- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Leukemia Journal
(Discontinued)
ISSN: 1876-8164 ― Volume 5, 2013
Effects of Epigenetic Drugs (Vorinostat, Decitabine) on Metabolism-Related Pathway Factors in Leukemic Cells
Heidrun Karlic1, 2, Julia Varga1, 2, Roman Thaler3, Cornelia Berger1, Silvia Spitzer3, Michael Pfeilstöcker1, 2, 4, Klaus Klaushofer3, Franz Varga3, *
Abstract
Novel tools for data-evaluation from gene-expression arrays allow analyses of biochemical pathways which may be influenced by treatment with epigenetic drugs which have originally been designed for re-activation of so-called tumour-suppressor genes that are known as key-molecules regulating differentiation or cell death in malignancy. Considering the fact that these processes are tightly associated with energy metabolism, this study evaluated the expression signature of prominently regulated pathways in the KG-1-leukemia cell line: Following a 3-day incubation, effects of pharmacologic concentrations from the histone-deacetylase inhibitor SAHA (suberoyl anilide hydroxamic acid, vorinostat) and the methylation-inhibitor desoxy-azacytidine (DAC) were comparatively analysed by transcriptional profiling (based on Affymetrix Human GeneChip Gene 1.0 ST microarrays) and quantitative real time PCR. Expression factors for pathways were calculated for comparative analyses. Epigenetic drugs SAHA and DAC had a downregulatory effect on metabolic pathway factors of carbohydrate metabolism and mitochondrial beta oxidation. Our data confirm SAHA-mediated downregulation of the histone deacetylase SIRT which regulates AKT-phosphatase. Associated pathways lead to regulation of numerous genes, including an upregulation of FOXO transcription factors. These regulatory networks are known for their crucial role in stem cell homeostasis and provide a mechanistic explanation for the fact that the number of SAHA-targeted genes (1392 up, 2651 down) exceeds the number of DAC-targeted genes (60 up, 15 down), besides known effects on cell-cycle-arrest and apoptosis induced by both drugs.
Thus, our data underline that epigenetic mechanisms are tightly associated with malignancy-associated metabolic control at least at 3 levels, starting from (i) glucose-uptake over (ii) mitochondrial pathways to (iii) AKT-PTEN-FOXO-signalling. All of them are known to be regulated by caloric restriction. We propose that these interactions should be carefully considered in clinical application, providing the basis for optimization of drug-combinations and complex treatment strategies
Article Information
Identifiers and Pagination:
Year: 2010Volume: 3
First Page: 34
Last Page: 42
Publisher Id: TOLEUKEMIAJ-3-34
DOI: 10.2174/1876816401003010034
Article History:
Received Date: 3/2/2010Revision Received Date: 25/3/2010
Acceptance Date: 25/3/2010
Electronic publication date: 17 /5/2010
Collection year: 2010
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Ludwig Boltzmann Institute of Osteology at the Hanusch Hospital of WGKK and AUVA Trauma Centre Meidling. 4th Medical Department, Hanusch Hospital, Heinrich Collin Str. 30, 1140 Vienna, Austria; Tel: +431 9102186933l Fax: +431 9102186929; E-mail: franz.varga@osteologie.at
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 3-2-2010 |
Original Manuscript | Effects of Epigenetic Drugs (Vorinostat, Decitabine) on Metabolism-Related Pathway Factors in Leukemic Cells | |
1 . INTRODUCTION
It is generally accepted that epigenetic active drugs such as aza-nucleosides and histone-deacetylase-inhibitors act via re-activation of a large panel of genes including so-called tumor suppressor genes responsible for cell cycle arrest, differentiation and apoptosis in hematologic malignancies [1Cameron EE, Bachman KE, Myohanen S. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer Nat Genet 1999; 21: 103-7., 2Bhalla KN. Epigenetic and chromatin modifiers as targeted therapy of hematologic malignancies J Clin Oncol 2005; 23: 3971-93.]. The drugs have been well characterized as inhibitors of DNMTs (DNA methyl transferase) and are known since more than 20 years (e.g. [3Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat development of this histone deacetylase inhibitor as an anticancer drug Nat Biotechnol 2007; 25: 84-90.-7Lubbert M. Combined targeting of the epigenetic silence in leukemia cooperating activities of DNA methylation and histone deacetylation inhibitors Leuk Res 2005; 29: 727-8.]). Their application is promising because of their low toxicity to normal cells and their specific biological effects on a large number of cellular processes.
