- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

Current Chemical Genomics and Translational Medicine
(Discontinued)
ISSN: 2213-9885 ― Volume 12, 2018
Evidence for Natural Selection in Nucleotide Content Relationships Based on Complete Mitochondrial Genomes: Strong Effect of Guanine Content on Separation between Terrestrial and Aquatic Vertebrates
Kenji Sorimachi1, 2, *, Teiji Okayasu3
Abstract
The complete vertebrate mitochondrial genome consists of 13 coding genes. We used this genome to investigate the existence of natural selection in vertebrate evolution. From the complete mitochondrial genomes, we predicted nucleotide contents and then separated these values into coding and non-coding regions. When nucleotide contents of a coding or non-coding region were plotted against the nucleotide content of the complete mitochondrial genomes, we obtained linear regression lines only between homonucleotides and their analogs. On every plot using G or A content purine, G content in aquatic vertebrates was higher than that in terrestrial vertebrates, while A content in aquatic vertebrates was lower than that in terrestrial vertebrates. Based on these relationships, vertebrates were separated into two groups, terrestrial and aquatic. However, using C or T content pyrimidine, clear separation between these two groups was not obtained. The hagfish (Eptatretus burgeri) was further separated from both terrestrial and aquatic vertebrates. Based on these results, nucleotide content relationships predicted from the complete vertebrate mitochondrial genomes reveal the existence of natural selection based on evolutionary separation between terrestrial and aquatic vertebrate groups. In addition, we propose that separation of the two groups might be linked to ammonia detoxification based on high G and low A contents, which encode Glu rich and Lys poor proteins.
Article Information
Identifiers and Pagination:
Year: 2015Volume: 9
First Page: 1
Last Page: 5
Publisher Id: CCGTM-9-1
DOI: 10.2174/2213988501509010001
Article History:
Received Date: 1/12/2014Revision Received Date: 31/12/2014
Acceptance Date: 10/1/2015
Electronic publication date: 27/2/2015
Collection year: 2015
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Life Science Research Center, Higashi-Kaizawa, Takasaki, Gunma 370-0041, Japan; Tel: +81-27-352-2955; E-mail: kenjis@jcom.home.ne.jp
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 1-12-2014 |
Original Manuscript | Evidence for Natural Selection in Nucleotide Content Relationships Based on Complete Mitochondrial Genomes: Strong Effect of Guanine Content on Separation between Terrestrial and Aquatic Vertebrates | |
INTRODUCTION
The concept of natural selection was established by Charles Darwin and Alfred Russell Wallace 150 years ago when genetic information was as yet unavailable. This theory was derived from specific differences or similarities in the phenotypes of organisms that lived on geologically isolated islands. Many complete genomes, including that of Homo sapiens [1Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome Nature 2001; 409: 860-921., 2Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome Science 2001; 291: 1304-51.], have now been analyzed. On the one hand, our knowledge of genomic and genetic changes is not enough to clarify all phenotypic changes in organisms related to natural selection. On the other hand, we showed that vertebrates can be classified into terrestrial and aquatic groups in phylogenetic trees based on Ward’s clustering analysis using amino acid composition or nucleotide content predicted from complete mitochondrial genomes as traits [3Sorimachi K, Okayasu T, Ohhira S, Masawa N, Fukasawa I. Natural selection in vertebrate evolution under genomic and biosphere biases based on amino acid content: primitive vertebrate hagfish (Eptatretus burgeri). Nat Sci 2013; 5: 221-7., 4Sorimachi K, Okayasu T. Phylogenetic tree construction based on amino acid composition and nucleotide content of complete vertebrate mitochondrial genomes. IOSR J Phamacy 2013; 3: 51-6.]. This result was consistent with that obtained from 16S RNA sequences [3Sorimachi K, Okayasu T, Ohhira S, Masawa N, Fukasawa I. Natural selection in vertebrate evolution under genomic and biosphere biases based on amino acid content: primitive vertebrate hagfish (Eptatretus burgeri). Nat Sci 2013; 5: 221-7., 4Sorimachi K, Okayasu T. Phylogenetic tree construction based on amino acid composition and nucleotide content of complete vertebrate mitochondrial genomes. IOSR J Phamacy 2013; 3: 51-6.].
Cytochrome C [5Dayhoff MO, Park CM, McLaughlin PJ. Building a phylogenetic trees: cytochrome C. In: Dayhoff MO, Ed. Atlas of protein sequence and structure National Biomedical foundation. Washington, D.C. 1977; Vol. 5: pp. 7-16.], t-RNA [6Maizels N, Weiner AM. Phylogeny from function: evidence from the molecular fossil record that tRNA originated in replication, not translation. Proc Natl Acad Sci USA 1994; 91(15): 6729-34.
