- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

Current Chemical Genomics and Translational Medicine
(Discontinued)
ISSN: 2213-9885 ― Volume 12, 2018
A Comparison of the Performance and Application Differences Between Manual and Automated Patch-Clamp Techniques
Xiao Yajuan, Liang Xin, Li Zhiyuan*
Abstract
The patch clamp technique is commonly used in electrophysiological experiments and offers direct insight into ion channel properties through the characterization of ion channel activity. This technique can be used to elucidate the interaction between a drug and a specific ion channel at different conformational states to understand the ion channel modulators’ mechanisms. The patch clamp technique is regarded as a gold standard for ion channel research; however, it suffers from low throughput and high personnel costs. In the last decade, the development of several automated electrophysiology platforms has greatly increased the screen throughput of whole cell electrophysiological recordings. New advancements in the automated patch clamp systems have aimed to provide high data quality, high content, and high throughput. However, due to the limitations noted above, automated patch clamp systems are not capable of replacing manual patch clamp systems in ion channel research. While automated patch clamp systems are useful for screening large amounts of compounds in cell lines that stably express high levels of ion channels, the manual patch clamp technique is still necessary for studying ion channel properties in some research areas and for specific cell types, including primary cells that have mixed cell types and differentiated cells that derive from induced pluripotent stem cells (iPSCs) or embryonic stem cells (ESCs). Therefore, further improvements in flexibility with regard to cell types and data quality will broaden the applications of the automated patch clamp systems in both academia and industry.
Article Information
Identifiers and Pagination:
Year: 2012Volume: 6
First Page: 87
Last Page: 92
Publisher Id: CCGTM-6-87
DOI: 10.2174/1875397301206010087
Article History:
Received Date: 4/2/2012Revision Received Date: 29/6/2012
Acceptance Date: 5/11/2012
Electronic publication date: 28/12/2012
Collection year: 2012
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Zhiyuan Li, Guangzhou Institute of Biomedicine and Health, Chinese Academy of Sciences, Kaiyuan Road 190, Guangzhou Science Park, China 510530; Tel: 20-32015241; Fax: 20-32015299; E-mail: li_zhiyuan@gibh.ac.cn
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 4-2-2012 |
Original Manuscript | A Comparison of the Performance and Application Differences Between Manual and Automated Patch-Clamp Techniques | |
INTRODUCTION
Ion channels play essential roles in neuronal signal transduction, neuronal cell excitement, electrolyte transportation, and muscle contraction [1 Ackerman MJ, Clapham DE. Ion channels--basic science and clinical disease N Engl J Med 1997; 336: 1575-86.]. The patch clamp technique, which is considered a gold standard in ion channel research, has been employed by various laboratories to characterize the ion channel properties of a variety of cells. This technique has also been used to study the pharmacological properties of the ion channel modulators that are used to treat many diseases, including epilepsy, diabetes, hypertension and pain [2 Kass RS. The channelopathies: novel insights into molecular and genetic mechanisms of human disease J Clin Invest 2005; 115: 1986-89.]. However, despite its high informational content, the patch clamp technique suffers from intensive labor requirements, a complex experimental procedure and low screen throughput.
To increase the screen throughput and reduce labor costs, several automated patch clamp systems have been developed and introduced in last ten years. These automated systems have primarily been used on compound screens to quickly quantify their affinities and to study the action mechanisms of on the compounds’ ion channels. Inhibition of activity in the hERG channel has been linked to cardiac safety issues for many approved drugs. To avoid such occurrences in the future, compound activity in the hERG channel should be examined early in the drug discovery process [3 Neher E, Sakmann B. Single-channel currents recorded from membrane of denervated frog muscle fibres Nature 1976; 260: 799-802.]. Additionally, the patch clamp technique has recently been used to verify the maturity of differentiated cells that were derived from stem cells because ion channels are the ultimate functional markers of matured cells, such as neurons and muscle cells. Below, we review the features of both manual and automated patch clamp experiments and their concomitant advantages and limitations.
MANUAL PATCH CLAMP TECHNIQUE
In the late 1970s, Bert Sakmann and Erwin Neher developed a patch clamp technique that accurately measured single channel activity [3 Neher E, Sakmann B. Single-channel currents recorded from membrane of denervated frog muscle fibres Nature 1976; 260: 799-802.]. Their work led to the 1991 Nobel Prize in Physiology or Medicine.
