- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Medical Informatics Journal
(Discontinued)
ISSN: 1874-4311 ― Volume 13, 2019
A Scalable Architecture for Incremental Specification and Maintenance of Procedural and Declarative Clinical Decision-Support Knowledge
Avner Hatsek*, 1, Yuval Shahar 1, Meirav Taieb-Maimon 1, Erez Shalom 1, Denis Klimov 1, Eitan Lunenfeld 2
Abstract
Clinical guidelines have been shown to improve the quality of medical care and to reduce its costs. However, most guidelines exist in a free-text representation and, without automation, are not sufficiently accessible to clinicians at the point of care. A prerequisite for automated guideline application is a machine-comprehensible representation of the guidelines. In this study, we designed and implemented a scalable architecture to support medical experts and knowledge engineers in specifying and maintaining the procedural and declarative aspects of clinical guideline knowledge, resulting in a machine comprehensible representation. The new framework significantly extends our previous work on the Digital electronic Guidelines Library (DeGeL) The current study designed and implemented a graphical framework for specification of declarative and procedural clinical knowledge, Gesher. We performed three different experiments to evaluate the functionality and usability of the major aspects of the new framework: Specification of procedural clinical knowledge, specification of declarative clinical knowledge, and exploration of a given clinical guideline. The subjects included clinicians and knowledge engineers (overall, 27 participants). The evaluations indicated high levels of completeness and correctness of the guideline specification process by both the clinicians and the knowledge engineers, although the best results, in the case of declarative-knowledge specification, were achieved by teams including a clinician and a knowledge engineer. The usability scores were high as well, although the clinicians’ assessment was significantly lower than the assessment of the knowledge engineers.
Article Information
Identifiers and Pagination:
Year: 2010Volume: 4
First Page: 255
Last Page: 277
Publisher Id: TOMINFOJ-4-255
DOI: 10.2174/1874431101004010255
Article History:
Received Date: 11/8/2009Revision Received Date: 16/7/2010
Acceptance Date: 6/8/2010
Electronic publication date: 14/12/2010
Collection year: 2010
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Department of Information System Engineering, Medical Informatics Research Center, Ben Gurion University of the Negev, P/O/B/ 653, Beer Sheva, 84105, Israel; Tel: 97286477160; Fax: 97286477161; E-mail hatsek@bgu.ac.il
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 11-8-2009 |
Original Manuscript | A Scalable Architecture for Incremental Specification and Maintenance of Procedural and Declarative Clinical Decision-Support Knowledge | |
1. INTRODUCTION
Clinical guidelines are evidence-based recommendations for diagnosing and/or treating patients with certain medical conditions. When adapted and implemented by medical care providers, clinical guidelines have been shown to improve the quality of medical care and to reduce its costs [1Grimshaw JM, Russel I T. Effect of clinical guidelines on medical practice: A systematic review of rigorous evaluations Lancet 1993; 342: 1317-22.-4Quaglini S, Cavallini A, Gerzeli S, Micieli G. Economic benefit from clinical practice guideline compliance in strok patient management Health Policy 2004; 69: 305-15.]. However, most clinical guidelines published by professional associations around the world exist only in a free-text representation and are not sufficiently accessible to clinicians at the point-of-care. Applying clinical guidelines using computerized methods for guideline retrieval, display and automatic application as part of computer-based clinical decision support (CDS) systems is expected to increase their utilization and thus improve the quality of medical care. A prerequisite for automated guideline application, however, is a machine-comprehensible (formal) representation of the guidelines.
The task of representing clinical guidelines in a machine-comprehensible format is complicated and has many different aspects. In the background section of the paper we summarize different approaches and studies in this field. These approaches often successfully solve a subset of the problems concerning guideline specification but typically do not address the full scope of these problems. The heart of the problem of creating and maintaining a scalable repository of formal medical knowledge is that, on the one hand, clinicians cannot and need not program in formal specification languages. On the other hand, computer programmers and knowledge engineers do not completely understand the semantics of medical knowledge and procedures. For both types of experts, time is limited and expensive.
In addition, another issue that requires special care when representing the knowledge embodied by clinical guidelines is that clinical guidelines include both procedural knowledge, such as workflows, as well as declarative knowledge, such as definition of clinically meaningful abstract terms and temporal patterns. Each type of knowledge requires a unique methodology and set of tools for its acquisition and specification, but both need to be integrated within the same, well-encapsulated guideline, while still supporting the option for reuse of the knowledge by future guidelines.
In our previous work, which we describe in detail in the Background section, we have proposed, in order to support an incremental guideline-specification process, a hybrid guideline-representation format, which includes several increasingly formal representation levels, from a semi-structured format, through a semi-formal representation, to a formal, machine-executable representation. We have implemented this concept in the Digital electronic Guideline Library (DeGeL) architecture [5Shahar Y, Young O, Shalom E, et al. A hybrid, multiple-ontology framework for specification and retrieval of clinical guidelines J Biomed Inform 2004; 37(5): 325-44.]. We have also evaluated in detail the ability of clinicians to understand the semi-structured and semi-formal formats and to use them to represent clinical guidelines [6Shalom E, Shahar Y, Taieb MM, et al. A quantitative evaluation of a methodology for collaborative specification of clinical guidelines at multiple representation levels J Biomed Inform 2008; 41(6): 889-903., 7Shalom E, Shahar Y, Taieb MM, et al. Ability of expert physicians to structure clinical guidelines: reality versus perception J Eval Clin Pract 2009 Dec; 15(6): 1043-53.]. However, the necessity for a comprehensive methodology for graphical guideline specification from free text down to the fully formal representation format, and the need for integration of the procedural and declarative aspects of the knowledge embodied by clinical guidelines, needed further addressing.
In the current study, we start by describing our design and implementation of a new knowledge-specification platform, called Gesher. We then focus on describing the detailed evaluation of three important services of that platform: Specification of procedural clinical knowledge, specification of declarative clinical knowledge, and exploration of the knowledge embodied by a given clinical guideline.
2. . OBJECTIVES
The objectives of the current study were to address the main issues that remain unresolved by existing guideline-specification approaches (as we describe in detail in the Background section) and to evaluate the proposed solutions. Thus, we focused on the following four main objectives:
- Develop a Central Repository for Hybrid Clinical Guidelines. The first objective of this study was to develop an integrated framework that supports all of the steps for an incremental guideline specification process. To support the basic requirements of sharing and reusing the formally represented medical knowledge and to provide a scalable infrastructure, the framework should include a central knowledge repository for storing, searching and retrieving guidelines.
- Propose a Methodology for Guideline Specification that Supports Collaboration between Expert Physicians and Knowledge Engineers. The second objective was to propose a comprehensive methodology for specifying clinical guidelines at all guideline-representation levels. The proposed methodology should take into consideration the collaboration of different types of participants, such as expert physicians, clinical [knowledge] editors, and knowledge engineers, all of whom are necessary for achieving a complete and valid guideline specification. The methodology should also address the required tasks of localization and customization of the clinical guidelines according to the special requirements of each specific clinical organization.
- Develop a Graphical Framework for Clinical Guideline Specification at Multiple Representation Levels. To support the incremental knowledge specification process, it is necessary to provide expert physicians, clinical editors, and knowledge engineers with usable tools for performing the gradual and methodological process of clinical guideline specification.
- Evaluate the Knowledge Specification Framework and the Suggested Methodology. Another major objective of this study was to evaluate the developed framework and to assess the quality of the specification products, the time required for completing the specification task, and the usability of the guideline specification tools. We wanted to assess these features in detail for both the declarative and procedural aspects of guideline-based knowledge. Finally, we wished to assess the functionality and usability of the knowledge browsing tool for clinical guidelines.
3. BACKGROUND
Over the past 20 years there have been many efforts to provide automated support to evidence-based medicine by formalizing guidelines into machine-interpretable formats that can be applied within CDS automated systems. Several guideline-specification ontologies that were developed represent guidelines in a formal and machine-interpretable format. A comprehensive comparison between most of those approaches can be found in [8Peleg M, Tu S, Bury J, et al. Comparing computer-interpretable guideline models: a case-study approach J Am Med Inform Assoc 2003; 10(1): 52-68., 9De Clercq P, Blom J, Korsten HHM , Hasman A. Approaches for creating computer-interpretable guidelines that facilitate decision support Artif Intell Med 2004; 31(1): 1-27.]. Existing major approaches for formally representing and applying guidelines vary in terms of the goals they were designed to achieve, in their representation model and in the knowledge specification tools they provide. To summarize these approaches and to understand some of the unresolved issues in the field, we describe them here divided into two groups: model- and document-centric. The model-centric approaches (e.g. PROforma, GLARE, GASTON, and GLIF) emphasize the formal representation of guidelines and aim for guideline application by runtime engines. The document-centric approaches (e.g. GEM, HGML)stress the original textual document and provides tools mainly for converting the guideline text into a structured representation, for such tasks as guideline retrieval, verification and presentation.