These drugs should actually target epigenetic processes like histone modifications (e.g. acetylation) and/or DNA-CpG methylation. However, recent studies using high sensitive microarray platforms for comparative evaluation of gene-methylation- and respective mRNA-expression-patterns indicate that, also in responders, these epigenetic drugs do not induce significant changes in gene expression driven by changes of the epigenetic DNA methylation status [8Robertson KD, Bhalla KN. Missteps in tango for epigenome targeting Blood 2009; 114: 2569-70., 9Fandy TE, Herman JG, Kerns P. Early epigenetic changes and DNA damage do not predict clinical response in an overlapping schedule of 5-azacytidine and entinostat in patients with myeloid malignancies Blood 2009; 114: 2764-73.], and therefore, the exact therapeutic molecular targets of epigenetic drugs still remain to be clarified.
It appears possible that aza-nucleosides and histone-deacetylase-inhibitors also target epigenetic processes which are tightly associated with malignancy-associated features of energy metabolism [10Karlic H, Varga F. Epigenetics and Tumorigenesis.In Haslberger AGl S editor. Epigenetics and Human Health. Linking Hereditary Environmental and Nutritional Aspects Hoboken NJ USA Wiley 2009; 179-94.]. The rationale for this is based on the fact that increased supply of energy from carbohydrate- and fat catabolism confers a significant growth advantage to leukemias and cancer. An increased glycolytic activity of leukemia- and cancer cells produces an acidic environment which is more toxic to the adjacent tissue compared with malignant cells themselves [11Gillies RJ, Robey I, Gatenby RA. Causes and consequences of increased glucose metabolism of cancers J Nucl Med 2008; 49( Suppl 2): 24S-42S.].
Enhanced energy metabolism in malignancy is closely associated with a changed availability of acyl groups from fatty acid catabolism. As fatty acids represent a source of acyl groups which are substrates both for cytoplasmic and nuclear acyl- (or acetyl-) coenzyme A, the latter play a role for histone acetylation and thus gene activation / regulation. Histone-deacetylases which may be stimulated in response to a reduced availablity of intranuclear acyl groups, induce a hypoacetylation of histones in malignancy and promote a tighter packing of chromatin and thus inhibit transcription of critical genes such as so-called tumor suppressors which is also associated with promoter-methylation of these genes. Recent data showed that histone-deacetylase (HDAC) - inhibitors of distinct chemotypes have the potential to reverse this by downregulating glucose transporter 1 (SLC2A1) - mediated glucose transport and inhibition of hexokinase (HK1) – enzymatic activity [12Wardell SE, Ilkayeva OR, Wieman HL. Glucose metabolism as a target of histone deacetylase inhibitors Mol Endocrinol 2009; 23: 388-401.]. In addition, several DNA-demethylating agents such as DAC exert this activity by promoting apoptosis and growth arrest at different direct or indirect pathways which may also regulate (de)differentiation of leukemic cells.
Array technologies have turned out as powerful tools for evaluating biological basal processes such as metabolism where reliability has been confirmed by comparative analyses of proteomic and transcriptomic arrays [13Adler P, Peterson H, Agius P. Ranking genes by their co-expression to subsets of pathway members Ann N Y Acad Sci 2009; 1158: 1-13.]. Comparative analyses of methylation-specific DNA-arrays with gene-expression arrays showed that elements of the forkhead box of transcription factors are hypermethylated and transcriptionally downregulated together with an aberrant upregulation of metabolic genes in KG-1 cells [14Gebhard C, Schwarzfischer L, Pham TH. Genome-wide profiling of CpG methylation identifies novel targets of aberrant hypermethylation in myeloid leukemia Cancer Res 2006; 66: 6118-28.] referring to array GSM73642 in the GEO dataset. As downregulation of these so-called FOXO genes plays a key role in aberrant upregulation of carbohydrate metabolism in malignancy [15Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism differentiation and transformation Cell 2004; 117: 421-6.], the aim of this study was to find out, if re-activation of these and other genes by epigenetic drugs might induce a downregulation of metabolic pathways to a level known for normal (stem) cells.
This study applies novel analytic tools for exploration of biochemical pathways [16vanIersel MP, Kelder T, Pico AR. Presenting and exploring biological pathways with PathVisio BMC Bioinformatics 2008; 9: 399.-18Pico AR, Kelder T, vanIersel MP. WikiPathways pathway editing for the people PLoS Biol 2008; 6: e184.] from gene expression arrays to visualize expression patterns from metabolic pathways in the KG-1 cell line treated with known epigenetic drugs SAHA and DAC.