The conventional patch clamp experiment, which consists of manual electrophysiology, uses glass microelectrodes that press against the cell surface to form a tight interaction with a giga-ohm (GΩ) seal resistance between the cell membrane and the rim of glass microelectrode. The current that flows across the plasma membrane can be directly measured by controlling the voltage applied to the membrane. This manual technique allows for a high degree of flexibility in recording configurations, including cell-attached patches, whole-cell patches, inside-out patches, and outside-out patches.
The cell-attached method, or on-cell patch, is often used to study ligand-gated ion channels, channels that are modulated by metabotropic receptors, or neurotransmitters. Compounds that will directly contact the external surface of the membrane are usually included in the pipette solution (Fig. 1A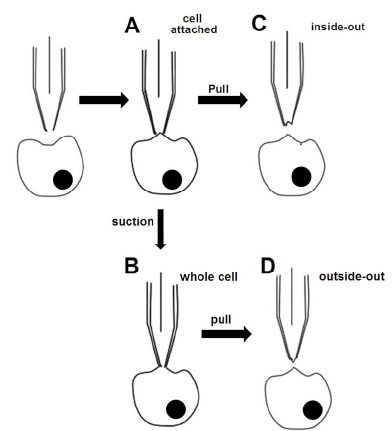 ). This contact allows the concentration-response curves of the compound on ion channels can be accurately determined. The main disadvantage of this method is that only one compound concentration in a concentration-response curve can be measured per patch.
). This contact allows the concentration-response curves of the compound on ion channels can be accurately determined. The main disadvantage of this method is that only one compound concentration in a concentration-response curve can be measured per patch.
The whole cell method records the currents of multiple channels at once through a V-clamp and measures the membrane potential variation under the current clamp (Fig. 1B ). The advantage of this method is that it allows better electrical access to the inside of a cell because the larger opening tip of the electrode provides lower resistance. The disadvantage is that intracellular contents may be dialyzed by a large volume of the pipette solution after 10 minutes of recording.
). The advantage of this method is that it allows better electrical access to the inside of a cell because the larger opening tip of the electrode provides lower resistance. The disadvantage is that intracellular contents may be dialyzed by a large volume of the pipette solution after 10 minutes of recording.
The inside-out patch is particularly useful for accessing the environment of the intracellular surfaces of the ion channels (Fig. 1C ). Using this method, the channels that are activated by the intracellular ligands can be studied and the concentration response curves of the ligands can be established.
). Using this method, the channels that are activated by the intracellular ligands can be studied and the concentration response curves of the ligands can be established.
The outside-out patch can be used to examine the properties of an ion channel that is isolated from the cell (Fig. 1D ). Compared with the cell-attached method, it is more convenient to perfuse the same patch with different solutions, and an entire concentration-response curve of a compound can be obtained in a single patch when the ion channels are activated from the extracellular face.
). Compared with the cell-attached method, it is more convenient to perfuse the same patch with different solutions, and an entire concentration-response curve of a compound can be obtained in a single patch when the ion channels are activated from the extracellular face.
The loose patch clamp is different from the patch clamps discussed above. This type of clamp forms a loose-seal interaction between the cell membrane and the rim of the glass microelectrode. A significant advantage of this technique is that the pipette can be used repeatedly and the cell membrane remains intact. Thus, the loose patch clamp allows the recording of measurements in different locations on the plasma membrane of the same cell without destroying the cell during the each measurement. The major disadvantage of this method is the larger leakage that occurs due to the loose seal.
The manual patch clamp has long being considered the gold standard for investigations of ion channel properties and compound activities because it usually generates high-quality data, but the experimental procedures are complicated and time consuming. Therefore, the screening throughput for the compounds is very limited. To address this bottleneck, several automated patch clamp systems have been developed and introduced to the research market.
AUTOMATED PATCH CLAMP TECHNIQUE
The currently available automated electrophysiology technologies can be divided into three categories: (A) the automated glass pipette-based patch clamp; (B) the micro-fabricated planar electrode-based patch clamp; (C) automated TEVC on Xenopus oocytes [4 Wang X, Li M. Automated electrophysiology: high throughput of art Assay Drug Dev Technol 2003; 1: 695-708.].