It should be noted that both approaches, the model-centric and to a lesser extent the document centric, imply predefined underlying guideline ontology based on a set of knowledge-roles, such as eligibility-conditions, exit-conditions, process objectives, and outcome goals. These knowledge-roles require explicit modeling and typically cannot be accommodated by a standard rule-based approach in which all rules are handled equally with uniform semantics. Furthermore, experience has shown that large rule-based systems become unwieldy and adding rules might actually degrade overall system performance due to the lack of an underlying, explicit semantics regarding the role of each rule [10Soloway E, Bachant J, Jensen K. Assessing the maintainability of XCON-in-RIME: coping with the problems of a very large rule-base AAAI-87 Proceedings 1987; 824-9., 11Stefik M. Introduction to Knowledge Systems. San Fransisco, California: Morgan Kaufmann 1995; pp. 626-35.]. Since the objectives of guideline specification also facilitate maintenance as knowledge is updated, a common phenomenon in medical domains, there is significant advantage in having clear underlying guideline ontology.
3.1. The Model-Centric Approaches
This group [12Svatek V, Ruzicka M. Step-by-step mark-up of medical guideline documents Int J Med Inform 2003; 70: 329-5.] comprises all the frameworks that were designed to achieve automatic guideline application within CDS systems. Most of these frameworks include sophisticated tools for specifying guidelines to create a formal format according to their underlying representation model. Since these methods do not attribute much importance to directly relating the formal representation to the original free-text, they often do not maintain references to the original text source. Many of the tools that these frameworks provide for guideline specification are based on the Protégé knowledge acquisition framework and are designed for use by knowledge engineers who are familiar with the complex structures of the underlying formal representation ontologies. None of these approaches suggests a gradual specification methodology that supports collaboration with medical experts who are not medical-informaticians. The collaboration of medical experts, however, is crucial for achieving a correct specification that is customized to local applications in specific medical centers. Most of the model-centric methods also do not include either a central digital repository with tools for guideline storage, retrieval, sharing, and versioning or a formal authorization model that determine access for various knowledge-base functionalities. Table 1 summarizes these approaches.
3.2. The Document-Centric Approaches
The document-centric approaches include frameworks that regard the original free-text guidelines, as the starting point of the specification and use it in the specification process. Several of these methods present and share the guideline's knowledge in a more structured, but still textual-based representation. The objective of some other document-centric approaches is to extract a more formal representation of the guideline knowledge in order to verify the guideline or as a later step towards formal specification. Table 2 summarizes these approaches.
3.3. Unresolved Issues in Guideline Specification
When examining existing approaches for automated specification and application of clinical guidelines, there are several unresolved issues. To achieve automated application of the knowledge of clinical guidelines, and embed it within automated CDS systems, it is necessary to create a formal model of the guideline, resolving ambiguities in the free-text representation and specifying relevant new knowledge that is only implicit in the original, free-text clinical guideline sources. To create a reliable specification based on evidence-based recommendations, and to enable updating of the formal representation of the guideline when new versions of the textual source are being published, it is necessary to maintain references from the formal model to the source text and vise-versa. None of the existing methods mentioned above fully supports both of these requirements. In addition, it is also necessary to integrate the guideline specification and runtime application tools with a central repository that makes it possible to share the guidelines as well as to control and restrict their access and to provide tools for reproducing and versioning existing guidelines. Most of the existing methods do not suggest an overall methodology for guideline specification, from its free-text source to a fully formal representation. In particular, such a methodology must support a collaboration of the knowledge engineers with the expert physicians, a collaboration that is crucial for achieving a complete and correct specification. Several additional unresolved issues are related to methods for supporting specification into multiple guideline representation languages and the explicit handling of procedural versus declarative knowledge.
In our research, we propose an overall methodology for the guideline specification process and provide a new framework to support the steps of this methodology. This framework also includes a central knowledge library and a graphical knowledge specification tool. As explained later in this paper, some aspects of this process are not specific to specific guideline ontology. For the formal representation format, we used the Asbru ontology [21Shahar Y, Miksch S, Johnson P. The Asgaard project: A task-specific framework for the application and critiquing of time-oriented clinical guidelines Artif Intell Med 1998; 14(1-2): 29-51.-23Seyfang A, Miksch S, Marcos M. Combining diagnosis and treatment using asbru Int J Med Inform 2002; 68(1): 49-57.], which is an expressive guideline-specification formal language, whose focus is on representing explicit intentions for the process and outcome in order to better support the quality assessment tasks. Asbru can be used to design specific plans as well as to support the performance of different reasoning and executing tasks. Asbru also provides a powerful mechanism to express extended time-oriented actions and plans caused by extended time- oriented states of an observed agent (e.g., many actions and plans need to be executed in parallel or at a very particular time point). These plans, which are combined with the intentions of the executing agent of the plan, are uniformly represented and organized in the guideline-specification library. During the execution phase, an applicable plan is instantiated with distinctive arguments and state-transition criteria are added to execute and reason about different tasks. Asbru is unique in its ability to represent explicitly different aspects of the guideline, each of which is useful to one or more guideline-support tasks and the computational mechanisms that perform these tasks.
In the following sections, we describe our approach and the tools we implemented. We also present three experiments that evaluate the functionality and usability of our approach's procedural knowledge specification, its declarative knowledge specification, and the knowledge exploration services it offers.
4. METHODS
Our study included four main phases: development of the infrastructure, including a central guideline repository; development of an incremental guideline specification methodology; development of graphical tools that support each step of this specification methodology; and performance of an evaluation of the knowledge specification tools.
4.1. A Central Repository for Hybrid Clinical Guidelines
To support the need for sharing and reusing medical knowledge, possibly represented in multiple formats, as well as the need for gradual specification of the knowledge, we implemented a new version of our Digital Guidelines Library (DeGeL), which had been developed previously [5Shahar Y, Young O, Shalom E, et al. A hybrid, multiple-ontology framework for specification and retrieval of clinical guidelines J Biomed Inform 2004; 37(5): 325-44.]. Since, as explained in the introduction, the DeGeL framework supports a hybrid, multiple representation-levels model; it enabled us to support a gradual specification process that combines and benefits from both the model-centric and from the document-centric approaches. The earlier version of the DeGeL framework also included a Web-based guideline-specification tool, a guideline-indexing tool, a concept-based and context-sensitive guideline search engine [38Moskovitch R, Martins SB, Behiri E, Weiss A, Shahar Y. Comparative Evaluation of Full-text, Concept-based, and Context-sensitive Search J Am Med Inform Assoc 2007; 14(2): 164-74., 39Moskovitch R, Shahar Y. Vaidurya: A multiple-ontology, concept-based, context-sensitive clinical-guideline search engine J Biomed Inform 2009; 42(1): 11-21.], an interface for visualizing the guideline search results, and a role-based access control (RBAC) authorization model.
The hybrid guideline representation model is a representation that includes several intermediate, increasingly formal formats. All intermediate and final formats are stored within the knowledge-base and include: (1) the original full text; (2) a structured-text representation (marked-up text); (3) a semi-formal representation that includes control structures such as sequential or parallel ordering of sub-plans; and (4) a fully formal, machine-comprehensible format.
The DeGeL library supports multiple guideline ontologies for representing guidelines. Each of these ontologies consists of knowledge-roles, which are semantic fields within the ontology such as, for example, “Eligibility Conditions”. DeGeL provides a hybrid meta-ontology that supports knowledge roles common to all specific guideline ontologies, such as classification axes by which the guideline can be indexed. In contrast to many of the existing guideline ontologies, such as GLIF or GEM, DeGeL's hybrid meta-ontology distinguishes between two major components: documentation ontology and a specification meta-ontology.
The documentation ontology includes documentary knowledge roles that are common to all guideline ontologies. The guideline's title, authors and semantic classification indices are examples of common elements. The documentation ontology distinguishes between source (free-text) guidelines and hybrid (structured at one or more levels) guidelines and provides different documentation elements for each of these guideline types. Source guidelines are stored as free-text (HTML) documents while hybrid guidelines are the products of the specification process. The knowledge roles of the documentation ontology were created according to knowledge roles existing in other guideline ontologies, for example, knowledge-roles describing the guideline's identity (e.g., title, date of publication, date of last revision); knowledge roles describing the guideline developers (e.g., developer name, committee name); and knowledge roles describing the guideline quality (e.g. strength of recommendation, level of evidence). A detailed description of the knowledge roles existing in several guideline ontologies can be found at [34Shiffman RN, Michel G, Essaihi A, Thornquist E. Bridging the guideline implementation gap: a systematic, document-centered approach to guideline implementation J Am Med Inform Assoc 2004; 11(5 ): 418-26.], where the GEM ontology is described in detail and compared to other ontologies. Although DeGeL's documentation ontology includes most of GEM's documentary knowledge roles, it is important to mention that the ontology can be easily extended and the change will be immediately reflected in the guidelines library (i.e., all existing and new guidelines will be extended with elements to retain the new knowledge roles).