[http://dx.doi.org/10.1073/pnas.91.15.6729] [PMID: 8041690] , 7Ribas de Pouplana L, Turner RJ, Steer BA, Schimmel P. Genetic code origins: tRNAs older than their synthetases? Proc Natl Acad Sci USA 1998; 95(19): 11295-300.
[http://dx.doi.org/10.1073/pnas.95.19.11295] [PMID: 9736730] ], 12S RNA [8Puslednik L, Serb JM. Molecular phylogenetics of the Pectinidae (Mollusca: Bivalvia) and effect of increased taxon sampling and outgroup selection on tree topology. Mol Phylogenet Evol 2008; 48(3): 1178-88.
[http://dx.doi.org/10.1016/j.ympev.2008.05.006] [PMID: 18579415] , 9Poulakakis N, Pakaki V, Mylonas M, Lymberakis P. Molecular phylogenetics of the Pectinidae (Mollusca: Bivalvia) and effect of increased taxon sampling and outgroup selection on tree topology. Mol Phylogenet Evol 2008; 47: 396-402.
[http://dx.doi.org/10.1016/j.ympev.2007.10.014] [PMID: 18494151] ], 16S RNA [3Sorimachi K, Okayasu T, Ohhira S, Masawa N, Fukasawa I. Natural selection in vertebrate evolution under genomic and biosphere biases based on amino acid content: primitive vertebrate hagfish (Eptatretus burgeri). Nat Sci 2013; 5: 221-7., 4Sorimachi K, Okayasu T. Phylogenetic tree construction based on amino acid composition and nucleotide content of complete vertebrate mitochondrial genomes. IOSR J Phamacy 2013; 3: 51-6., 8Puslednik L, Serb JM. Molecular phylogenetics of the Pectinidae (Mollusca: Bivalvia) and effect of increased taxon sampling and outgroup selection on tree topology. Mol Phylogenet Evol 2008; 48(3): 1178-88.
[http://dx.doi.org/10.1016/j.ympev.2008.05.006] [PMID: 18579415] -10Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 1991; 173(2): 697-703.
[PMID: 1987160] ] and 18S RNA genes [11Aguinaldo AM, Turbeville JM, Linford LS, et al. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature 1997; 387(6632): 489-93.
[http://dx.doi.org/10.1038/387489a0] [PMID: 9168109] ] have been used to construct phylogenetic trees by various analytical methods based on amino acid or nucleotide sequence changes. Although these methods are applicable for single gene(s) or small genome fragments, they are not suitable for whole genomes that consist of a huge number of genes. However, using normalization of amino acid compositions or nucleotide contents, we can use whole genomes to compare various organisms. Indeed, biological evolution has been investigated based on cellular amino acid composition obtained from cell hydrolysates [11Aguinaldo AM, Turbeville JM, Linford LS, et al. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature 1997; 387(6632): 489-93.
[http://dx.doi.org/10.1038/387489a0] [PMID: 9168109] ], and amino acid composition [12Sorimachi K. Evolutionary changes reflected by the cellular amino acid composition. Amino Acids 1999; 17(2): 207-26.
[http://dx.doi.org/10.1007/BF01361883] [PMID: 10524278] ] or nucleotide contents [14Sorimachi K, Okayasu T. An evaluation of evolutionary theories based on genomic structures in Saccharomyces cerevisiae and Encephalitozoon cunicli. Mycoscience 2004; 45: 345-50.
[http://dx.doi.org/10.1007/S10267-004-0192-X] ] predicted from whole genomes. The ratios of amino acids to the total amino acids, or those of nucleotides to the total nucleotides can characterize whole genomes [15Sorimachi K. Evolution from primitive life to Homo sapiens based on visible genome structures: the amino acid world. Nat Sci 2009; 1: 107-19.], and these indices were used to construct phylogenetic trees [3Sorimachi K, Okayasu T, Ohhira S, Masawa N, Fukasawa I. Natural selection in vertebrate evolution under genomic and biosphere biases based on amino acid content: primitive vertebrate hagfish (Eptatretus burgeri). Nat Sci 2013; 5: 221-7., 4Sorimachi K, Okayasu T. Phylogenetic tree construction based on amino acid composition and nucleotide content of complete vertebrate mitochondrial genomes. IOSR J Phamacy 2013; 3: 51-6.]. The basis of this concept is the homogenous structure of the genome, composed of small units encoding similar amino acid compositions [13Sorimachi K, Okayasu T. Gene assembly consisting of small units with similar amino acid composition in the Saccharomyces cerevisia. Mycoscience 2003; 44: 415-7.