(A). Automated Glass Pipette-Based Patch Clamp
Several automated patch-clamp systems have been developed that use conventional electrodes to simplify the time-consuming patching procedure and to achieve a higher throughput and reproducibility with batter data quality. The techniques used by these systems [4 Wang X, Li M. Automated electrophysiology: high throughput of art Assay Drug Dev Technol 2003; 1: 695-708.] are represented in (Fig. 2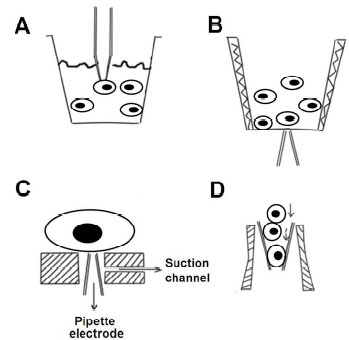 ), except for the Apachi-1 system, which uses imaging for pipette positioning [5 Dunlop J, Bowlby M, Peri R, Vasilyev D, Arias R. Highthroughput electrophysiology: an emerging paradigm for ionchannel screening and physiology Nat Rev Drug Discov 2008; 7: 358-68.].
), except for the Apachi-1 system, which uses imaging for pipette positioning [5 Dunlop J, Bowlby M, Peri R, Vasilyev D, Arias R. Highthroughput electrophysiology: an emerging paradigm for ionchannel screening and physiology Nat Rev Drug Discov 2008; 7: 358-68.].
In one approach, a pipette is moved to contact the surface of a randomly chosen cell that is suspended in one or more cell layers in a density-gradient solution (Fig. 2A , Bristol-Myers Squibb Company) or at the air/liquid interface (Fig. 2B
, Bristol-Myers Squibb Company) or at the air/liquid interface (Fig. 2B , AutoPatchTM). The gigaohm seal can be achieved and whole cell access or other patch clamp configurations may be achieved in the same fashion as the conventional manual patch clamp technique. In a second approach, a tiny pipette tip is vertically positioned upwards in the center of a polyimide sheet that is set at the bottom of a well. Suspension cells are added, and one cell becomes attracted to and then attaches onto the opening of the pipette tip by negative pressure applied through the pipette tip (Fig. 2C
, AutoPatchTM). The gigaohm seal can be achieved and whole cell access or other patch clamp configurations may be achieved in the same fashion as the conventional manual patch clamp technique. In a second approach, a tiny pipette tip is vertically positioned upwards in the center of a polyimide sheet that is set at the bottom of a well. Suspension cells are added, and one cell becomes attracted to and then attaches onto the opening of the pipette tip by negative pressure applied through the pipette tip (Fig. 2C , an automated patch-clamp system based on the “Cytocentering” technique for mammalian cells) [6 Stett A, Burkhardt C, Weber U, van Stiphout P, Knott T. Cytocentering: a novel technique enabling automated cell-by-cell patch clamping with the Cytopatch chip Receptors Channels 2003; 9: 59-66.]. In a third approach, suspended cells flow through a glass pipette to the inside of the pipette’s tip, achieving the gigaohm seal and whole-cell patch configuration inside the micropipette tip (Fig. 2D
, an automated patch-clamp system based on the “Cytocentering” technique for mammalian cells) [6 Stett A, Burkhardt C, Weber U, van Stiphout P, Knott T. Cytocentering: a novel technique enabling automated cell-by-cell patch clamping with the Cytopatch chip Receptors Channels 2003; 9: 59-66.]. In a third approach, suspended cells flow through a glass pipette to the inside of the pipette’s tip, achieving the gigaohm seal and whole-cell patch configuration inside the micropipette tip (Fig. 2D , the Flyscreen system) [7 Zhao Y, Inayat S, Dikin DA, Singer JH, Ruoff RS, Troy JB. Patch clamp technique: Review of the current state of the art and potential contributions from nanoengineering Proc Inst Mech Eng Part N: J Nanoeng Nanosys 2008; 222: 1-11.].
, the Flyscreen system) [7 Zhao Y, Inayat S, Dikin DA, Singer JH, Ruoff RS, Troy JB. Patch clamp technique: Review of the current state of the art and potential contributions from nanoengineering Proc Inst Mech Eng Part N: J Nanoeng Nanosys 2008; 222: 1-11.].
These systems significantly reduce the amount of labor and attention required of the experimental operators. Some of the techniques are semi-automated, such as the Apachi-1, while the Robopatch performs entirely automatically, thus eliminating all of the manual operation steps from the traditional patch clamp technique [8 Vasilyev D, Merrill T, Iwanow A, Dunlop J, Bowlby M. A novel method for patch-clamp automation Pflugers Arch: Eur J Physiol 2006; 452: 240-7.].
However, there are still some drawbacks to the glass pipette-based automated patch clamp systems.
- Suspension cells are required, similar to the planar-based patch clamp technique that will be discussed later. Thus, these techniques are not applicable for tissue slices and differentiated cells derived from iPSCs or ESCs. Additionally, none of these automated systems are capable of studying neural network dynamics [7 Zhao Y, Inayat S, Dikin DA, Singer JH, Ruoff RS, Troy JB. Patch clamp technique: Review of the current state of the art and potential contributions from nanoengineering Proc Inst Mech Eng Part N: J Nanoeng Nanosys 2008; 222: 1-11.].
