Phosphorylation of histone H3 at threonine 11 establishes a novel chromatin mark for transcriptional regulation Nat Cell Biol 2008; 10(1 ): 53.]; in contrast, phosphorylation of H3S10 prevents demethylation of H3K9 by the JMJD2 demethylases [15Ng SS, Kavanagh KL, McDonough MA, et al. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity Nature 2007; 448(7149 ): 87.].
DISEASE LINKS
Cancer
Aberrations in levels of histone methylation are frequently correlated with tumorigenesis, presumably resulting from an imbalance between histone methyltransferases and demethylases [1Portela A, Esteller M. Epigenetic modifications and human disease Nat Biotechnol 2010; 28(10 ): 1057-68.]. Common changes include loss of activating marks (H3K4Me3), loss of certain repressive marks (H4K20Me3), and gain of other repressive marks (H3K9Me3 and H3K27Me3). Several demethylases are specifically implicated in the pathogenesis of a variety of cancer types. Both LSD1 and JARID1B are overexpressed in prostate cancer, while LSD1 expression correlates with tumor recurrence during therapy [18Kahl P, Gullotti L, Heukamp LC, et al. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence Cancer Res 2006; 66(23 ): 11341-7.]. LSD1 also demethylates p53, repressing p53-mediated transcriptional activation and inhibiting the role of p53 in promoting apoptosis [19Huang J, Berger SL. The emerging field of dynamic lysine methylation of non-histone proteins Curr Opin Genet Dev 2008; 18(2 ): 152-8.]. LSD1 inhibition by treatment of colon cancer cells with the oligoamine inhibitor SL111144 led to increases in H3K4Me3, restoring expression of secreted frizzled-related proteins (SFRP) Wnt signaling pathway antagonist genes [20Huang Y, Stewart TM, Wu Y, et al. Novel oligoamine analogues inhibit lysine-specific demethylase 1 and induce reexpression of epigenetically silenced genes Clin Cancer Res 2009; 15(23 ): 7217-28.]. In neuroblastoma cells, siRNA–mediated knockdown of LSD1 decreased cellular growth, induced expression of differentiation-associated genes, and increased target gene–specific H3K4 methylation [21Schulte JH, Lim S, Schramm A, et al. lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy Cancer Res 2009; 69(5 ): 2065-71.]. These effects were recapitulated by LSD1 inhibition using monoamine oxidase inhibitors, which further demonstrated growth inhibition of neuroblastoma cells in vitro and reduced neuroblastoma xenograft growth in vivo. JARID1B and JMJD2C are overexpressed in breast and testis cancer and esophageal squamous carcinoma [22Yamane K, Tateishi K, Klose RJ, et al. PLU-1 Is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation Mol Cell 2007; 25(6 ): 801-12.], and RNAi inhibition of JMJD2C resulted in the inhibition of cell proliferation, which highlights this isoform as a potential therapeutic target [11Cloos PAC, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease Genes Dev 2008; 22(9 ): 1115-40.]. Systematic sequencing of renal carcinomas has identified inactivating mutations in UTX and JARID1C [23Dalgliesh GL, Furge K, Greenman C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes Nature 2010; 463(7279 ): 360-.].
Immuno-Inflammation
In addition to classical genetic susceptibilities, the etiologies of a variety of immuno-inflammatory diseases including asthma have been associated with early life programming of immune T-cell response, dendritic cell function, and macrophage activation mediated by epigenetic responses to environmental cues [24Ho S-K. Environmental epigenetics of asthma: An update J Allergy Clin Immunol 2010; 126(3 ): 453-65.]. Global mapping of histone H3K4Me3 and H3K27Me3 has revealed specificity and plasticity in lineage fate determination of differentiating CD4+ T cells, suggesting that lineage fates might be manipulated by modulators of lysine demethylase enzymes targeting these marks [25Wei G, Wei L, Zhu J, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells Immunity 2009; 30(1 ): 155-67.]. Importantly, expression of the demethylase JMJD3, which targets repressive H3K27Me3 marks, is induced in macrophages by the inflammatory transcription factor NFκB in response to stimuli including LPS and the proinflammatory cytokines IL4, IL13 and CCL17 [26Ishii M, Wen H, Corsa CAS, et al. Epigenetic regulation of the alternatively activated macrophage phenotype Blood 2009; 114(15 ): 3244-54.]. Some 70% of lipopolysaccharide (LPS)-inducible genes have been shown to be JMJD3 targets, suggesting that JMJD3 is situated at a key position in inflammatory signalling cascades [27De Santa F, Narang V, Yap ZH, et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages EMBO J 2009; 28: 3341-52.].