[http://dx.doi.org/10.1007/S10267-003-0131-2] , 14Sorimachi K, Okayasu T. An evaluation of evolutionary theories based on genomic structures in Saccharomyces cerevisiae and Encephalitozoon cunicli. Mycoscience 2004; 45: 345-50.
[http://dx.doi.org/10.1007/S10267-004-0192-X] ], despite each gene’s different nucleotide sequence. Chargaff’s parity rules [G = C, A = T and (A + G) = (T + C)] are inter and intra molecular rules, and the first and second parity rules represent double DNA strand [16Chargaff E. Chemical specificity of nucleic acids and mechanism of their enzymatic degradation. Experientia 1950; 6(6): 201-9.
[http://dx.doi.org/10.1007/BF02173653] [PMID: 15421335] ] and single DNA strand [17Rudner R, Karkas JD, Chargaff E. Separation of B. subtilis DNA into complementary strands. 3. Direct analysis. Proc Natl Acad Sci USA 1968; 60(3): 921-2.
[http://dx.doi.org/10.1073/pnas.60.3.921] [PMID: 4970114] ] molecules, respectively. The first parity rule is understandable based on Watson and Crick’s DNA model [18Watson JD, Crick FH. Genetical implications of the structure of deoxyribonucleic acid. Nature 1953; 171(4361): 964-7.
[http://dx.doi.org/10.1038/171964b0] [PMID: 13063483] ], whereas the second parity rule is not understandable because of the structure of single strand DNA. However, this question was recently solved using a simple mathematical calculation [19Sorimachi K. A proposed solution to the historic pazzle of Chargaff’s second parity rule. Open Genomics J 2009; 2: 12-4.
[http://dx.doi.org/10.2174/1875693X00902010012] ]. Mitchell and Bridge showed that Chargaff’s second parity rule is applicable for inter species research using a large data set of whole genomes, and that inter species nucleotide relationships are expressed with linear regression lines [20Mitchell D, Bridge R. A test of Chargaff’s second rule. Biochem Biophys Res Commun 2006; 340(1): 90-4.
[http://dx.doi.org/10.1016/j.bbrc.2005.11.160] [PMID: 16364245] ]. We also showed that inter species nucleotide relationships were expressed with linear regression lines among not only whole genomes but also coding or non-coding regions [21Sorimachi K, Okayasu T. Codon evolution is governed by linear formulas. Amino Acids 2008; 34(4): 661-8.
[http://dx.doi.org/10.1007/s00726-007-0024-3] [PMID: 18180868] ]. In our previous study, we constructed phylogenetic trees based on amino acid composition or nucleotide content as traits, and we classified vertebrates into terrestrial and aquatic groups [3Sorimachi K, Okayasu T, Ohhira S, Masawa N, Fukasawa I. Natural selection in vertebrate evolution under genomic and biosphere biases based on amino acid content: primitive vertebrate hagfish (Eptatretus burgeri). Nat Sci 2013; 5: 221-7., 4Sorimachi K, Okayasu T. Phylogenetic tree construction based on amino acid composition and nucleotide content of complete vertebrate mitochondrial genomes. IOSR J Phamacy 2013; 3: 51-6.]. Consequently, this study was designed to show that simple plotting of nucleotide content predicted from complete mitochondrial genomes can also classify vertebrates into these two groups.
MATERIALS AND METHODS
The same data set that was used in our previous studies on nucleotide relationships [22Sorimachi K, Okayasu T. Universal rules govering genome evolution expressed by linear formulas. Open Genomics J 2008; 1: 33-43.
[http://dx.doi.org/10.2174/1875693X00801010033] -25Sorimachi K, Okayasu T, Ohhira S, Fukasawa I, Masawa N. Evidence for the independent divergence of vertebrate and high C/G ratio invertebrate mitochondria from the same origin. Nat Sci 2012; 4: 479-83.] and phylogenetic tree construction [3Sorimachi K, Okayasu T, Ohhira S, Masawa N, Fukasawa I. Natural selection in vertebrate evolution under genomic and biosphere biases based on amino acid content: primitive vertebrate hagfish (Eptatretus burgeri). Nat Sci 2013; 5: 221-7., 4Sorimachi K, Okayasu T. Phylogenetic tree construction based on amino acid composition and nucleotide content of complete vertebrate mitochondrial genomes. IOSR J Phamacy 2013; 3: 51-6., 26Sorimachi K, Okayasu T, Ebara Y, Furuta E, Ohhira S. Phylogenetic position of Xenoturbella bocki and Hemichordates Balanoglossus carnosus and Saccoglossus kowalevskii based on amino acid composition or nucleotide content of complete mitochondrial genomes. Int J Biol 2014; 6: 82-94.] were investigated in this study. Thus, these organisms were chosen according to their alphabetical order without any preconception of the organisms’ characteristics [21Sorimachi K, Okayasu T. Codon evolution is governed by linear formulas. Amino Acids 2008; 34(4): 661-8.