- The cells are selected blindly, thus the quality of the suspension cells must be both high quality and uniform. This blind selection means that automated methods may not be used on the differentiated cells derived from iPSCs or ESCs because a mixture of different cell types is present in these differentiated cells. The manual selection of a cell type by operators is usually required [9 Miles GB, Yohn DC, Wichterle H, Jessell TM, Rafuse VF, Brownstone RM. Functional properties of motoneurons derived from mouse embryonic stem cells J Neurosci : Off J Soc Neurosci 2004; 24: 7848-58.].
- Parallel experiments of multiple cells for high throughput are very challenging because the precise control of multiple simultaneous pipettes is complicated.
(B). Micro-Fabricated Planar Electrode-Based Patch Clamp
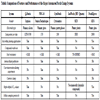
This technique utilizes microfabrication such as silicon or plastic–based planar arrays that have micron-size holes that allow loose or tight seal formations (Fig. 3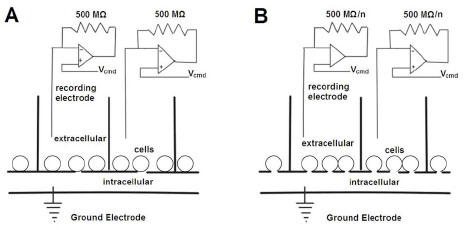 ) [10 Fertig N, Farre C. Renaissance of ion channel research and drug discovery by patch clamp automation Future Med Chem 2010; 2: 691-5.]. Several systems that employ this technique are commercially available, including Q-patch, NPC-16, CytoPatchTM, IonWorks (IonWorks HT and IonWorks Quatto), and PatchXpress. Each technique system has its specific advantages and weaknesses (Table 1) that have been reviewed before [4 Wang X, Li M. Automated electrophysiology: high throughput of art Assay Drug Dev Technol 2003; 1: 695-708., 5 Dunlop J, Bowlby M, Peri R, Vasilyev D, Arias R. Highthroughput electrophysiology: an emerging paradigm for ionchannel screening and physiology Nat Rev Drug Discov 2008; 7: 358-68., 8 Vasilyev D, Merrill T, Iwanow A, Dunlop J, Bowlby M. A novel method for patch-clamp automation Pflugers Arch: Eur J Physiol 2006; 452: 240-7., 10 Fertig N, Farre C. Renaissance of ion channel research and drug discovery by patch clamp automation Future Med Chem 2010; 2: 691-5.]. The major advantages of the planar electrode-based patch clamp include a large increase in throughput, exemplified by IonWorks HT, and very low compound consumption due to the low volume of planar chips. This method is especially suitable for experiments that use rare and expensive compounds or limited amounts of compounds are limited. Additionally, the electronic noise interference from the environment is reduced because of the compact size of the recording chips. However, there are still some drawbacks within these systems.
) [10 Fertig N, Farre C. Renaissance of ion channel research and drug discovery by patch clamp automation Future Med Chem 2010; 2: 691-5.]. Several systems that employ this technique are commercially available, including Q-patch, NPC-16, CytoPatchTM, IonWorks (IonWorks HT and IonWorks Quatto), and PatchXpress. Each technique system has its specific advantages and weaknesses (Table 1) that have been reviewed before [4 Wang X, Li M. Automated electrophysiology: high throughput of art Assay Drug Dev Technol 2003; 1: 695-708., 5 Dunlop J, Bowlby M, Peri R, Vasilyev D, Arias R. Highthroughput electrophysiology: an emerging paradigm for ionchannel screening and physiology Nat Rev Drug Discov 2008; 7: 358-68., 8 Vasilyev D, Merrill T, Iwanow A, Dunlop J, Bowlby M. A novel method for patch-clamp automation Pflugers Arch: Eur J Physiol 2006; 452: 240-7., 10 Fertig N, Farre C. Renaissance of ion channel research and drug discovery by patch clamp automation Future Med Chem 2010; 2: 691-5.]. The major advantages of the planar electrode-based patch clamp include a large increase in throughput, exemplified by IonWorks HT, and very low compound consumption due to the low volume of planar chips. This method is especially suitable for experiments that use rare and expensive compounds or limited amounts of compounds are limited. Additionally, the electronic noise interference from the environment is reduced because of the compact size of the recording chips. However, there are still some drawbacks within these systems.