The specification meta-ontology defines multiple target specification ontologies (e.g. GLIF, Asbru) that can be used for guideline representation. It enables knowledge engineers to structure the guideline ontology (i.e., when adding a new ontology and when maintaining an existing one). The meta-ontology makes the following assumptions: the specification ontology consists of a hierarchical structure of plans and sub-plans as is common in all major ontologies (e.g., Asbru, GLIF, Prodigy, Proforma and others); several action and plan types exist, such as "medication" or "procedure," which are also common to all ontologies; and each guideline (which is composed from multiple plans and sub-plans) can relate to multiple declarative knowledge elements that describe the medical concepts.
When defining a new ontology within the DeGeL knowledge base, it will inherit the mentioned elements, which will then be extended to include the specific knowledge-roles of the new target ontology. For example, to define the Asbru ontology, we used DeGeL's tools to define the structure of the knowledge roles of this ontology, including the plan's conditions (e.g., filter, setup, abort); the plan-body; the plan intentions; (e.g., process, outcome); the actors; and the plan's effects, preferences and clinical-settings.
According to the hybrid representation model, when specifying new guideline plans into the library, each of the knowledge-roles of the target ontology can be specified according to the intermediate and formal levels of representation. In the following paragraph we clarify the hybrid representation model and describe in more detail the various levels of representation and the motivation behind each.
The Structured-Text representation level is achieved by textually describing the knowledge-role of a specific plan. For example, the filter condition of a guideline for the treatment of hypertension, can be specified with the following text "Adult patient 18 years and over, with blood pressure of more than 140 systolic or more than 90 diastolic". This textual annotation can completely originate from the source guideline (in this case, back pointers to the source can be mentioned) or may be created by the medical expert. There are three major benefits in providing this level of representation: (1) it can be created by the clinical editor or the medical expert without any knowledge about any formal specification language; (2) it clarifies the definition to the knowledge engineer who will be responsible for describing this annotation in a formal representation; and (3) it can be used by the context-sensitive search engine that can retrieve guidelines according to textual queries which are computed according to the guidelines text, but within specific knowledge-roles that can be selected by the users.
The Semi-Formal representation level is achieved by describing the knowledge in a more detailed manner that includes major elements from specific target ontology. Thus, the semi-formal representation, which has a different schema for each of the guideline ontologies, is determined by the knowledge engineers when deciding to support semi-formal representation for that ontology. The major motivation of providing semi-formal representation is to support a gradual representation process where the medical experts can take part in the more formal representation, but will not have to understand the complex structure of the formal specification language. Another benefit of this kind of representation is that it can support semi-automatic application of the guideline for guideline simulation, verification or even at the point-of-care in scenarios where an electronic medical record (EMR) is not available [40Young O, Shahar Y, Lunenfeld E, et al. Runtime application of hybrid-Asbru clinical guidelines J Biomed Inform 2007; 40: 507-26.].
The Formal representation level is achieved by a complete specification of the knowledge according to syntax and semantics of the guideline ontology. A fully formal representation of the guideline will include a standard description of the knowledge elements within the guideline according to terms from standard vocabularies (such as SNOMED, ICD, LOINC), and include all necessary mappings between the knowledge items of the guideline to the data items within a specific EMR. This makes it possible to apply the knowledge of different types of application engines that achieve several tasks such as point-of-care recommendations, point-of-care critiquing and retrospective quality assessment. The motivation behind this level of representation is clear; usually it will be specified by a knowledge engineers assisted by a clinical editor.
4.1.1. DeGeL-II: A New Version of the DeGeL Library
To improve the Web-based architecture of the previous version of DeGeL, we developed a new version (DeGeL-II) of the digital library. When designing the new version, our goal was to create a distributed, Web service- based, open architecture implementation according to the service oriented architecture (SOA) [41Michael N, Munindar P. Service-Oriented Computing: Key Concepts and Principles IEEE Internet Comput 2005; 09(1): 75-81.] design specification. The SOA architecture, which is also used by other initiatives such as SEBASTIAN [32Kawamoto K, Lobach DF. Design, implementation, use, and preliminary evaluation of SEBASTIAN, a standards-based web service for clinical decision support Proc AMIA Annu Symp 2005; 380-4.], SAGE [29Ram P, Berg D , Tu S , et al. Executing clinical practice guidelines using the SAGE execution engine Medinfo 2004; 11: 251-5.] SANDS [42Wright A, Sittig F D. SANDS: An Architecture for Clinical Decision Support in a National Health Information Network Proc AMIA Annu Symp 2007; 816-20.] and CDS consortium [43Middleton B. The clinical decision support consortium Stud Health Technol Inform 2009; 150: 26-30.], has the power to distinguish between different parts of the system such as the knowledge repository, the knowledge specification tools and the run-time applications engines.
This new design provides the ability to develop a suite of tools for guideline specification, retrieval and application. These tools use the services of the central digital repository but are not dependent on it. The open architecture may also host alternative tools for guideline specification and application. DeGeL-II's server allows development of rich client tools by using Web-service methods to retrieve and edit guidelines in the knowledge-base.
The internal architecture of DeGeL-II server was assembled from the following five modules: (1) a guideline database that contains the overall schema to support the hybrid multiple ontology representation; (2) a module responsible for content management; (3) a new guideline search engine that replaces the previous search engine [39Moskovitch R, Shahar Y. Vaidurya: A multiple-ontology, concept-based, context-sensitive clinical-guideline search engine J Biomed Inform 2009; 42(1): 11-21.] and supports full-text, context-sensitive and concept-based searches for enhanced guideline retrieval; (4) an authorization and authentication module that supports the group-based authorization model; and (5) a Web-service API that enables the guideline knowledge-base server to accept client requests and to orchestrates multiple steps in order to perform the requested transactions.
As DeGeL supports storing and retrieving guidelines from multiple ontologies, it uses several standards to represent the specification languages. All entities within DeGeL, such as documentation ontologies, specification ontologies and guidelines are stored within a central database using a relational schema. The schema allows the library administrators to define a hierarchical structure for each of the ontologies. This hierarchical structure is used to represent the ontology's hierarchy of knowledge-roles. For example, the Asbru ontology includes hierarchy nodes for the guideline's intentions, plan-body, effect and conditions. The conditions node includes sub-nodes for filter, setup, complete, abort and other conditions. When a new guideline is created the schema stores data to describe each of its plans, which can have details according to all knowledge roles at multiple representation levels. The internal structure of the data that is stored in each knowledge role is defined by the Semi-Formal and Formal schemas of each specification language. Each specification language is defined by an XML schema that is used by the knowledge specification tool to validate the content of the knowledge roles.
In addition to the library server, we have developed several external tools to administer and maintain the guideline library. These tools include: (1) An ontology-builder tool for the specifying and maintaining the hybrid ontologies stored within the knowledge-base server. (2) An authorization-specification tool which was developed to allow the library administrators to create and manage groups of users with different profiles consisting of a set of library roles. (3) An axes-builder tool which was developed to create and maintain the semantic axes of medical concepts used to classify (index) guidelines. These semantic axes are needed to support the enhanced concept-based retrieval mechanism of the guideline search engine.
The two main applications that are part of the overall architecture and use the library server are the guideline runtime application engine [40Young O, Shahar Y, Lunenfeld E, et al. Runtime application of hybrid-Asbru clinical guidelines J Biomed Inform 2007; 40: 507-26.], which is outside of the scope of the current paper and the guideline specification platform, Gesher, which was developed in the current study and is described in detail in the following sections.
4.2. Methodology and Graphical Framework for a Gradual Specification Process
To achieve a high quality formal representation of medical knowledge, the specification process should be performed gradually, in a methodological fashion, and be supported by graphical tools that are used by different types of users during the process. In the following section we describe the methodology and tools we developed to support a gradual specification process. The methodology we propose combines the ideas of the two major approaches for guideline specification: (1) the document-centric approach, in which the specification process begins from the original free-text guideline and therefore elements of the formal representation relate to some part of the source document, and (2) the model-centric approach, which is focused mainly on the full formal representation. Therefore, we defined a gradual methodology to convert the guideline from its original text to a fully formal representation.