[http://dx.doi.org/10.1007/s00726-007-0024-3] [PMID: 18180868] ]. All complete mitochondrial genomes (45 species) that were available were collected in our previous study [22Sorimachi K, Okayasu T. Universal rules govering genome evolution expressed by linear formulas. Open Genomics J 2008; 1: 33-43.
[http://dx.doi.org/10.2174/1875693X00801010033] ] from the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/sites). The list of organisms examined is shown in Table 1. Nucleotide contents were normalized to 1. (G + C + A + T = 1). Statistical analyses were carried out using Student’s t-tests with Microsoft Excel 2010.
RESULTS AND DISCUSSION
Coding Region Nucleotide Relationships
In our previous study, plotting nucleotide contents of the coding region against nucleotide contents of the complete mitochondrial genome, linear regression lines were obtained between homonucleotides and their analogs, whereas relationships were heteroskedastic between heteronucleotides and their analogs [22Sorimachi K, Okayasu T. Universal rules govering genome evolution expressed by linear formulas. Open Genomics J 2008; 1: 33-43.
[http://dx.doi.org/10.2174/1875693X00801010033] ]. When G content of the coding region was plotted against G content of the complete mitochondrial genome, their relationships were expressed by linear regression lines (terrestrial; y = 1.0851 × −0.0222, R2 = 0.6596 and aquatic; y = 0.8409 × + 0.0133, R2 = 0.8527). This is consistent with our previous result [22Sorimachi K, Okayasu T. Universal rules govering genome evolution expressed by linear formulas. Open Genomics J 2008; 1: 33-43.
[http://dx.doi.org/10.2174/1875693X00801010033] ]. Vertebrates were separated into two groups, terrestrial and aquatic, based on G content of the mitochondrial coding region or that of the complete mitochondrial genome (Fig. 1A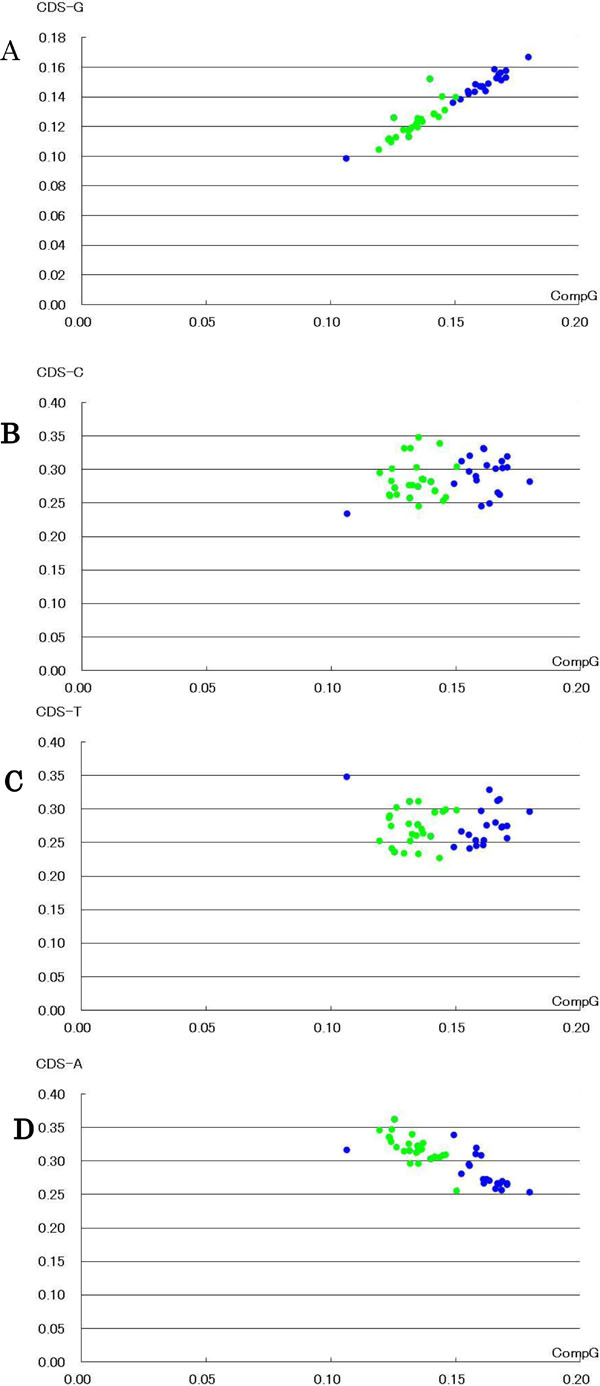 ). The G content of the complete mitochondrial genome in terrestrial vertebrates was lower than that in aquatic vertebrates. External biosphere bias might lead to the separation of vertebrates into terrestrial and aquatic groups as a result of natural selection [3Sorimachi K, Okayasu T, Ohhira S, Masawa N, Fukasawa I. Natural selection in vertebrate evolution under genomic and biosphere biases based on amino acid content: primitive vertebrate hagfish (Eptatretus burgeri). Nat Sci 2013; 5: 221-7.]. The hagfish (Eptatretus burgeri), whose G content of the complete mitochondrial genome and of the coding region is the lowest among vertebrates examined, was separated from aquatic and terrestrial vertebrates (Fig. 1