Metabolic Disorders & Diabetes
The histone H3K9 demethylase, JMJD1A has recently been associated with metabolic dysregulation: loss of function resulted in decreased expression of metabolically active genes (e.g. peroxisome proliferator–activated receptor-α and medium-chain acyl-CoA dehydrogenase) in skeletal muscle, and impaired expression of cold-induced uncoupling protein 1 in brown adipose tissue in rodents [28Tateishi K, Okada Y, Kallin EM, Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance Nature 2009; 458(7239 ): 757-61.]. This study provides support for a causal relationship between epigenetic mechanisms and obesity (it has long been known that famine exposure in utero and in early infancy is linked to obesity in young men [29Ravelli G-P, Stein ZA, Susser MW. Obesity in young men after famine exposure in utero and early infancy New Eng J Med 1976; 295(7 ): 349-53.]). However, it is not evident that JMJD1A is a good target for the treatment of obesity, since the same authors have shown other essential roles for JMJD1A in mice, including in spermatogenesis [30Okada Y, Scott G, Ray MK, et al. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis Nature 2007; 450(7166 ): 119-23.].
Although there are as yet no published links between demethylase function and the etiology of diabetes, data from several recent diabetes complication trials have shown that in patients who have returned to glycemic control for over 5 years, altered gene expression profiles persist that are linked to eventual complications including blindness, end-stage renal failure, and peripheral neuropathy [31Brasacchio D, Okabe J, Tikellis C, et al. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail Diabetes 2009; 58(5): 1229-36.]. This “hyperglycemic memory” has been attributed to changes in epigenetic information including H3K4 and H3K9 modifications at the NFκB-p65 promoter mediated by the histone methyltransferases (Set7 and SuV39h1) and the lysine-specific demethylase (LSD1).
Neuroscience
Epigenetic abnormalities, which may be introduced during embryogenesis, puberty, or adulthood, have been noted in several psychiatric disorders, including drug addiction, depression and schizophrenia [32Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders Nat Rev Neurosci 2007; 8(5 ): 355-67.]. In rats, acute stress has been shown to increase levels of the repressive H3K9Me3 mark in the dentate gyrus and hippocampal CA1 region, while reducing levels of H3K27Me3 in the same regions, with no effect on levels of H3K4Me3 [33Hunter RG, McCarthy KJ, Milne TA, et al. Regulation of hippocampal H3 histone methylation by acute and chronic stress Proc Nat Acad Sci 2009; 106(49 ): 20912-7.]. Interestingly, treatment with the anxiolytic SSRI antidepressant fluoxetine reversed the decrease in dentate gyrus H3K9Me3, but had no effect on the other marks. Mutations of the human H3K9/27 demethylase PHF8 cluster within its JmjC encoding exons, and are linked to mental retardation (MR) and a cleft lip/palate phenotype [34Loenarz C, Ge W, Coleman ML, et al. PHF8, a gene associated with cleft lip/palate and mental retardation, encodes for an N?-dimethyl lysine demethylase Hum Mol Genet 2010; 19(2 ): 217-2.].
Antiviral
Invading viral pathogens that depend upon the host cell's transcriptional machinery are also subject to the regulatory impact of histone modifications, and this has been specifically demonstrated for LSD1: depletion or inhibition of its activity with monoamine oxidase inhibitors (MAOIs) results in blockade of herpes simplex virus (HSV) and varicella zoster virus (VZV) gene expression [35Liang Y, Vogel JL, Narayanan A, et al. Inhibition of the histone demethylase LSD1 blocks [alpha]-herpesvirus lytic replication and reactivation from latency Nat Med 2009; 15(11 ): 1312-7.].