2 . MATERIALS AND METHODS
2.1 . Cell Culture
KG-1 cells (obtained from the American Type Culture Collection, ATCC, Manassas, VA) were seeded in petri dishes (8 cm in diameter) or culture flasks in RPMI 1640 supplemented with 5% glutamine, 10% FCS, 100 units/mL penicillin G sodium and 100 µg/ml streptomycin sulphate at a concentration of 300.000 cells/ml medium. Cells were incubated with either 0.5 µM SAHA or 5 µM DAC for 72h in comparison to untreated controls.
2.2 . RNA-Isolation and Definition of Gene-Expression-Profiles
Isolated RNA (using eg the PROMEGA Z-3105 RNA-Isolation system) was used to determine, which genes and associated regulative pathways are affected by treatments with specific drugs including SAHA or desoxy-azacytidine (DAC).
Analysis and data evaluation for such Affymetrix Arrays (Type Human Gene 1.0 ST Array) was commercially obtained from an internationally certified institution (Kompetenzzentrum für Biofluoreszenz, Regensburg). “Pathvisio” [16vanIersel MP, Kelder T, Pico AR. Presenting and exploring biological pathways with PathVisio BMC Bioinformatics 2008; 9: 399., 18Pico AR, Kelder T, vanIersel MP. WikiPathways pathway editing for the people PLoS Biol 2008; 6: e184.] and “iPath” [17Letunic I, Yamada T, Kanehisa M. iPath interactive exploration of biochemical pathways and networks Trends Biochem Sci 2008; 33: 101-3.] software packages were applied for specific analyses of defined pathways from Affymetrix Arrays (Type Human Gene 1.0 ST Array).
2.3 . Calculation of Expression Factor for a Pathway (PEF)
To get a factor, which describes the regulation of a pathway in a qualitative manner, we adapted published algorithms [19Tian L, Greenberg SA, Kong SW. Discovering statistically significant pathways in expression profiling studies Proc Natl Acad Sci USA 2005; 102: 13544-9., 20Su J, Yoon BJ, Dougherty ER. Accurate and reliable cancer classification based on probabilistic inference of pathway activity PLoS One 2009; 4: e8161.] for defining the following formula:
Log2 signals represent the intensity of gene-expression on the Human Gene 1.0 ST Array: Low expression levels are represented by log2signals <5, median basic expression levels have log2signals >5, and high expression levels refer to log2signals. Basic expression levels (BELs) are shown in Suppl. Figs. (1-3). Multiplication of BELs with PEF indicates the effect of each treatment on gene expression.
2.4 . Gene Expression Analysis by qRT-PCR (Quantitative Reverse Transcriptase PCR)
For comparative analyses cDNA was synthesized from 0.1 µg RNA using the 1st Strand cDNA Synthesis Kit as described by the supplier (Roche and/or Promega). The obtained cDNA was subjected to PCR amplification with a real-time cycler (Corbett Research). TaqMan gene expression probes & primers-sets (all from Applied Biosystems) in the respective master mix were used for amplification according to the suppliers suggested conditions. For normalization of expression we used the GAPDH TaqMan primer & probe-set in the same reaction vial (4310884E, Applied Biosystems). Relative quantification of mRNA within the samples was examined using the comparative Ct method:
(ΔCtuntreated control - ΔCttreated cells= ΔΔCt; relative quantity = 2-DDCt) according to Livak & Schmittgen [21Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method Methods 2001; 25: 402-8.].