The actors involved in the methodology are expert physicians from the guideline's medical domain; clinical editors with general medical knowledge and familiarity with the guideline specification language and tools; and knowledge engineers who thoroughly understand the guideline ontology and the technical implications of using computer systems to apply the guideline. From our experience, since the time available to the expert physicians is very limited, it is necessary to use it efficiently. Therefore, the first steps of the specification involve the expert physicians, while the later steps involve mainly the clinical editors and the knowledge engineers.
As noted above, the graphical framework we developed is called Gesher. It is a client application designed to support the process of incremental guideline specification at multiple representation levels according to multiple target specification ontologies. To achieve a high quality specification in a reasonable time, Gesher supports the collaboration of the expert physicians, clinical editors and knowledge engineers. Gesher supports this gradual specification process through all of the steps of creating a formal representation of the guideline. Note that the capability for effective gradual specification also supports maintenance and modification of the clinical-guideline’s knowledge. Although Gesher supports specification into multiple target ontologies and is not restricted to a specific one, we used the Asbru ontology for the formal level of representation and the components we developed for this level are used to generate a representation according to the Asbru language.
An important preliminary step for the guideline specification process is to form a clinical consensus about the semantics of the guideline. Previous research [6Shalom E, Shahar Y, Taieb MM, et al. A quantitative evaluation of a methodology for collaborative specification of clinical guidelines at multiple representation levels J Biomed Inform 2008; 41(6): 889-903.] has emphasized the importance of this step, which includes customizing the guideline knowledge needed for its adaptation in a local clinical setting. The knowledge within each clinical guideline includes procedural aspects with detailed descriptions of the actions taken during the patient care process, and declarative aspects with details about the medical concepts, definitions, and patterns of the patient's state. When adapting clinical guidelines into specific clinical settings, both the procedural and declarative aspects should be examined and, if necessary, modified by the local medical experts. Thus, a preliminary step to the specification is to conduct meetings with senior physicians who are expert in the guideline's domain. In these meetings, the experts construct a clinical consensus of the guideline based on the recommendations from the original source guidelines. The expert physicians should include the required local customizations. Since further specifications of the guideline will be based on the clinical consensus, the expert physician who participates in structuring the consensus using the graphical tools should take part in these meetings. Usually, the output of this step is documented in text and used in the first step of the specification.
In the following sub-sections we describe the various components of Gesher that are used in the gradual process. Each of these components supports different phases in the overall methodology, as illustrated in steps 2-5 of the following figure (Fig. 1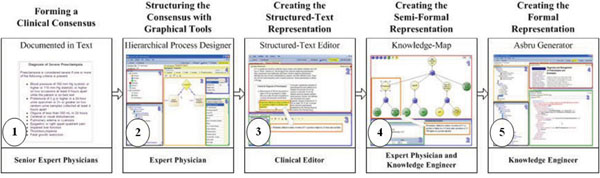 ).
).
4.2.1. Specifying the Consensus in Gesher: The Decomposition of the Guideline
To support the task of structuring the consensus, Gesher provides a graphical interface (see Fig. 2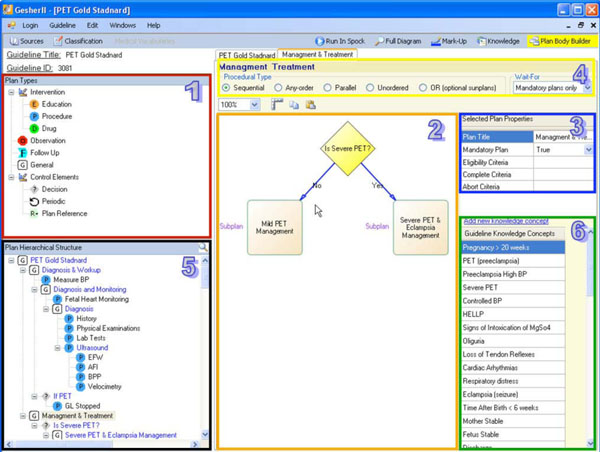 ) that is used by an expert physician, assisted by a knowledge engineer, to roughly structure the main procedural aspects of the guideline consensus. When using the tool, the users can choose plans (i.e., Asbru's notification for a clinical step) from different semantic types and add them to the guideline's procedural flow. These clinical semantic types of plans include procedures, drugs, observations, educational steps, follow-ups, decisions (e.g., if-then-else plan in Asbru), general plans and reference-plans (i.e., reference to existing plans). In addition, the expert can also mark a clinical step as a periodic step, meaning that the step should be performed more than once.
) that is used by an expert physician, assisted by a knowledge engineer, to roughly structure the main procedural aspects of the guideline consensus. When using the tool, the users can choose plans (i.e., Asbru's notification for a clinical step) from different semantic types and add them to the guideline's procedural flow. These clinical semantic types of plans include procedures, drugs, observations, educational steps, follow-ups, decisions (e.g., if-then-else plan in Asbru), general plans and reference-plans (i.e., reference to existing plans). In addition, the expert can also mark a clinical step as a periodic step, meaning that the step should be performed more than once.
Since most of the approaches for guideline specification contain a hierarchical structure to represent the overall complex guideline, Gesher provides the user with the ability to transform each plan into a complex plan consisting of a set of sub-plans. A new diagram will then be created for specifying the new complex plan. When sub-plans are being added to the diagram, the hierarchical structure of the guideline is being constructed. Gesher provides the ability to explore this hierarchy of plans and sub-plans in several visual ways such as by navigating through the tree-structure. This makes it possible to search for plans by their titles, or by navigating through the control flow diagrams. The hierarchical structure of the guideline is immediately stored in DeGeL (using the Web-service based API).
The consensus includes specifying the declarative aspects regarding the clinical algorithm. These aspects include the procedural type of a complex plan (i.e., one that has more than one sub-plan) including: "Parallel" execution of sub-plans; "Sequential"; "Any-Order" (i.e., plans are executed in parallel but no order defined); "Unordered" where the execution of sub-plans may overlap; and "Or" where not all sub-plans are mandatory. The declarative aspects also include specifying the eligibility criteria for starting each sub-plan; the abort criteria to cancel the plan's execution; the complete criteria for successfully finishing the plan; a notation whether the success of completing the sub-plan is mandatory for the success of completing its parent plan; and for each of the complex plans, a notifications of the number of the sub-plan that is mandatory for completing successfully the parent plan. In order to assist the user in structuring the consensus, default values were selected for some of these declarative properties. For example, the default value for the procedural type of complex plans is sequential while sub-plans are mandatory by default (except when the parent's procedural type is "OR").
Another task performed in this step is to create a list of the declarative medical concepts that are related by all the sub-plans comprising the guideline and to provide a textual description for each of these concepts. This list of medical concepts is called the “Guideline Declarative Knowledge”, and will be further specified and defined, in the next step of the specification methodology.
4.2.2. Creating the Structured Text Representation in Gesher
In this step, a clinical editor uses Gesher to create a complete Structured-Text representation of the guideline, according to the knowledge-roles of the specification ontology. Although we used the Asbru language, this step is not restricted to particular guideline ontology. Using Gesher, the clinical editor refines the plans created in the consensus and links these plans to portions of text in the source guideline. This creates a textual representation that is detailed according to the knowledge-roles of the target ontology and is performed for each of the guideline's sub-plans.
When the clinical editor marks-up text from the original guideline, the reference to the particular text segment in the source guideline is saved in the guideline library. These back-pointers that link the hybrid content to the sources are in the library and used by the interface to mark the location in the source when editing a knowledge role of a specific plan. Marking the text can assist the user when creating the specification or when updating a guideline at a later point in time. To edit the content of the structured-text representation (Fig. 3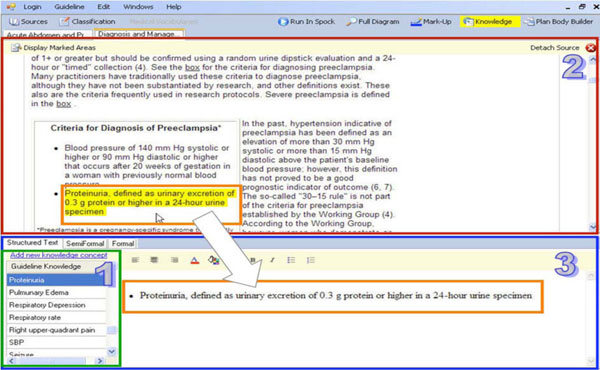 ), Gesher provides a rich HTML editor, and help the user to mark-up a text by dragging portions of labeled content from one or more source guidelines into the selected knowledge role frames.