Regenerative Medicine
Epigenetic landscapes are implicitly involved in the differentiation of stem cells [36Chung H, Sidhu KS. Epigenetic modifications of embryonic stem cells: current trends and relevance in developing regenerative medicine Stem Cells Cloning: Adv Appl 2008; 1: 11-21., 37Kim K, Doi A, Wen B, et al. Epigenetic memory in induced pluripotent stem cells Nature 2010; 467(7313 ): 285-90.], and modulation of the enzymes mediating epigenetic marks might be expected to allow manipulation of stem cell fates, an approach of great interest in regenerative medicine. Among the histone modifying enzymes, evidence is emerging to implicate lysine demethylases in maintenance or progression of stem cell states. The H3K9 demethylases JMJD1a and JMJD2c regulate self-renewal in embryonic stem cells: depletion of either enzyme using shRNA results in progression to ES cell differentiation, accompanied by a reduction in the expression of ES cell-specific genes and an induction of lineage marker genes [38Loh Y, Zhang W, Chen X, et al. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells Genes Dev 2007; 21(20 ): 2545-57.]. Progression of neural stem cells to neurons is regulated by the nuclear receptor co-repressors N-CoR and SMRT (aka N-CoR2), which repress expression of the H3K27 demethylase JMJD3, preventing activation of specific components of the neurogenic program [39Jepsen K, Solum D, Zhou T, et al. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron Nature 2007; 450(7168 ): 415-9.]. The H3K4 demethylase LSD1 is recruited by nuclear receptor TLX, an essential neural stem cell regulator, to the promoters of TLX target genes to repress the expression of these genes, which are known regulators of cell proliferation: inhibition or knockdown of LSD1 was reported to dramatically reduce neural stem cell proliferation [40Sun G, Alzayady K, Stewart R, et al. Histone demethylase LSD1 regulates neural stem cell proliferation Mol Cell Biol 2010; 30(8 ): 1997-2005.].
STRUCTURAL BIOLOGY
Both classes of lysine demethylase are well characterized structurally, including substrate complexes that aid understanding of their mechanisms: a selection of representative structures are summarized in Table 1. The structure of the LSD1-CoREST (corepressor for element-1-silencing transcription factor) complex containing a covalent adduct between FAD and a suicide substrate based on the target H3K4Me2 histone peptide (Fig. 4 ) shows positioning of the lysine methyl groups in suitable proximity for FAD-mediated hydride abstraction to form the iminium intermediate, as per Fig. (2a
) shows positioning of the lysine methyl groups in suitable proximity for FAD-mediated hydride abstraction to form the iminium intermediate, as per Fig. (2a ) [41Yang M, Culhane JC, Szewczuk LM, et al. Structural basis of histone demethylation by LSD1 revealed by suicide inactivation Nat Struct Mol Biol 2007; 14(6
): 535-9.]. The structure also provides an explanation for the specificity of demethylation at H3K4: the terminal amino group of Ala1 inserts into an anionic pocket comprized of Asn, Trp, and two Asp residues, a binding mode not possible with substrates with more than three residues on the N-terminal side of the target methyllysine.
) [41Yang M, Culhane JC, Szewczuk LM, et al. Structural basis of histone demethylation by LSD1 revealed by suicide inactivation Nat Struct Mol Biol 2007; 14(6
): 535-9.]. The structure also provides an explanation for the specificity of demethylation at H3K4: the terminal amino group of Ala1 inserts into an anionic pocket comprized of Asn, Trp, and two Asp residues, a binding mode not possible with substrates with more than three residues on the N-terminal side of the target methyllysine.
 |
Fig. (4) left: X-ray crystal structure of LSD1 (blue) in complex with FAD-H3K4Me2-suicide substrate peptide (red). H3KMe3 peptide (red). right: close-up view of active site, showing positioning of the lysine methyl groups in close proximity for FAD-mediated hydride abstraction to form the iminium intermediate, as per Fig. (2a |
Crystal structures for several members of the 2-OG-dependent histone demethylase family show a common double-stranded β-helix (DSBH) fold typical of 2-OG oxygenases, which supports the common Fe(II)-binding facial triad of a single glutamate or aspartate and two histidine residues (Fig. 5 ). The cofactor 2-OG coordinates to Fe(II) in a bidentate manner through its carboxylate and ketone moieties at C-1 and C-2, while the C-5 carboxylate is tethered by forming a salt bridge to a lysine residue at the other end of the cofactor binding site. X-ray structures of a series of complexes between the JMJD2A demethylase and histone H3 peptides differentially methylated at K9 showed that the peptide chain lies across a shallow groove, presenting the methylated lysine side-chain into a relatively large cavity bounded by several aromatic residues, with the methylamino group sufficiently close to the Fe(II) for hydroxylation [15Ng SS, Kavanagh KL, McDonough MA, et al. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity Nature 2007; 448(7149
): 87.]. These structures also provide a rationale for the sequence preference of JMJD2 enzymes for methylation at H3K9: the preferred peptide conformation is stabilized by an intramolecular H-bond between the backbone carbonyl of K9 and the side chain hydroxyl group of S10, which is not possible with other flanking residues or upon phosphorylation of S10 (vide infra).