). The G content of the complete mitochondrial genome in terrestrial vertebrates was lower than that in aquatic vertebrates. External biosphere bias might lead to the separation of vertebrates into terrestrial and aquatic groups as a result of natural selection [3Sorimachi K, Okayasu T, Ohhira S, Masawa N, Fukasawa I. Natural selection in vertebrate evolution under genomic and biosphere biases based on amino acid content: primitive vertebrate hagfish (Eptatretus burgeri). Nat Sci 2013; 5: 221-7.]. The hagfish (Eptatretus burgeri), whose G content of the complete mitochondrial genome and of the coding region is the lowest among vertebrates examined, was separated from aquatic and terrestrial vertebrates (Fig. 1 ). Similarly, plotting A content of the coding region against G content of the complete mitochondrial genome, linear regression lines were obtained, although their regression coefficients were barely reduced (terrestrial; y = −2.0201 × + 0.5893, R2 = 0.63 and aquatic; y = −1.3314 × + 0.4973, R2 = 0.5873) (Fig. 1D
). Similarly, plotting A content of the coding region against G content of the complete mitochondrial genome, linear regression lines were obtained, although their regression coefficients were barely reduced (terrestrial; y = −2.0201 × + 0.5893, R2 = 0.63 and aquatic; y = −1.3314 × + 0.4973, R2 = 0.5873) (Fig. 1D ). In addition, vertebrates were separated into two groups, terrestrial and aquatic, based on A content in the mitochondrial coding region and the complete mitochondrial genome, although a slight overlap between two groups was observed (Fig. 1D
). In addition, vertebrates were separated into two groups, terrestrial and aquatic, based on A content in the mitochondrial coding region and the complete mitochondrial genome, although a slight overlap between two groups was observed (Fig. 1D and Supplementary Fig.
1). The A content of the complete mitochondrial genome in terrestrial vertebrates was higher than that in aquatic vertebrates versus the G content. G and A purine contents played an important role in separating vertebrates into terrestrial and aquatic groups. When C or T content pyrimidine of the coding region was plotted against G content of the complete mitochondrial genome, their relationships were heteroskedastic (Supplementary Fig.
1). The hagfish was separated from both groups in every plot (Fig. 1
and Supplementary Fig.
1). The A content of the complete mitochondrial genome in terrestrial vertebrates was higher than that in aquatic vertebrates versus the G content. G and A purine contents played an important role in separating vertebrates into terrestrial and aquatic groups. When C or T content pyrimidine of the coding region was plotted against G content of the complete mitochondrial genome, their relationships were heteroskedastic (Supplementary Fig.
1). The hagfish was separated from both groups in every plot (Fig. 1 ).
).