3 . RESULTS
3.1 . Epigenetic Drugs Downregulate Cell Proliferation
Drug modulated inhibition of cell multiplication was used to find an optimal concentration for drug treatment. For this purpose, cells were seeded and cultured for 72 hours with increasing concentrations of either SAHA or DAC. Fig. (1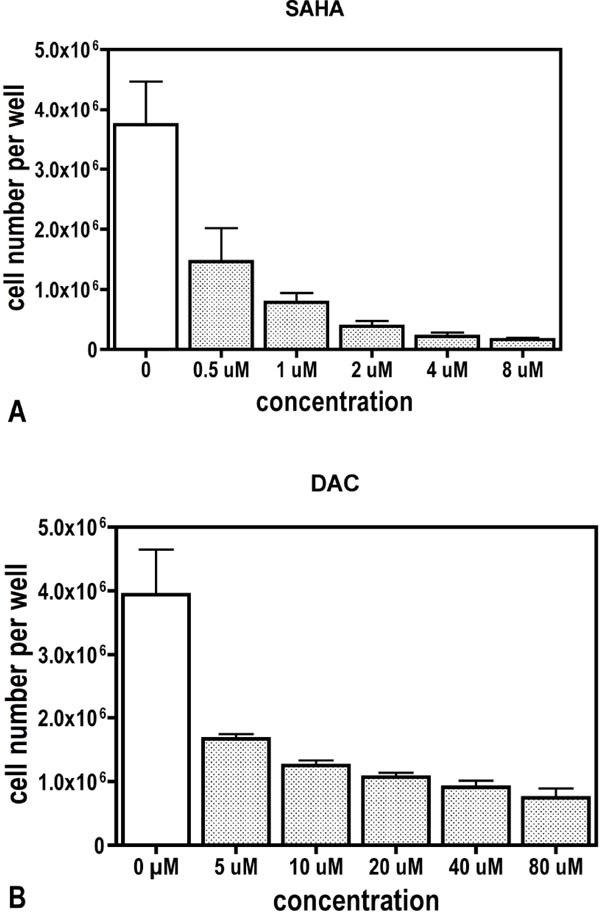 ) demonstrates a dose dependent attenuation of cell multiplication. 0.5 µM and 5 µM, respectively, reduced the proliferation rate to about 40% and were, therefore, choosen for experiments.
) demonstrates a dose dependent attenuation of cell multiplication. 0.5 µM and 5 µM, respectively, reduced the proliferation rate to about 40% and were, therefore, choosen for experiments.
3.2 . Epigenetic Drugs Downregulate Key Pathways of Energy Metabolism
As shown in Tables 1-3, pathways which are associated with cancer-related metabolic dysregulations were targeted by the epigenetic drugs. As a consequence of drug-induced inhibition of glucose-uptake [12Wardell SE, Ilkayeva OR, Wieman HL. Glucose metabolism as a target of histone deacetylase inhibitors Mol Endocrinol 2009; 23: 388-401.], genes regulating catabolism in both cytoplasm as well as in the mitochondria were down-regulated more efficiently by SAHA compared to DAC. Table 1 and Suppl. Fig. (1) demonstrate that of genes involved in glycolysis, 15 of 16 genes whose signal was strong enough to suggest a significant rate of mRNA expression, were down-regulated by SAHA; thereof 12 genes were strictly down-regulated (more than 2-fold), 3 weak (between 1.1-fold and 2-fold) and one was up-regulated. DAC regulated the expression similarily but at the used concentration it was less effective (Table 1, Suppl. Fig. 1). Catabolism is continued in the mitochondria by the enzymes of the “three carbonic acid” (TCA) cycle. In this pathway, 20 of 30 genes responsible for the catabolism of Acetyl-CoA were strongly down-regulated to less than 20% by SAHA, 3 weak or not regulated, and only 6 were up-regulated. Not surprisingly, out of the 6 up-regulated genes, three genes (PDK1, 2, 3) were inhibitors of enzymes catalysing the first step of this cycle (Table 2 and Suppl. Fig. 2). Again, DAC had a weaker effect on gene regulation of those genes that led to the fact that less genes were regulated (19/30). Another pathway which is localized in the mitochondria is the “fatty acid beta oxidation” (FABO), for which 18 genes are suggested (Table 3, Suppl. Fig. 3, [16vanIersel MP, Kelder T, Pico AR. Presenting and exploring biological pathways with PathVisio BMC Bioinformatics 2008; 9: 399., 18Pico AR, Kelder T, vanIersel MP. WikiPathways pathway editing for the people PLoS Biol 2008; 6: e184.]). Twelve genes were strongly down-regulated by SAHA while 5 were weakly attenuated and one was up-regulated. Similiarily as found with the other pathways DAC had a weaker effect on the down-regulation (Table 3 and Suppl. Fig. 3). Evaluation of gene array results with qRT-PCR as shown in Fig. (2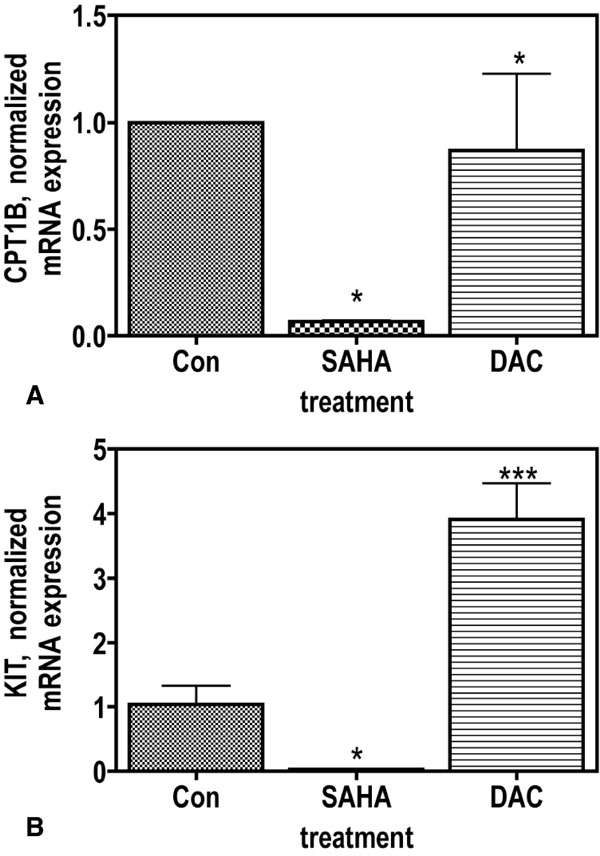 ) analyzing expression of CPT1B by qRT-PCR yielded a comparable result.