). The cofactor 2-OG coordinates to Fe(II) in a bidentate manner through its carboxylate and ketone moieties at C-1 and C-2, while the C-5 carboxylate is tethered by forming a salt bridge to a lysine residue at the other end of the cofactor binding site. X-ray structures of a series of complexes between the JMJD2A demethylase and histone H3 peptides differentially methylated at K9 showed that the peptide chain lies across a shallow groove, presenting the methylated lysine side-chain into a relatively large cavity bounded by several aromatic residues, with the methylamino group sufficiently close to the Fe(II) for hydroxylation [15Ng SS, Kavanagh KL, McDonough MA, et al. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity Nature 2007; 448(7149
): 87.]. These structures also provide a rationale for the sequence preference of JMJD2 enzymes for methylation at H3K9: the preferred peptide conformation is stabilized by an intramolecular H-bond between the backbone carbonyl of K9 and the side chain hydroxyl group of S10, which is not possible with other flanking residues or upon phosphorylation of S10 (vide infra).
INHIBITOR DESIGN
The druggability of both FAD-dependent and 2-OG-dependent demethylases is supported by the recent development of low micromolar inhibitors of members of both these protein families, a selection of which are highlighted in Table 2. Small molecule LSD1 inhibitors include the non-specific monoamine oxidase inhibitors, tranylcypromine [42Culhane JC, Wang D, Yen PM, Cole PA. Comparative analysis of small molecules and histone substrate analogues as LSD1 lysine demethylase inhibitors J Am Chem Soc 2010; 132(9
): 3164-76.] and pargyline [14Metzger E, Wissmann M, Yin N, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription Nature 2005; 437(7057
): 436-9.], and thalidomide derivatives pomalidomide and lenalidomide [43Escoubet-Lozach L, Lin IL, Jensen-Pergakes K, et al. Pomalidomide and lenalidomide induce p21 WAF-1 expression in both lymphoma and multiple myeloma through a LSD1-mediated epigenetic mechanism Cancer Res 2009; 69(18
): 7347-56.]. These inhibitors show effects on histone H3K9 and H3K4 methylation levels in cells, mediated by inhibition of LSD1-catalysed demethylation. The inhibition by tranylcypromine derivatives has been shown to occur by formation of a covalent adduct with the FAD cofactor [42Culhane JC, Wang D, Yen PM, Cole PA. Comparative analysis of small molecules and histone substrate analogues as LSD1 lysine demethylase inhibitors J Am Chem Soc 2010; 132(9
): 3164-76.]. Structure-guided design has led to substituted variants of tranylcypromine with potency enhancements of over 100-fold, exemplified by the homoserine derivative Compound 2, which is selective against MAO-A and B, and shows recovery of H3K4Me2 levels in HEK293T cells and growth inhibition against human cancer cell lines at concentrations ranging from 6-67 µM [44Ueda R, Suzuki T, Mino K, et al. Identification of cell-active lysine specific demethylase 1-selective inhibitors J Am Chem Soc 2009; 131(48
): 17536-7.]. The simpler analogue S2101, which shows a Ki of 0.6 µM, was able to increase levels of H3K4Me2 in HEK293T cells at concentrations as low as 1 µM [45Mimasu S, Umezawa N, Sato S, et al. Structurally designed trans-2-phenylcyclopropylamine derivatives potently inhibit histone demethylase LSD1/KDM1 Biochem 2010; 49(30
): 6494-503.]. The co-structure of a related analogue S1201 with LSD1 confirms the covalent adduct formed with FAD (Fig. 6 ), and shows how the additional benzyloxy substituent occupies an additional active site cavity, proposed to enhance the potency for LSD1 and selectivity over MAOs for this inhibitor series. Polyamines such as PG-11150 have also been shown to be inhibitors of LSD1 in vitro; in cells, these compounds show increased H3K4Me levels and, as described above, elicit reexpression of aberrantly silenced genes in human colon cancer cells [20Huang Y, Stewart TM, Wu Y, et al. Novel oligoamine analogues inhibit lysine-specific demethylase 1 and induce reexpression of epigenetically silenced genes Clin Cancer Res 2009; 15(23
): 7217-28.].