- Specific cell lines are required. Due to the blind capturing of cells in the pipette-based patch clamp technique, most of the systems have been designed to use stable cell lines that express a single ion channel type in large quantities. These cells used in the automatic techniques usually produce well-behaved patch clamp characteristics such as membrane seal, stability, and sufficient amplitude of currents. The primary cells, differentiated cells derived from iPSCs/ESCs or transiently transfected cells with low expression levels of ion channels may not feasibly be used in most of the systems on the market. It is also possible that the cells that working well in the manual patch clamp experiments may not work in the automated patch clamp systems.
- Right-shifting occurs for some of the compound activities. The IC50 values of compounds have been reported to shift to the right (e.g., reduced compound activities were reported) due to the nonspecific stickiness of lipophilic compounds to the plastic surfaces of the tiny chips that were used in the IonWorks HT system, although the use of glass material has mitigated this issue to some degree [11 Sorota S, Zhang XS, Margulis M, Tucker K, Priestley T. Characterization of a hERG screen using the Ion Works HT: comparison to a hERG rubidium efflux screen Assay Drug Dev Technol 2005; 3: 47-57.].
- The capability for agonist washout during experiments is limited. This limited capability not only constrains the number of data points that can be obtained from a single cell patch but also limits the recording of fast-desensitizing ligand-gated channels. Only a few systems have addressed these problems. The sophisticated fluidics in Dynaflow HT and IonWorks can allow drug washout during the recording process as well as the collection of full-dose responses from these cells [10 Fertig N, Farre C. Renaissance of ion channel research and drug discovery by patch clamp automation Future Med Chem 2010; 2: 691-5.]. The fluidics of platforms such as PatchXpress, Q-patch, and Nanion may be adequate for all but the fastest ligand-gated channels.
- These techniques produce lower quality data. Some systems such as Dynaflow HT and IonWorks sacrifice data quality to pursue higher throughput. The low-quality recording (usually with high leaking currents) imposes limitations on some channels, such as inwardly rectifying potassium (Kir) channels and transient receptor channels (TRP), which are difficult to distinguish from nonspecific “leak” currents [12 Priest BT, Swensen AM, McManus OB. Automated electrophysiology in drug discovery Curr Pharm Des 2007; 13: 2325-37.].
- There is less flexibility with cell types. Some technically demanding experiments such as precise patching onto multiple small structures in a preparation (e.g., the axon, dendrites, and soma of neurons) still largely depend on experienced electrophysiologists’ manual skills using the conventional patch clamp system. The development of these automated systems caters to industrial requirements but undoubtedly neglects the flexibility required by academic researchers. For example, the neurons differentiated from iPSCs usually contain small and/or uncharacterized currents that cannot be measured using these automated patch clamp systems.
(C). Automated, TEVC on Xenopus Oocytes
The automated systems of two-electrode voltage clamps (TEVC), including Robocyte and OpusXpressTM 6000A, have also been developed for studying ion channels expressed in Xenopus oocytes. These systems provide an option for multiple recordings in parallel or serial in Xenopus oocytes, although the development of automated patch clamp systems has mainly focused on mammalian cells using the planar-array-based approaches, as discussed above. Oocyte-based systems are not widely used for drug discovery for several reasons, including the non-mammalian nature of oocytes, for the need to inject RNA or cDNA into each oocyte and the potential differences in IC50 values between the TEVC and patched mammalian cells due to the compound lipophilicity [13 Wood C, Williams C, Waldron GJ. Patch clamping by numbers Drug Discov Today 2004; 9: 434-1.].
PERSPECTIVE
The automating of patch clamp systems greatly simplifies the procedures of electrophysiology experiments and significantly increases compound screening throughput. This automation has broad applications in the ion channel drug discovery process. The systems that are currently commercially available to study ion functions possess a high throughput capability (represented by IonWorks HT) and produce high-quality data (represented by Patch-Xpress). The conventional patch clamp technique is not as suitable for screening large amounts of compounds as the automated patch clamp systems because the former has low throughput and requires extensive manual labor. However, the conventional patch clamp technique is still not replaceable by these automated patch clamp systems due to its unique features of high data quality and flexibility with cell types. For example, the conventional technique can be used to characterize the ion channels in differentiated cells (such as neurons and muscle cells) derived from iPSCs/ESCs. It can also be used for the in-depth characterization of ion channel properties and mechanism of action of a compound. Therefore, the automated patch clamp systems must be further improved and optimized to accommodate a variety of tasks, especially in terms of the flexibility required to meet the needs of academic researchers.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
ACKNOWLEDGEMENT
Declared none.