Non-coding Region Nucleotide Relationships
Using nucleotide content of the non-coding region, the relationships between non-coding G content and complete mitochondrial G content were expressed by linear regression lines (terrestrial; y = 0.7203x + 0.0609, R2 = 1.488: aquatic; y = 1.3523x − 0.0295, R2 = 0.7444) (Fig. 2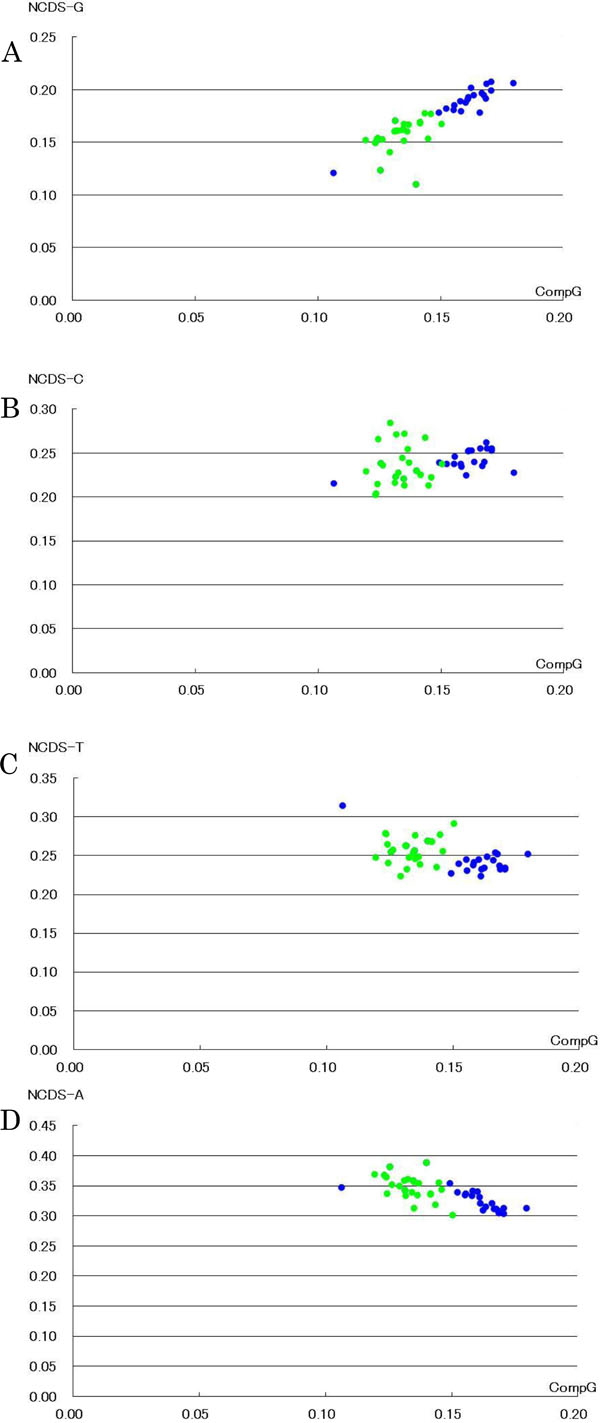 ), as observed in the coding region (Fig. 1
), as observed in the coding region (Fig. 1 ). Clear separation between terrestrial and aquatic vertebrates was observed based on both G contents of the non-coding region and of the complete mitochondrial genome. G content in aquatic vertebrates was higher than that in terrestrial vertebrates in the non-coding region as well as in the complete mitochondrial genome. By contrast, A content in aquatic vertebrates was lower than that in terrestrial vertebrates, and vertebrates were separated into two groups based on A content of the non-coding region, as observed in the coding region (Fig. 1
). Clear separation between terrestrial and aquatic vertebrates was observed based on both G contents of the non-coding region and of the complete mitochondrial genome. G content in aquatic vertebrates was higher than that in terrestrial vertebrates in the non-coding region as well as in the complete mitochondrial genome. By contrast, A content in aquatic vertebrates was lower than that in terrestrial vertebrates, and vertebrates were separated into two groups based on A content of the non-coding region, as observed in the coding region (Fig. 1 and Supplementary Fig.
1). Relationships between C or T nucleotide content of the non-coding region and G content of the complete mitochondrial genome were heteroskedastic, and no significant separation between the two groups was observed (Fig. 2
and Supplementary Fig.
1). Relationships between C or T nucleotide content of the non-coding region and G content of the complete mitochondrial genome were heteroskedastic, and no significant separation between the two groups was observed (Fig. 2 ).
).
The hagfish was separated from both terrestrial and aquatic vertebrates even in the case of using the non-coding region nucleotide content. The characteristics of the hagfish seem to be of a primitive vertebrate [27Janvier P. microRNAs revive old views about jawless vertebrate divergence and evolution. Proc Natl Acad Sci USA 2010; 107(45): 19137-8.
[http://dx.doi.org/10.1073/pnas.1014583107] [PMID: 21041649] ]. In our previous phylogenetic study of the sample containing both vertebrates and high C/G-invertebrates, the hagfish belonged to the vertebrate group, while it belonged to invertebrate group, using a mixture of vertebrates, high C/G- and low C/G-invertebrates [26Sorimachi K, Okayasu T, Ebara Y, Furuta E, Ohhira S. Phylogenetic position of Xenoturbella bocki and Hemichordates Balanoglossus carnosus and Saccoglossus kowalevskii based on amino acid composition or nucleotide content of complete mitochondrial genomes. Int J Biol 2014; 6: 82-94.]. This result could be attributed to the primitive characteristics of hagfish.
Regarding nucleotide contents of the non-coding region, G and A purine contents played an important role in separating vertebrates into aquatic and terrestrial groups, whereas C and T pyrimidine did not (Fig. 2 and Supplementary Fig.
2), as observed in the coding region (Fig. 1
and Supplementary Fig.
2), as observed in the coding region (Fig. 1 and Supplementary Fig.