), and shows how the additional benzyloxy substituent occupies an additional active site cavity, proposed to enhance the potency for LSD1 and selectivity over MAOs for this inhibitor series. Polyamines such as PG-11150 have also been shown to be inhibitors of LSD1 in vitro; in cells, these compounds show increased H3K4Me levels and, as described above, elicit reexpression of aberrantly silenced genes in human colon cancer cells [20Huang Y, Stewart TM, Wu Y, et al. Novel oligoamine analogues inhibit lysine-specific demethylase 1 and induce reexpression of epigenetically silenced genes Clin Cancer Res 2009; 15(23
): 7217-28.].
 |
Fig. (6) X-ray crystal structure of LSD1active site containing FAD-S1201 suicide inhibitor covalent adduct. |
JMJD2 demethylases, the most studied subfamily of 2-OG dependent enzymes, are inhibited by co-factor analogues including N-oxalylamino acids such as N-oxalyl glycine, which coordinates to the catalytic Fe(II) in a similar orientation to 2-OG itself, via its carboxylate and amide carbonyl moieties at C-1 and C-2 [46Rose NR, Ng SS, Mecinovic J, et al. Inhibitor scaffolds for 2-oxoglutarate-dependent histone lysine demethylases J Med Chem 2008; 51(22
): 7053-6., 47Hamada S, Kim T-D, Suzuki T, et al. Synthesis and activity of N-oxalylglycine and its derivatives as Jumonji C-domain-containing histone lysine demethylase inhibitors Bioorg Med Chem Lett 2009; 19(10
): 2852-5.]. Across the JMJD2A, FBXL11 and PHF8 enzymes the C-5 carboxylate of NOG is tethered by forming a salt bridge to a lysine residue at the other end of the cofactor binding site (Table 2, Fig. 7a-c ). The similarity in the binding mode suggests that targeting 2-OG analogues might bring a significant challenge in achieving selectivity; however, these enzymes display significant differences in active site structures that might be exploited in selective inhibitor design, and indeed the FBXL11/2-OG complex structure indicates a degree of plasticity in iron coordination geometry as compared with JMJD2A and PHF8. Recently, the protein crystal structure of JMJD2A was used with a dynamic combinatorial chemistry approach to derive a series of substituted oxalyltyrosines that exploit a subpocket of this enzyme to improve potency into the single-digit micromolar range and enhance the selectivity over the 2-OG-dependent prolyl hydroxylase PHD2 (Table 2, Fig. 7d
). The similarity in the binding mode suggests that targeting 2-OG analogues might bring a significant challenge in achieving selectivity; however, these enzymes display significant differences in active site structures that might be exploited in selective inhibitor design, and indeed the FBXL11/2-OG complex structure indicates a degree of plasticity in iron coordination geometry as compared with JMJD2A and PHF8. Recently, the protein crystal structure of JMJD2A was used with a dynamic combinatorial chemistry approach to derive a series of substituted oxalyltyrosines that exploit a subpocket of this enzyme to improve potency into the single-digit micromolar range and enhance the selectivity over the 2-OG-dependent prolyl hydroxylase PHD2 (Table 2, Fig. 7d ) [48Rose NR, Woon EC, Kingham GL, et al. Selective inhibitors of the JMJD2 histone demethylases: combined nondenaturing mass spectrometric screening and crystallographic approaches J Med Chem 2010; 53(4
): 1810-8.]. Interestingly, 2,4-pyridine dicarboxylate is a relatively potent inhibitor with high ligand efficiency (Table 2); this inhibitor mimics 2-OG but adopts an alternative iron coordination geometry whilst maintaining polar interactions between the 4-carboxyl group and the active site lysine residue (Fig. 7e
) [48Rose NR, Woon EC, Kingham GL, et al. Selective inhibitors of the JMJD2 histone demethylases: combined nondenaturing mass spectrometric screening and crystallographic approaches J Med Chem 2010; 53(4
): 1810-8.]. Interestingly, 2,4-pyridine dicarboxylate is a relatively potent inhibitor with high ligand efficiency (Table 2); this inhibitor mimics 2-OG but adopts an alternative iron coordination geometry whilst maintaining polar interactions between the 4-carboxyl group and the active site lysine residue (Fig. 7e ) [46Rose NR, Ng SS, Mecinovic J, et al. Inhibitor scaffolds for 2-oxoglutarate-dependent histone lysine demethylases J Med Chem 2008; 51(22
): 7053-6.]. Fragment growing of 2,4-PDCA generated the related bipyridyl template (Table 2). Other inhibitor chemotypes that are also presumed to bind to the active site Fe(II) include catechols, hydroxamic acids (including the clinically used HDAC inhibitor SAHA/Vorinostat), and TCA cycle intermediates, such as succinate and fumarate) [46Rose NR, Ng SS, Mecinovic J, et al. Inhibitor scaffolds for 2-oxoglutarate-dependent histone lysine demethylases J Med Chem 2008; 51(22
): 7053-6., 49Sakurai M, Rose NR, Schultz L, et al. A miniaturized screen for inhibitors of Jumonji histone demethylases Mol Biosyst 2010; 6(2
): 357-64.].