) analyzing expression of CPT1B by qRT-PCR yielded a comparable result.
Possibly as a kind of “rescue mechanism” initiated from deprival of energy from glycolysis, the key enzyme of gluconeogenesis, fructose-1,6-bisphosphatase 1 and 2 (FBP1 and FBP2), which is also responsible for prevention of metabolic acidosis during conditions of starving, was stimulated with SAHA and DAC.
Considering glycolysis most of the genes were down regulated by SAHA and a comparable effect was found with
DAC, although, the drug was again less effective. The calculated expression factors for the pathways (PEF) were -1.21 for SAHA and -0.19 for DAC (Table 1 and Suppl. Fig. 1). This was also evident for the TCA circle where PEFs were -1.20 for SAHA and -0.17 for DAC (Table 2 and Suppl. Fig. 2) and for mitochondrial beta oxidation were PEFs were -1.15 for SAHA and -0.23 for DAC (Table 3 and Suppl. Fig. 3).
3.3 . Epigenetic Drugs Target KIT- and FOXO- Associated Pathways in KG-1 Cells
As demonstrated in Table 4a, oncogenic KIT signalling is exclusively suppressed by SAHA as indicated by downregulation of KIT (confirmed by qPCR as seen in Fig. 2 ) and stimulation of its inhibitor PTEN, thus confirming this specific feature of HDAC-inihibitors [22Guo W, Lasky JL, Chang CJ. Multi-genetic events collaboratively contribute to Pten-null leukaemia stem-cell formation Nature 2008.]. The serine /threonine kinase AKT1 regulates both KIT and the family of forkhead transcription factors (FOXO1, 3, 4, Table 4b). The strong stimulation of FOXO resulting from epigenetic downregulation of SIRT1, was also exclusively observable by SAHA [23Gross DN, vandenHeuvel AP, Birnbaum MJ. The role of FoxO in the regulation of metabolism Oncogene 2008; 27: 2320-36.-25Yang Y, Hou H, Haller EM. Suppression of FOXO1 activity by FHL2 through SIRT1-mediated deacetylation Embo J 2005; 24: 1021-32.].
) and stimulation of its inhibitor PTEN, thus confirming this specific feature of HDAC-inihibitors [22Guo W, Lasky JL, Chang CJ. Multi-genetic events collaboratively contribute to Pten-null leukaemia stem-cell formation Nature 2008.]. The serine /threonine kinase AKT1 regulates both KIT and the family of forkhead transcription factors (FOXO1, 3, 4, Table 4b). The strong stimulation of FOXO resulting from epigenetic downregulation of SIRT1, was also exclusively observable by SAHA [23Gross DN, vandenHeuvel AP, Birnbaum MJ. The role of FoxO in the regulation of metabolism Oncogene 2008; 27: 2320-36.-25Yang Y, Hou H, Haller EM. Suppression of FOXO1 activity by FHL2 through SIRT1-mediated deacetylation Embo J 2005; 24: 1021-32.].
3.4 . Comparative Analyses Explain Common Regulatory Mechanisms of SAHA and DAC
Venn diagrams (Fig. 3 ) showed that the majority of significantly (= more than 2-fold) regulated mRNAs were targeted by SAHA: Downregulation (Fig. 3A
) showed that the majority of significantly (= more than 2-fold) regulated mRNAs were targeted by SAHA: Downregulation (Fig. 3A ) was observed in 2651 (pseudo)genes with SAHA as compared to 15 with DAC. Upregulation (Fig. 3B
) was observed in 2651 (pseudo)genes with SAHA as compared to 15 with DAC. Upregulation (Fig. 3B ) was evidenced for 1392 mRNAs with SAHA, but only 60 with DAC.