), Gesher provides a rich HTML editor, and help the user to mark-up a text by dragging portions of labeled content from one or more source guidelines into the selected knowledge role frames.
In addition to specifying the optional text of each knowledge role of the sub-plans comprising the guideline, the clinical editor performs additional tasks such as classifying the guideline according to the semantic indices of the digital library and connecting raw data concepts and declarative the knowledge concepts, to concepts from available, standard medical vocabularies, such as LOINC, CPT and ICD.
4.2.3. Specifying the Semi-Formal Representation in Gesher
In this step, the expert physician and the knowledge engineer use a graphical interface within Gesher to further specify the consensus for the declarative aspects of the guideline. These aspects, which were textually described in the previous step of the process, are now specified in a detailed manner that we call semi-formal representation. The idea of this representation is to describe the declarative concepts in a complete manner that will later allow the knowledge engineer to create the formal and computer interpretable representation. To implement this action, we developed a specific interface called the Knowledge Map that allows the medical expert, in a reasonable amount of time, to examine and confirm all the concepts described in the guideline knowledge.
The knowledge map tool provides the ability to organize and describe the concepts within a specific guideline, at a level of detail that will later allow creating the formal representation. Each concept in the guideline is described by several attributes that are available in the graphical interface. These attributes include a textual description; a description of the type of concepts; a description of the possible values; and a definition of temporal aspects such as the period during which a certain measurement of the concept value is valid in the context of applying this guideline (i.e., for how long is a specific measurement valid).
To keep the knowledge map easy to understand and usable for expert physicians, we had to consider the trade-off between the expressiveness and simplicity of the underlying semi-formal model. To support cases in which the concepts needed to be described, complex constraints that exceed the expressiveness provided by the semi-formal model (e.g. composite temporal patterns or complex mathematical calculations), the expert can use a textual explanation in which these complex constraints should be described. The textual description allows the knowledge engineer to later specify these constraints in the formal representation.
The semi-formal model includes several types of concepts that can be added for describing the guideline's knowledge. The following section describes these types:
Primitives describe the raw data which is collected and examined during a clinical procedure. The primitive concepts may be numeric, Boolean (true/false) or symbolic (e.g., low/normal/high). The white blood cell count is an example of a primitive parameter. Most of the primitives will also include a time-stamp to specify the date and time of the measurement.
Dates of Events describe raw data that includes a time stamp without any scalar value. "Date of Birth" or "Last Menstrual Period" are examples of this type of concepts.
Abstractions describe composite concepts that relate or derive from one or more other concepts. Most of the concepts within free-text source guidelines are abstract concepts. For example, a concept such as "Thrombocytopenia" is a Boolean concept (i.e., it can be evaluated as true or false for a given patient in a given point of time) that is abstracted from "Platelet count". In the PET guideline, Thrombocytopenia is defined as Platelet count < 100,000 per mm3. When describing an abstract concept, it is necessary to describe the relation(s), which we call "Abstracted-From", that involve other concepts and logical constraints on their value or temporal constraints on their time-stamps. When the abstraction is composed from more than one concept, it should also include a description of the logical operators that should be applied. Another example of an abstract concept from the PET guideline is "Elevated Liver Enzymes", which is defined as SGOT>60 AND SGPT>60. Notice the value constraints (">60") and the "And" logic operator.
Logical Operators and Functions. Another type of knowledge element, available in the knowledge map, is the Logical Operators and Functions. Elements of these types are used to describe the logical operator to apply when a concept is abstracted from more than one concept. The logical operator types include elements to describe the relation of AND (e.g., "all of the following"); the relation of OR (e.g., "one or more existing of the following"); and the relation of K-From-N (e.g. at least two are present from all of the following). The function type is used to describe mathematical functions to compute on the "Abstracted-From" concepts. When adding a function to the knowledge map, it is necessary to express it in the textual description.
4.2.4. Example: Specifying the Concept "Mild PreEclampsia"
To illustrate the knowledge map interface and its underlying semi-formal model, we use the following example which is taken from the PET guideline. Fig. (4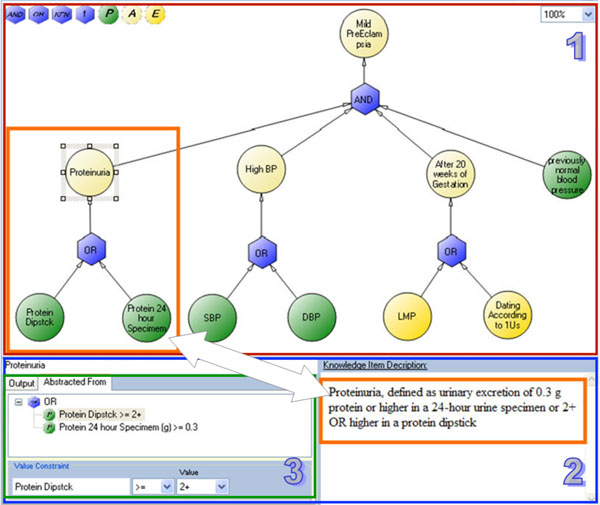 ) illustrates the graph for specifying the concept “Mild PreEclampsia”, which, according to the guideline’s text, is defined as: “Blood pressure of 140 mm Hg systolic or higher, or 90 mm Hg diastolic or higher, that occurs after 20 weeks of gestation in a woman with previously normal blood pressure and Proteinuria, defined as the urinary excretion of 0.3 g or more, in a 24-hour urine specimen, OR a level of +2 or higher in a protein dipstick”.
) illustrates the graph for specifying the concept “Mild PreEclampsia”, which, according to the guideline’s text, is defined as: “Blood pressure of 140 mm Hg systolic or higher, or 90 mm Hg diastolic or higher, that occurs after 20 weeks of gestation in a woman with previously normal blood pressure and Proteinuria, defined as the urinary excretion of 0.3 g or more, in a 24-hour urine specimen, OR a level of +2 or higher in a protein dipstick”.
4.2.5. Specification of the Formal Representation in Gesher
In this step, the knowledge engineer uses Gesher to create the formal representation in the final executable, target guideline ontology. Because we have chosen in this study to use the Asbru ontology as the underlying formal specification language, we developed a module that automatically generates Asbru guidelines (see Fig. 5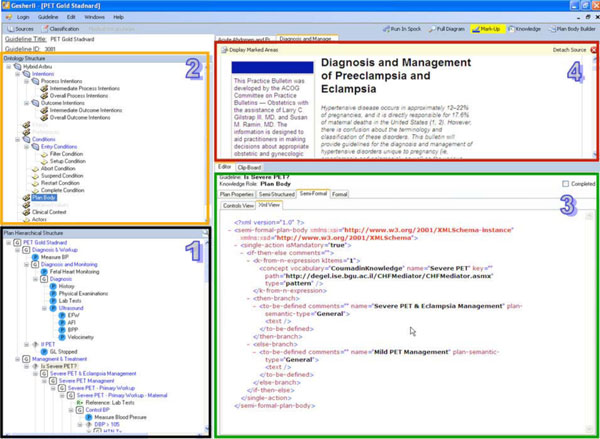 ) from the inputs of the previous steps of the specification. By using the products of the former steps of the methodology, this module creates the representations of the guideline according to the syntax of the Asbru language and immediately stores them in the library.
) from the inputs of the previous steps of the specification. By using the products of the former steps of the methodology, this module creates the representations of the guideline according to the syntax of the Asbru language and immediately stores them in the library.
The task of the knowledge engineer when using this module is to link between the procedural and declarative aspect of the guidelines and then to generate and validate the formal representation, which, in case of Asbru, is represented in XML. To further validate and, if necessary, to correct the guideline, the knowledge engineer can run a simulation of it by using the runtime application engine that is integrated within Gesher in a mode that we call "debug mode". To examine the behavior of the multiple paths within the guideline, the user utilizes the interface of the runtime application engine to provide simulated results regarding the different medical concepts and to observe the corresponding behavior of the system.
Despite the fact that this step is specific to a single ontology and that the module we developed supports only the Asbru ontology, Gesher can be easily extended to support the generation of formal representations according to other target ontologies. For example, a plan that was declared in the consensus as a decision plan is translated to Asbru as an “if-then-else” plan. However, if GLIF is being used, it can be translated into a decision-step plan. Although the design of the internal architecture of the system supports such extensions, nevertheless, an additional development will still be needed.