1). These results indicate that nucleotide alternations including mutations occurred randomly over the mitochondrial genome, as reported in whole chromosomal genomes based on amino acid compositions [13Sorimachi K, Okayasu T. Gene assembly consisting of small units with similar amino acid composition in the Saccharomyces cerevisia. Mycoscience 2003; 44: 415-7.
and Supplementary Fig.
1). These results indicate that nucleotide alternations including mutations occurred randomly over the mitochondrial genome, as reported in whole chromosomal genomes based on amino acid compositions [13Sorimachi K, Okayasu T. Gene assembly consisting of small units with similar amino acid composition in the Saccharomyces cerevisia. Mycoscience 2003; 44: 415-7.
[http://dx.doi.org/10.1007/S10267-003-0131-2] ] or nucleotide contents [14Sorimachi K, Okayasu T. An evaluation of evolutionary theories based on genomic structures in Saccharomyces cerevisiae and Encephalitozoon cunicli. Mycoscience 2004; 45: 345-50.
[http://dx.doi.org/10.1007/S10267-004-0192-X] ]. Natural selection seems to contribute to evolution after genotype alternations.
In the codon table, Gly is encoded with GGT, GGC, GGA and GGG, and Glu is encoded with GAG and GAA, while Lys is encoded with AAA and AAG. These relationships indicate that high G and low A contents produce acidic proteins rather than basic proteins, because Gly is a neutral amino acid. Indeed, Glu content in fish cell hydrolysates was higher than that in rat or human cell hydrolysates [11Aguinaldo AM, Turbeville JM, Linford LS, et al. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature 1997; 387(6632): 489-93.
[http://dx.doi.org/10.1038/387489a0] [PMID: 9168109] ]. In many fish that lack urea production in nitrogen metabolism, ammonia (which is fatally toxic to organisms) is excreted directly out of the body. Terrestrial vertebrates including mammals have a urea cycle, and excrete urea as a final nitrogen metabolite. As acidic proteins contribute to neutralize ammonia in the bodies of fish, high G and low A content must be linked with aquatic vertebrate evolution. In the dark-spotted frog (Rana nigromaculata) tadpoles are aquatic and ammonotelic, whereas after metamorphosis, the adult, which has a urea cycle, is terrestrial. This amphibian was grouped with aquatic vertebrates based on Ward’s clustering analysis using amino acid composition or nucleotide contents predicted from complete mitochondrial genomes as traits [25Sorimachi K, Okayasu T, Ohhira S, Fukasawa I, Masawa N. Evidence for the independent divergence of vertebrate and high C/G ratio invertebrate mitochondria from the same origin. Nat Sci 2012; 4: 479-83.], and its G content was much closer to that of aquatic vertebrates in this study. Thus, these results seem to be based on genomic characteristics of R. nigromaculata. We have proposed that separation between aquatic and terrestrial vertebrates might be linked with nitrogen metabolism in biological evolution.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s web site along with the published article.
ACKNOWLEDGEMENTS
Declared none.
REFERENCES
| [1] | Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome Nature 2001; 409: 860-921. |
| [2] | Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome Science 2001; 291: 1304-51. |
| [3] | Sorimachi K, Okayasu T, Ohhira S, Masawa N, Fukasawa I. Natural selection in vertebrate evolution under genomic and biosphere biases based on amino acid content: primitive vertebrate hagfish (Eptatretus burgeri). Nat Sci 2013; 5: 221-7. |
| [4] | Sorimachi K, Okayasu T. Phylogenetic tree construction based on amino acid composition and nucleotide content of complete vertebrate mitochondrial genomes. IOSR J Phamacy 2013; 3: 51-6. |
| [5] | Dayhoff MO, Park CM, McLaughlin PJ. Building a phylogenetic trees: cytochrome C. In: Dayhoff MO, Ed. Atlas of protein sequence and structure National Biomedical foundation. Washington, D.C. 1977; Vol. 5: pp. 7-16. |
| [6] | Maizels N, Weiner AM. Phylogeny from function: evidence from the molecular fossil record that tRNA originated in replication, not translation. Proc Natl Acad Sci USA 1994; 91(15): 6729-34. [http://dx.doi.org/10.1073/pnas.91.15.6729] [PMID: 8041690] |