) was evidenced for 1392 mRNAs with SAHA, but only 60 with DAC.
Considering downregulated genes, there are remarkable common features between DAC and SAHA as shown by the fact that 5/15 DAC-downregulated (pseudo)genes were also downregulated by SAHA (Fig. 3A ) and 37/60 DAC-upregulated (pseudo)genes were also upregulated by SAHA. However, as all of them belong to the large group of small nuclear and nucleolar RNAs, a functional interpretation of this observation awaits to be delineated.
) and 37/60 DAC-upregulated (pseudo)genes were also upregulated by SAHA. However, as all of them belong to the large group of small nuclear and nucleolar RNAs, a functional interpretation of this observation awaits to be delineated.
4 . DISCUSSION
Results of this study show that chromatin-targeting drugs (DAC and SAHA) have the potential to target key metabolic pathways which are tightly associated with malignancy (Tables 1-3 and Suppl. Figs. 1-3). Our data underline that SAHA targets malignancy-associated metabolic control at least at 3 levels, starting from (i) glucose-uptake over (ii) mitochondrial pathways to (iii) AKT-PTEN-FOXO-signaling. DAC-mediated targeting of mitochondrial pathways appears to be related to its apoptosis-promoting activity [26Stresemann C, Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine Int J Cancer 2008; 123: 8-13.] and a slight stimulation of forkhead transcription factors by DAC may be promoted by demethylation of these hypermethylated genes [14Gebhard C, Schwarzfischer L, Pham TH. Genome-wide profiling of CpG methylation identifies novel targets of aberrant hypermethylation in myeloid leukemia Cancer Res 2006; 66: 6118-28.].
Considering the crucial role of targeting these pathways in treatment of cancers and leukemias [11Gillies RJ, Robey I, Gatenby RA. Causes and consequences of increased glucose metabolism of cancers J Nucl Med 2008; 49( Suppl 2): 24S-42S., 12Wardell SE, Ilkayeva OR, Wieman HL. Glucose metabolism as a target of histone deacetylase inhibitors Mol Endocrinol 2009; 23: 388-401., 27Gogvadze V, Orrenius S, Zhivotovsky B. Mitochondria as targets for cancer chemotherapy Semin Cancer Biol 2009; 19: 57-66.], this may provide an explanation for the clinical efficiency of those drugs, and may indicate, besides their known effects on epigenetic re-activation of (so-called tumor suppressor) genes involved in regulation of cell cycle and apoptosis [3Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat development of this histone deacetylase inhibitor as an anticancer drug Nat Biotechnol 2007; 25: 84-90., 4Momparler RL. Epigenetic therapy of cancer with 5-aza-2'-deoxycytidine (decitabine) Semin Oncol 2005; 32: 443-51.], the systemic consequences of epigenetic regulations.
The observed downregulation of energy metabolism may also be related to inhibition of cytoplasmic genes encoding mitochondrial proteins, as prominently observed with epigenetic drugs (Table 1 and Suppl. Figs. 1-3). Recent reviews indicate that mitochondria also play a key role in treatment-resistance of many cancers, thus confirming their increasing attractiveness as targets in cancer-chemotherapy [27Gogvadze V, Orrenius S, Zhivotovsky B. Mitochondria as targets for cancer chemotherapy Semin Cancer Biol 2009; 19: 57-66.], in addition to their known role in energy metabolism [28Weinberg F, Chandel NS. Mitochondrial metabolism and cancer Ann N Y Acad Sci 2009; 1177: 66-73.]. Mitochondrial energy genes are dispersed across the chromosomes plus the maternally inherited mitochondrial DNA. Moreover, the cells and tissues most affected by aging and malignancy are those most reliant on mitochondrial energy. The mitochondrion lies at the interface between environmental calories and human physiology, as well as metabolic diseases. Both the signal transduction pathways and the epigenome are regulated by ATP mediated protein phosphorylation, acetylation via acetyl-CoA, and methylation by S-adenosylmethionine, all driven by mitochondrial high energy substrates modulated by available energy. Furthermore, mitochondrial redox chemistry regulates reactive oxygen production and thiol/disulfide chemistry and these also regulate cellular signaling and function.