4.2.6. Additional Functionality of the Gesher System
Guideline Classification
The DeGeL library contains structures of semantic indices for classifying guidelines (of both original sources and formal mark-ups). These indices provide better retrieval abilities when users are searching for guidelines in the library. Currently, these axes of semantic indices include thousands of medical concepts that can be used to classify each guideline. In order to classify a guideline with the relevant indices, the clinical editors use a graphical interface (see Fig. 6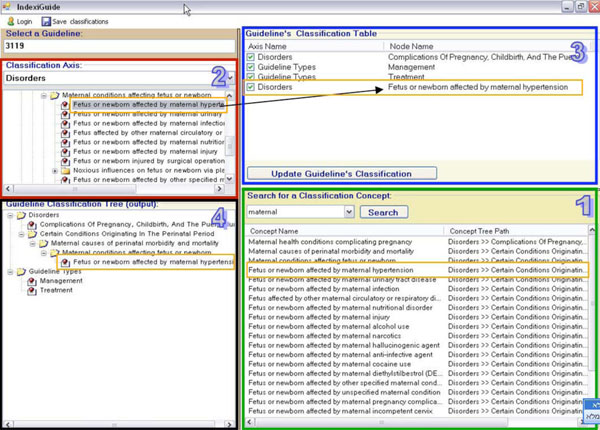 ), which is part of Gesher. The interface of this component enables the editors to navigate through the hierarchical axes and to select the relevant concepts for the classification. It also provides a capability for searching the hierarchical axes of concepts using an automatic module that retrieves all the relevant results for a given key word and allows the user to allocate the position of each result in the hierarchical structures. The interface displays the current classifications of the guideline as a flat list of classifications; a second display presents the location of each assigned concept in the hierarchical structure.
), which is part of Gesher. The interface of this component enables the editors to navigate through the hierarchical axes and to select the relevant concepts for the classification. It also provides a capability for searching the hierarchical axes of concepts using an automatic module that retrieves all the relevant results for a given key word and allows the user to allocate the position of each result in the hierarchical structures. The interface displays the current classifications of the guideline as a flat list of classifications; a second display presents the location of each assigned concept in the hierarchical structure.
Guideline Retrieval
To allow users to search and retrieve sources and formal guidelines from DeGeL, we developed a component that uses the API of DeGeL's search engine to retrieve guidelines from the library and to load them into the knowledge specification tool. The interface of this component provides the users with the ability to use the special functionalities of the search engine such as a context-based search within selected elements in the ontology and/or a concept-based search using the concept axes of DeGeL library.
To simplify the interface of the search functionality of DeGeL's search engine, the user can choose from three separate interfaces. The first interface is simple and provides the user with the option of searching with key words contained within the relevant guidelines (content and meta-data). The second interface enables the user to expand his query using concepts from the semantic axes within the library and to conduct context-sensitive searches. Such searches make it possible for the user to restrict a search to selected knowledge-roles of the relevant ontology (from the multiple ontologies within DeGeL). Providing these advanced search options in the search interface results in a complex interface that can only be used by those who are familiar with DeGeL, In order to allow less experienced users to benefit from the advanced functionality, we developed a third interface that allows experienced users to create search templates which are stored in the library server. These can then be used by less-experienced users who are required to only input their selected key words into the pre-made interface. We also developed a rich visual interface to display the search result. This interface provides visualizations of the guidelines from multiple ontologies; results for a given query; and their relations to the classification indices.
Guideline Exploration
The guideline exploration module of Gesher is used for exploring the hierarchical structure of clinical guidelines (see Fig. 8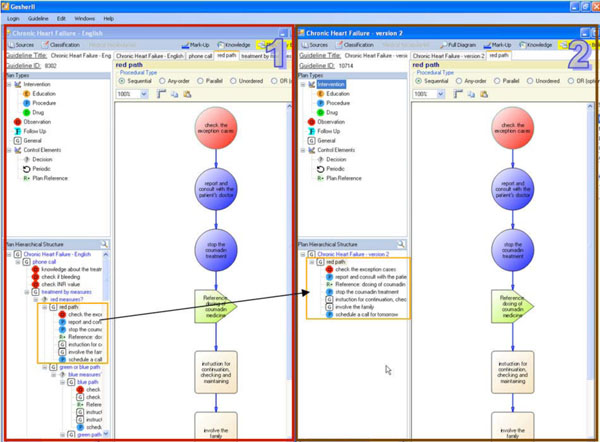 ). This module allows navigation through the hierarchical structure of the guideline and provides visualizations of the procedural aspects. To support the display of guidelines from multiple ontologies, a generic structure based on DeGeL's meta-ontology is used for hierarchically representing the procedural aspects.
). This module allows navigation through the hierarchical structure of the guideline and provides visualizations of the procedural aspects. To support the display of guidelines from multiple ontologies, a generic structure based on DeGeL's meta-ontology is used for hierarchically representing the procedural aspects.
Reproduction and Versioning of Guidelines
To reuse existing knowledge, we have implemented a mechanism for reproducing existing guidelines. This mechanism allows the user to easily create from an existing guideline a new version that includes a deep copy of all knowledge elements of the original one. When a user creates a new version, she is automatically granted an ownership authorization on the new guideline and any changes made to the new version do not affect the original one. This mechanism supports such common requirements as modifying and customizing a guideline when implemented for a certain medical institution other than the one it was originally developed for.
When creating a new version of a guideline, the relation to the original one is saved, along with meta-data such as the date of creation and the editor's name, and the new version can be used for retrieving all versions for a specific guideline. The elements within the new version will be directly connected to the source guidelines that were referenced by the original one. If the content is modified, the editor is responsible for referencing the new sources. Currently, the system does not support automatic comparison between guidelines, but this functionality can be added using the pointers between guideline elements, the relations between sources and versions, and the dates of modifications.
In addition to reproducing a complete guideline, the system also supports the ability to copy sub-plans and declarative knowledge concepts between guidelines. This functionality (see Fig. 7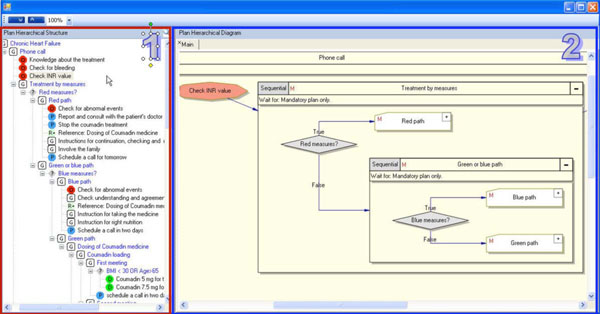 ) enhances the reuse of existing knowledge and assists in minimizing the overall duration of the specification process. In principle, when copying a plan or a declarative concept, all levels of the representation, from structured text through semi-formal and formal representations, are reused and can then be modified in the new guideline. There are still many outstanding questions about what level of representation is mostly being reused. However, from our experience, we believe that the semi-formal representation level is the one that would actually be reused most since the semi-structured representation level serves primarily for documenting the semantics of the reused knowledge and would usually be modified during the reuse process. The formal level, on the other hand, is directly derived from the semi-formal level.
) enhances the reuse of existing knowledge and assists in minimizing the overall duration of the specification process. In principle, when copying a plan or a declarative concept, all levels of the representation, from structured text through semi-formal and formal representations, are reused and can then be modified in the new guideline. There are still many outstanding questions about what level of representation is mostly being reused. However, from our experience, we believe that the semi-formal representation level is the one that would actually be reused most since the semi-structured representation level serves primarily for documenting the semantics of the reused knowledge and would usually be modified during the reuse process. The formal level, on the other hand, is directly derived from the semi-formal level.
Guideline Maintenance
The combination of the ontology-based, guideline specification methodology and its implementation as the Gesher framework significantly facilitates maintaining existing clinical guidelines in the DeGeL library as medical knowledge is updated. For example, if eligibility conditions for a particular guideline change, the clinical editor simply modifies the appropriate knowledge-role, for example, the filter condition in the case of the Asbru ontology. Similarly, if the outcome goals of the guideline with respect to the patient are updated by a professional society, the clinical editor modifies the outcome intentions in the Asbru ontology. In both cases, the knowledge engineer then modifies the formal representation level of the respective knowledge-role. Note that the existence of explicit knowledge roles significantly facilitates version maintenance since consecutive versions can now be differentiated on the basis of exactly which knowledge-role(s) was modified.
4.2.7. . Summary and Illustration of the Overall Specification Process
To summarize the specification process and to illustrate the overall methodology, we describe here the cycles of the guideline specification for diagnosing and managing preeclampsia/eclampsia toxemia (see section 4.1.1). The guideline was selected by the head of the OB/Gyn division at the Soroka Medical Center due to its high occurrence and risk, and the specification process was performed in collaboration with experts from Soroka. To form a clinical consensus two meetings were held with senior medical staff from the hospital, including the participation of three ward managers and two senior experts. The consensus, which included some customizing to the original source, was documented and used in the specification steps.