) [46Rose NR, Ng SS, Mecinovic J, et al. Inhibitor scaffolds for 2-oxoglutarate-dependent histone lysine demethylases J Med Chem 2008; 51(22
): 7053-6.]. Fragment growing of 2,4-PDCA generated the related bipyridyl template (Table 2). Other inhibitor chemotypes that are also presumed to bind to the active site Fe(II) include catechols, hydroxamic acids (including the clinically used HDAC inhibitor SAHA/Vorinostat), and TCA cycle intermediates, such as succinate and fumarate) [46Rose NR, Ng SS, Mecinovic J, et al. Inhibitor scaffolds for 2-oxoglutarate-dependent histone lysine demethylases J Med Chem 2008; 51(22
): 7053-6., 49Sakurai M, Rose NR, Schultz L, et al. A miniaturized screen for inhibitors of Jumonji histone demethylases Mol Biosyst 2010; 6(2
): 357-64.].
A series of N-propionyl hydroxamic acids were recently designed to incorporate a methyllysine mimetic in addition to their 2-OG mimetic features [50Hamada S, Suzuki T, Mino K, et al. Design, synthesis, enzyme-inhibitory activity, and effect on human cancer cells of a novel series of jumonji domain-containing protein 2 histone demethylase inhibitors J Med Chem 2010; 53(15 ): 5629-38.]. The most active analogue, Compound 8, showed low micromolar inhibition of JMJD2A/C with selectivity over prolyl hydroxylases PHD1 and 2 (Table 2); interestingly, ester prodrugs of this compound showed no effects on human cancer cells, but showed growth inhibition in combination with LSD1 inhibitors.
A functional high-throughput screen of JMJD2E identified the 8-hydroxyquinoline template, which coordinates the catalytic Fe(II) via the hydroxy group and pyridyl nitrogen [51King ONF, Li XS, Sakurai M, et al. Quantitative high-throughput screening identifies 8-hydroxyquinolines as cell-active histone demethylase inhibitors PLoS ONE 2010; 5(11
): e15535.]. A carboxylic acid group was added to mimic the distal carboxylate of 2-OG that interacts with the active site lysine, resulting in the micromolar inhibitor SID85736331 (Table 2, Fig. 7f ). The methyl ester of this compound showed reversal of H3K9Me3 depletion in JMJD2A-overexpressing cells, as measured by immunofluorescence.
). The methyl ester of this compound showed reversal of H3K9Me3 depletion in JMJD2A-overexpressing cells, as measured by immunofluorescence.
OUTLOOK
The recent explosion in research into the roles of histone modifying enzymes in healthy and diseased cell function has provided a strong impetus for new drug target discovery. While it is clear that many of these enzymes are likely to be important for normal healthy function, the recent approval of epigenetic modulators such as HDAC inhibitors for cancer treatment demonstrates the potential for therapeutic benefit with acceptable safety and tolerability.
Emerging data support a potential role for lysine demethylases as therapeutic targets in a variety of disease areas including oncology, immunoinflammation, metabolic disorders, neuroscience and virology. While tractability for screening and ligand discovery has been demonstrated for these enzymes, significant challenges remain, notably in identifying chemotypes that show potent and selective inhibition of isoforms of interest whilst retaining physicochemical properties suitable for the intracellular site of action. Further work to identify subtype-selective, cell penetrant inhibitors will be essential in providing tools for chemical biology experiments to further characterize the roles of individual enzymes and their potential as therapeutic targets.
CONFLICT OF INTEREST
The author is an employee of Astex Therapeutics Ltd and declares no competing interests.
ACKNOWLEDGEMENTS
The author wishes to thank Dr Chas Bountra, Prof Udo Oppermann, Prof Chris Schofield, Dr Nathan Rose and Dr Akane Kawamura for helpful discussions, and Dr Wen-Hwa Lee for assistance in the preparation of Fig. (1 ).
).
The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research and the Wellcome Trust.