Drug-induced downregulation of glycolysis was associated with stimulation of energy storage by gluconeogenesis (Table 1 and Suppl. Fig. 1). Enhanced gluconeogenesis is a hallmark of aging eukaryotic cells [29Lin SS, Manchester JK, Gordon JI. Enhanced gluconeogenesis and increased energy storage as hallmarks of aging in Saccharomyces cerevisiae J Biol Chem 2001; 276: 36000-7.], but may also be a “rescue-mechanism” in case of starvation. In mammals, a fasting-inducible switch consisting of the histone-acetyltransferase p300 (EP300) and the nutrient-sensing deacetylase sirtuin1 (SIRT1) which is also targeted by SAHA, is essential for maintenance of energy balance during fasting by activation of gluconeogenesis [25Yang Y, Hou H, Haller EM. Suppression of FOXO1 activity by FHL2 through SIRT1-mediated deacetylation Embo J 2005; 24: 1021-32., 30Liu Y, Dentin R, Chen D. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange Nature 2008; 456: 269-73.].
From a therapeutic standpoint, the increasing knowledge on acetylation-dependent mechanisms of FOXO regulation expands the potential repertoire of pharmacological HDAC inhibitors [23Gross DN, vandenHeuvel AP, Birnbaum MJ. The role of FoxO in the regulation of metabolism Oncogene 2008; 27: 2320-36., 31Burgering BM, Medema RH. Decisions on life and death: FOXO Forkhead transcription factors are in command when PKB/Akt is off duty J Leukoc Biol 2003; 73: 689-701.-33Hedrick SM. The cunning little vixen: Foxo and the cycle of life and death Nat Immunol 2009; 10: 1057-63.]. Yang et al. [25Yang Y, Hou H, Haller EM. Suppression of FOXO1 activity by FHL2 through SIRT1-mediated deacetylation Embo J 2005; 24: 1021-32.] showed that SIRT1 downregulates FOXO by deacetylation. Results of this study showed that three FOXO genes (FOXO1, FOXO3 and FOXO4 are significantly stimulated when SIRT1 was inhibited in KG-1 cells with SAHA (Table 4).
Although, the present report appears to be the first study, showing the metabolic effects of epigenetically active drugs, downregulatory effects on carbohydrate metabolism have been expected both with SAHA and DAC as a consequence of induced mitochondrial damage [34Chandra J. Oxidative stress by targeted agents promotes cytotoxicity in hematologic malignancies Antioxid Redox Signal 2009; 11: 1123-37.-37Almenara J, Rosato R, Grant S. Synergistic induction of mitochondrial damage and apoptosis in human leukemia cells by flavopiridol and the histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA) Leukemia 2002; 16: 1331-43.]. In addition, both these pharmacologic compounds are also known for their antiproliferative effects resulting from cell cycle inhibition [3Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat development of this histone deacetylase inhibitor as an anticancer drug Nat Biotechnol 2007; 25: 84-90., 4Momparler RL. Epigenetic therapy of cancer with 5-aza-2'-deoxycytidine (decitabine) Semin Oncol 2005; 32: 443-51.].
The mechanism for the antiproliferative effect of SAHA which targets the majority of expressed mRNAs as seen in the VENN diagrams (Fig. 3 ) is believed to be the result of inhibition of HDAC activity, resulting in the accumulation of acetylated proteins, including histones. Inhibition of HDAC activity by SAHA has multiple cellular effects. These effects include an alteration in the transcription of a finite number of genes (2–5% of expressed genes) via acetylation of histones and transcription factors, as well as effects such as cell cycle arrest via inhibition of mitosis. As an antimetabolite DAC also induces cell cycle arrest by stimulating cell cycle inhibitors as well as by inhibiting DNA-repair in the G2 phase of the cell cycle because of its structural similarity to desoxycytosine [26Stresemann C, Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine Int J Cancer 2008; 123: 8-13.].
) is believed to be the result of inhibition of HDAC activity, resulting in the accumulation of acetylated proteins, including histones. Inhibition of HDAC activity by SAHA has multiple cellular effects. These effects include an alteration in the transcription of a finite number of genes (2–5% of expressed genes) via acetylation of histones and transcription factors, as well as effects such as cell cycle arrest via inhibition of mitosis. As an antimetabolite DAC also induces cell cycle arrest by stimulating cell cycle inhibitors as well as by inhibiting DNA-repair in the G2 phase of the cell cycle because of its structural similarity to desoxycytosine [26Stresemann C, Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine Int J Cancer 2008; 123: 8-13.].