After a training session, one of the medical experts decomposed a skeleton of the guideline using the Gesher tool in an iterative fashion that included four meetings of a medical expert and a knowledge engineer. Each meeting focused on separate sections of the guideline (diagnosis, mild preeclampsia management, severe preeclampsia management, and eclampsia management). The decomposition of the guideline in Gesher was completed, and then validated in a meeting with senior medical managers.
In the next step, an intern was trained as a clinical editor and used Gesher to create the structured-text representation and to link between elements in the structured guideline and the original source. To complete this task, the clinical editor consulted with the knowledge engineer during the guideline structuring process to further understand Gesher and the semantics of the representation model.
During the next step, the medical expert was trained to use the knowledge-map interface for creating the Semi-Formal representation. To shorten the time for completing this part of the process, the semi-formal representation was actually created with the graphical tool by the knowledge engineer using the structured text representation. However, after only two one-hour sessions the medical expert was able to validate and approve all elements in the specification. Because the specification of the structured-text comprising the recommendations and customizations was very clear, there was no need to involve again the senior ward managers.
The formal level was completed by the knowledge engineer who used Gesher to generate the guideline in its formal Asbru-based XML representation.
5. EVALUATION
We decided to evaluate three distinct aspects of the guideline specification framework developed in this study. The experiments we conducted were designed to evaluate the quality of the specification products; the usability of the tools; and the usability of the overall methodology. The quality of the specification products were measured in terms of the correctness and completeness of the semi-structured and semi-formal representation levels appearing in the output guideline specification. These measures were defined in collaboration with the senior domain experts (further details are provided below), and the products of each participant were compared to those of the senior expert.
These aspects were evaluated in three major experiments:
- The first experiment was conducted with the cooperation of physicians from the Obstetrics and Gynecology (OB/Gyn) Division at the Soroka Medical Center. In this experiment we evaluated the feasibility and usability of completing two phases of the specification methodology: structuring the consensus in Gesher and creating the structured-text representation. This experiment focused on the OB/Gyn guideline for the treatment of preeclampsia and involved clinicians from several levels using Gesher to create the guideline specification.
- The second experiment focused on the semi-formal representation of clinical guidelines. In this experiment, users (both clinicians and knowledge engineers) utilized Gesher to create the declarative knowledge specification of the preeclampsia guideline.
- The third, somewhat less formal experiment evaluated the functionality and usability of the exploration tool. In this experiment we created a guideline specification for treating patients with congestive heart failure with the anticoagulant, Coumadin. Then, we asked participants to use the exploration interface to answer several questions that we presented to them.
In the first two experiments we evaluated the capability of the users to use the tools to perform the guideline specification. In addition, we also tested the products of the specification process by using them as an input for both the runtime application engine as well as for the exploration component used in the third experiment.
5.1. Experiment 1: Evaluation of the Interface for the Specification of Procedural Aspects of Clinical Guidelines
In this experiment, we applied Gesher to specify an important obstetric guideline-the diagnosis and management of preeclampsia/eclampsia toxemia. We assessed the feasibility (i.e., functionality and usability) of: (1) representing a clinical consensus customized for a particular medical center and (2) structuring the full content of the guideline. In addition, we assessed in a preliminary fashion the possibility of using a less experienced clinician as a markup editor by asking both a senior obstetrics and gynecology clinician and a general intern to represent the same guideline using Gesher. The results demonstrated the functionality and usability of Gesher in performing these tasks, at least for these two editors. Indeed, the intern's performance was at least as good as the senior physician with respect to the specific task of structuring the guideline in Gesher according to the hybrid Asbru ontology.
5.1.1. The Preeclampsia/Eclampsia Toxemia Guideline
To evaluate the Gesher system and the guideline specification methodology, we conducted a study that included specifying the guideline for diagnosing and managing preeclampsia/eclampsia toxemia (PET) [44American College of Obstetricians and Gynecologists. Diagnosis and management of preeclampsia and eclampsia. Washington (DC) 2002; p. 33.]. PET is a condition occurring in pregnancy characterized by high blood pressure and the appearance of protein in the urine. The condition is highly dangerous both to the fetus and to the mother. PET occurs in approximately 12-22% of pregnancies, and is directly responsible for 17.6% of maternal deaths in the United States. We created a structured representation of the PET guideline, which includes the required customizations for applying this guideline in a real clinical setting, in this case, the OB/GYN ward of the Soroka Medical Center.
5.1.2. Experiment 1: Objectives
The objectives of this experiment were to evaluate the feasibility of the Gesher system in structuring a guideline consensus and performing the mark-up process. We were also interested in assessing, in a very preliminary fashion, the clinical editing performance of a senior expert physician and an expert in the guideline’s domain compared to that of an intern with only general medical knowledge. We were also interested in assessing the time and human effort required for completing the specification process, and in discovering common mistakes made during the specification process.
5.1.3. Experiment 1: Evaluation Methods
In following section we describe the methodological specification of the PET guideline. Several participants took place in the process. (1) The expert physician and the knowledge engineer together created the ontology-specific consensus that was structured in Gesher. They also created the "Gold Standard" specification in Gesher that was used in the evaluation. (2) The senior OB/GYN expert physician and a general intern (referred to below respectively as the expert and intern) who participated as the clinical editors who used Gesher to create the structured consensus and the structured representation of the guideline. (3) A group of senior expert physicians that took part in creating the clinical consensus based on the recommendations of the American Congress of Obstetricians and Gynecologists (ACOG) guideline for diagnosis and management of preeclampsia and eclampsia.
Creating a Clinical Consensus
After choosing the guideline for the specification, we conducted two meetings with senior expert physicians from the Soroka Medical Center. In these meetings, they created a clinical consensus that included the recommendations from the original source guideline together with modifications and additions required for the customization and implementation of the guideline in this specific medical institution.
Creating Ontology Specific Consensus
Following the clinical consensus achieved in the first step, the expert physician, in collaboration with the knowledge engineer, created the ontology specific consensus, which is a detailed document describing both the procedural aspects of the clinical algorithm and the declarative definitions of the medical concepts within the guideline (e.g. the criteria for diagnosing severe PET). The ontology specific consensus was created as a printed flow-chart format.
Creating a Structured Consensus in Gesher
The clinical editors used Gesher to structure the clinical flow chart specified in the ontology specific consensus. The structured consensus includes all the sub-plans composing the guideline and specifies the semantic type of each sub-plan, such as drug administration or performance of an observation. The procedural type of each composite plan was determined in this step (e.g. sequential or parallel), and each plan was defined as mandatory or optional, with respect to whether it needs to be completed in order to allow the process to continue to the next sub-plan. For each sub-plan several properties were defined as structured text (e.g. “abort condition”). The plan properties included optional annotations describing the following procedural aspects: the eligibility criteria to enter the plan; the conditions for completing the plan successfully or for aborting the execution of the plan; and, in the case of periodic (repeating) plans, a specification of aspects such as frequency and number of iterations. Another task performed in this step was the creation of the list of all medical concepts defined in the ontology specific consensus. This list is called the knowledge guideline and was later further refined by the physicians.
Creating a Structured Text Representation in Gesher
The clinical editors were provided with the gold-standard structured consensus and then used Gesher to create the complete structured representation of the guideline according to the knowledge-roles of the Asbru specification ontology. Each of the sub-plans in an Asbru-based guideline is composed from knowledge-roles such as “abort-condition” and “clinical-settings”. The physicians used Gesher to markup text from the source guideline to create knowledge-roles for each of the sub-plans. The reference to the source was saved in the guideline library. Although the structured consensus includes informal specifications of all knowledge-roles, in this study the markup phase included only the following knowledge-roles: (1) filter-condition (2) setup-condition (3) complete-condition (4) abort-condition (5) actors (6) clinical-context.
5.1.4. Experiment 1: Evaluation Measures
The structured consensus and the structured representation created by the intern and by the expert were compared to a gold-standard structured consensus and a gold-standard structured representation. The gold-standard representation was created by the expert physician together with the knowledge engineer and was assumed to be the most detailed and correct specification, clinically and semantically. The comparison of the specifications created by the experiment’s participants to the gold-standard was also performed by the expert physician together with the knowledge engineer, using a software tool specifically developed for this purpose. The evaluation tool enabled users to navigate through the complex structure of guidelines which included more than 115 sub-plans, and to compare and grade each part of the structured consensus and the structured representation.