| [7] | Ribas de Pouplana L, Turner RJ, Steer BA, Schimmel P. Genetic code origins: tRNAs older than their synthetases? Proc Natl Acad Sci USA 1998; 95(19): 11295-300. [http://dx.doi.org/10.1073/pnas.95.19.11295] [PMID: 9736730] |
| [8] | Puslednik L, Serb JM. Molecular phylogenetics of the Pectinidae (Mollusca: Bivalvia) and effect of increased taxon sampling and outgroup selection on tree topology. Mol Phylogenet Evol 2008; 48(3): 1178-88. [http://dx.doi.org/10.1016/j.ympev.2008.05.006] [PMID: 18579415] |
| [9] | Poulakakis N, Pakaki V, Mylonas M, Lymberakis P. Molecular phylogenetics of the Pectinidae (Mollusca: Bivalvia) and effect of increased taxon sampling and outgroup selection on tree topology. Mol Phylogenet Evol 2008; 47: 396-402. [http://dx.doi.org/10.1016/j.ympev.2007.10.014] [PMID: 18494151] |
| [10] | Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 1991; 173(2): 697-703. [PMID: 1987160] |
| [11] | Aguinaldo AM, Turbeville JM, Linford LS, et al. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature 1997; 387(6632): 489-93. [http://dx.doi.org/10.1038/387489a0] [PMID: 9168109] |
| [12] | Sorimachi K. Evolutionary changes reflected by the cellular amino acid composition. Amino Acids 1999; 17(2): 207-26. [http://dx.doi.org/10.1007/BF01361883] [PMID: 10524278] |
| [13] | Sorimachi K, Okayasu T. Gene assembly consisting of small units with similar amino acid composition in the Saccharomyces cerevisia. Mycoscience 2003; 44: 415-7. [http://dx.doi.org/10.1007/S10267-003-0131-2] |
| [14] | Sorimachi K, Okayasu T. An evaluation of evolutionary theories based on genomic structures in Saccharomyces cerevisiae and Encephalitozoon cunicli. Mycoscience 2004; 45: 345-50. [http://dx.doi.org/10.1007/S10267-004-0192-X] |
| [15] | Sorimachi K. Evolution from primitive life to Homo sapiens based on visible genome structures: the amino acid world. Nat Sci 2009; 1: 107-19. |
| [16] | Chargaff E. Chemical specificity of nucleic acids and mechanism of their enzymatic degradation. Experientia 1950; 6(6): 201-9. [http://dx.doi.org/10.1007/BF02173653] [PMID: 15421335] |
| [17] | Rudner R, Karkas JD, Chargaff E. Separation of B. subtilis DNA into complementary strands. 3. Direct analysis. Proc Natl Acad Sci USA 1968; 60(3): 921-2. [http://dx.doi.org/10.1073/pnas.60.3.921] [PMID: 4970114] |
| [18] | Watson JD, Crick FH. Genetical implications of the structure of deoxyribonucleic acid. Nature 1953; 171(4361): 964-7. [http://dx.doi.org/10.1038/171964b0] [PMID: 13063483] |
| [19] | Sorimachi K. A proposed solution to the historic pazzle of Chargaff’s second parity rule. Open Genomics J 2009; 2: 12-4. [http://dx.doi.org/10.2174/1875693X00902010012] |
| [20] | Mitchell D, Bridge R. A test of Chargaff’s second rule. Biochem Biophys Res Commun 2006; 340(1): 90-4. [http://dx.doi.org/10.1016/j.bbrc.2005.11.160] [PMID: 16364245] |
| [21] | Sorimachi K, Okayasu T. Codon evolution is governed by linear formulas. Amino Acids 2008; 34(4): 661-8. [http://dx.doi.org/10.1007/s00726-007-0024-3] [PMID: 18180868] |
| [22] | Sorimachi K, Okayasu T. Universal rules govering genome evolution expressed by linear formulas. Open Genomics J 2008; 1: 33-43. [http://dx.doi.org/10.2174/1875693X00801010033] |
| [23] | Sorimachi K. Codon evolution in double-stranded organelle DNA: strong regulation of homonucleotides and their analog alternations. Nat Sci 2010; 2: 846-54. |
| [24] | Sorimachi K. Genomic data provides simple evidence for a single origin of life. Nat Sci 2010; 2: 519-25. |
| [25] | Sorimachi K, Okayasu T, Ohhira S, Fukasawa I, Masawa N. Evidence for the independent divergence of vertebrate and high C/G ratio invertebrate mitochondria from the same origin. Nat Sci 2012; 4: 479-83. |
| [26] | Sorimachi K, Okayasu T, Ebara Y, Furuta E, Ohhira S. Phylogenetic position of Xenoturbella bocki and Hemichordates Balanoglossus carnosus and Saccoglossus kowalevskii based on amino acid composition or nucleotide content of complete mitochondrial genomes. Int J Biol 2014; 6: 82-94. |
| [27] | Janvier P. microRNAs revive old views about jawless vertebrate divergence and evolution. Proc Natl Acad Sci USA 2010; 107(45): 19137-8. [http://dx.doi.org/10.1073/pnas.1014583107] [PMID: 21041649] |