Taken together, epigenetic drugs (especially SAHA) have the potential to simulate a situation of caloric restriction by (i) inhibiting glucose uptake, which results in (ii) a downregulation of carbohydrate catabolism, which is further stimulated by mitochondrial damage as evidenced by a downregulated beta oxidation and (iii) a stimulation of FOXO transcription factors as a consequence of downregulated SIRT and AKT resulting from SAHA-mediated HDAC-inhibition and possibly also by demethylation of these hypermethylated forkhead genes [14Gebhard C, Schwarzfischer L, Pham TH. Genome-wide profiling of CpG methylation identifies novel targets of aberrant hypermethylation in myeloid leukemia Cancer Res 2006; 66: 6118-28.]. In addition to these 3 effects on the transcriptome-level, there is a 4th effect on the proteome-level as shown by regulation of STAT (signal transducer and activator of transcription). Signaling by STAT1 requires HDAC-activity [38Klampfer L, Huang J, Swaby LA. Requirement of histone deacetylase activity for signaling by STAT1 J Biol Chem 2004; 279: 30358-68.]. Recent data indicate that STAT1- dependence of energy-metabolism links tumour growth and radioresistance to the Warburg effect [39Pitroda SP, Wakim BT, Sood RF. STAT1-dependent expression of energy metabolic pathways links tumour growth and radioresistance to the Warburg effect BMC Med 2009; 7: 68.].
Thus, our data confirm epigenetic effects on differentiation by re-activation of tumor-suppressors in leukemic cells [3Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat development of this histone deacetylase inhibitor as an anticancer drug Nat Biotechnol 2007; 25: 84-90., 4Momparler RL. Epigenetic therapy of cancer with 5-aza-2'-deoxycytidine (decitabine) Semin Oncol 2005; 32: 443-51., 40Dore BT, Chomienne C, Momparler RL. Effect of 5-aza-2'-deoxycytidine and vitamin D3 analogs on growth and differentiation of human myeloid leukemic cells Cancer Chemother Pharmacol 1998; 41: 275-80.-42Momparler RL, Dore BT, Momparler LF. Effect of 5-aza-2'-deoxycytidine and retinoic acid on differentiation and c-myc expression in HL-60 myeloid leukemic cells Cancer Lett 1990; 54: 21-8.]. SAHA and DAC are well established for therapy protocols in myelodysplastic syndromes and leukemias (e.g. [5Gurion R, Vidal L, Gafter-Gvili A. 5-azacitidine prolongs overall survival in patients with myelodysplastic syndrome - systematic review and meta-analysis Haematologica 2010; 95(2 ): 303-10., 43Leone G, Voso MT, Teofili L. Inhibitors of DNA methylation in the treatment of hematological malignancies and MDS Clin Immunol 2003; 109: 89-102.-45Gore SD, Hermes-DeSantis ER. Future directions in myelodysplastic syndrome newer agents and the role of combination approaches Cancer Control 2008; 15 ( Suppl): 40-9.]). However, it is intriguing that additive or synergistic effects against tumor cell growth have been observed in most reported preclinical studies where SAHA was combined with chemotherapeutic agents or radiation [45Gore SD, Hermes-DeSantis ER. Future directions in myelodysplastic syndrome newer agents and the role of combination approaches Cancer Control 2008; 15 ( Suppl): 40-9., 46Zhang G, Park MA, Mitchell C. Vorinostat and sorafenib synergistically kill tumor cells via FLIP suppression and CD95 activation Clin Cancer Res 2008; 14: 5385-99.].
Since these compounds are coming to the fore as antineoplastic agents, the present findings raise the question of whether they could be pressed into service against the raising tide of metabolic diseases and the associated risks for malignancies [47Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases aging and cancer a dawn for evolutionary medicine Annu Rev Genet 2005; 39: 359-407., 48Coughlin SS, Calle EE, Teras LR. Diabetes mellitus as a predictor of cancer mortality in a large cohort of US adults Am J Epidemiol 2004; 159: 1160-7.]. Furthermore, data presented in this study indicate that novel tools have the potential to illustrate the multitargeted effects of epigenetic drugs in treatment of malignancies including leukemias. Visualization of drug-effects on such complex processes requires a paradigm-change from „single-parameter-analyses“ to comparative evaluation of pathways, thus illustrating the innovative potential of the present study.
SUPPLEMENTARY MATERIAL
This article also contains supplementary material and it can be viewed online along with the article at publisher’s website.
ACKNOWLEDGEMENT
This work was funded by “Jubiläumsfonds der Österreichischen Nationalbank” (Project No. 13068) and “Ludwig Boltzmann Gesellschaft”.