We used objectives measures of completeness and soundness (correctness) in order to measure the level of success of the physicians in completing the specification tasks. The completeness of the specification was defined as the number of knowledge elements (such as sub-plans or plan properties) that exist in the physician-created structured consensus and the structured representation compared to the number of knowledge elements existing in the gold-standard. In order to measure the soundness of the specification products, each of the knowledge elements was assigned a discrete grade describing its correctness; If the content of the element was correct and similar to the gold-standard, the element was graded as “Correct”. If the content was not similar to the gold-standard, the element was assigned one of the following grades; “Not correct, worsening the patient outcome” or “Not correct, not worsening the patient outcome”. The overall level of correctness of a guideline or a group of knowledge-roles is the proportion of knowledge elements assigned with the grade “Correct”.
To evaluate the usability of the Gesher, we used the standard system usability scale (SUS) questionnaire [45Brooke J. SUS-A quick and dirty usability scale, http://www.usability.serco.com/trump/documents/Suschapt.doc 1996.] that was presented to the users after performing each of the tasks. To assess the effort in terms of time, we measured the time required for the expert and the intern to complete the specification process.
5.1.5. Experiment 1: Results
Results for Creating a Structured Consensus in Gesher
We started by assessing the feasibility of structuring, by the two clinical editors, of the existing ontology-specific consensus document in Gesher. Both the expert and the intern achieved a high level of completeness in this task. The proportions of plans that exist both in the gold-standard structured consensus and in the structured consensus were 96% and 97% for the expert and the intern respectively. The proportions of completeness of the plan properties of each of the existing sub-plans (e.g. periodic specification and complete condition) were 99.6% and 100% for the expert physician and the intern respectively.
The correctness of the structured consensus created by the physicians was measured by the proportion of the knowledge elements that was scored as “Correct” in the evaluation. The expert and the intern achieved very high levels of correctness, 97.78% and 99.22% respectively.
Results for Performing the Markup in Gesher
Both clinical editors performed the markup based on the gold-standard structured consensus that was provided to them. Table 3 summarizes the results for the completeness and correctness of the structured representation. The knowledge-roles of the Asbru specification ontology that were used in the evaluation were partitioned into three classes: the condition knowledge-roles (e.g. complete, abort) the Context knowledge-roles (“actor” and “clinical-settings”) and knowledge definition knowledge-roles.
The completeness of the structured representation that was created was defined as the proportion of the knowledge-roles with complete content from all knowledge-roles of all sub-plans. Both the expert physician and the intern achieved a very high level of completeness in structuring the conditional knowledge-roles, 99.79% and 100% respectively. The “actors” and “clinical-context” knowledge-roles were specified at a lower level of completeness, 63.25% and 94.44%, by the expert and the intern respectively.
The soundness of the structured representation was measured by the proportion of knowledge-roles that were judged as being similar to the gold-standard and to have the correct (clinically and semantically) content. Both clinical editors achieved a high level of soundness for structuring all classes of knowledge-roles.
Results for Amount of References to the Source Guideline
Another interesting measure we examined is the completeness of the references from the knowledge-roles of each sub-plan to the text of the source guideline. Each of the knowledge elements can include back pointers to the text of the source guideline where the recommendations originated. It is interesting to note in both markups the low proportion (about 50%) of elements that actually exist in the source guideline. To explain the result in Table 4, the expert structured 278 knowledge-roles, from which 134 had back pointers to the source according to the gold-standard. He completed full references to 107 knowledge-roles, partial references to 4 knowledge-roles and missing references to 23 knowledge-roles.
Results for the Usability of Gesher
The following results for assessing the usability of the interface of Gesher were achieved by using the SUS that was presented to the clinical editors after performing each task. The clinical editors gave a mean score of 85% for the interface for structuring the consensus, and a mean score of 77.5% for the interface for performing the markup. Both clinical editors thus considered the Gesher tool quite usable.
Results Regarding Time Measures
The overall time effort for completing both parts of the specification process, were measured for both clinical editors. The expert worked for a total of 32 hours which were composed of 16 short episodes and the intern worked for a total of 46 hours, which that were composed of 8 longer sessions.
5.2. Experiment 2: Evaluation of the Interface for Specifying the Declarative Concepts
5.2.1. Experiment 2: Objectives
The objectives of the following experiment were to evaluate our collaborative methodology and the knowledge map interface in terms of the quality of the specification product and the level of usability of the user interface. In addition to assessing the time needed to complete the specification tasks, we were also interested in identifying subjective insights and challenges that arise when representing guideline knowledge.
5.2.2. Experiment 2: Evaluation Methods
To accomplish these objectives we designed the following experiment. We asked three knowledge engineers, three physicians, and three combined teams (a physician working together with an engineer) to complete, after receiving a short training in the knowledge map tool, the declarative knowledge specification of the PET guideline. We then asked them to answer a SUS questionnaire. The results of the user's specifications were compared to a gold standard specification that a senior expert physician and a knowledge engineer created.
The training session included some background on the research and its objectives; a presentation of the theory behind the semi-formal representation of declarative knowledge; an explanation about the PET guideline; and an explanation about the knowledge map and the specification task. The training session also included a demonstration of the knowledge map tool and an explanation of the functions and of the display of the interface, which covered the various options available for the user when working with the knowledge map interface. In addition to the explanation, to improve user training, each participant used the tool in a self-training session to gain practice in specifying several medical concepts that were previously demonstrated. It was important to let the users work with the graphical tool and to complete the training session. Feedback from the participants indicated that they found the self-training session to be helpful. At the end of each training session, the participants completed a short questionnaire to determine if the training had been successful and to ensure that they all had the knowledge needed to complete the specification task. The questionnaire included ten multiple-choice questions about the main subjects of the training. After answering each question, the participants were told whether it was the correct answer and mistakes were corrected.
After the training, each participant was asked to create a full semi-formal specification of all of the knowledge concepts from the PET guideline. The guideline includes twenty abstract concepts that were defined, by a senior medical expert and a knowledge engineer during the consensus formation phase on the basis of the PET guideline. Each of these concepts was defined with a textual description (i.e. the structured text representation), and the task of the users was to achieve the most detailed semi-formal representation. To achieve the semi-formal representation, the users used the knowledge map interface to add additional concepts according to the textual definition; to assign them the correct properties (such as the data type of the concept); and to specify the correct "abstracted-from" relations which included specification of value and of temporal constraints. It was pointed out to the users that they should try to avoid redundancy in the knowledge specification i.e., primitive concepts should not appear more than once in the guideline's knowledge description. Redundancy was to be avoided to eliminate misunderstanding by the knowledge engineers when creating, at the next step of the specification, the formal representation. The participants were provided with access to the Gesher system and asked to complete the specification task in their own time. Most participants completed their work in several separate sessions; the time for completing the tasks was measured. For assessing the knowledge map tool's usability, the participant answered a SUS questionnaire, after completing the specification.
5.2.3. Experiment 2: Evaluation Measures
In the beginning of the specification, the participants were given a textual description of twenty abstract concepts of the PET guideline knowledge. "Severe PreEclampsia" or "HELLP syndrome", are two examples of these abstract concepts. The abstract concepts included one or more sub-concepts, which were either raw-data concepts or were themselves abstracted from lower level concepts.
To evaluate the completeness of the specification of each of these concepts and to decide on the sub-elements that should exist for each abstract concept, we used a gold standard specification created by a senior physician and a knowledge engineer. Each specification was given a completeness score based on calculating the proportion of the number of elements existing in the specification compared to the number of elements in the gold standard. We examined 57 sub-elements to evaluate the completeness of each participant's specification; thus, the nine participating individuals or teams examined in total 513 (57x9) elements.
To evaluate the correctness of each of the abstract concepts, we examined the gold standard specification and decided on the elements that would be used to measure the correctness. These elements were based on the properties required for creating the semi-formal specification. Examples of these properties included constraints on parameter values, logical operators and parameter data-types (e.g. selection of "Numeric" or "Boolean" data type for a parameter). In order to evaluate the correctness of each specification, we marked each element as "Correct" or "Not Correct", and calculated the correctness as the proportion of elements that were marked "Correct". We examined 221 elements to evaluate the correctness of each participant's specification. (The nine participating individuals and joint teams handled an overall number of 1989 elements ).
5.2.4. Experiment 2: Results
Results for Completeness
The overall result for completeness of the elements included within the nine specifications was 93.18% (478 existing sub-elements from an overall number of 513 sub-elements). The knowledge engineers all had the same mean level of completeness, 94.74%. The mean completeness scores of the three expert physicians were 87.72%, 89.47%, and 87.72%. All the collaborative teams had the same level of completeness, 96.49%.
When we compared the completeness measures of all of the participant pairs in each of the three groups of users, no significant differences were found (p>0.05). Therefore, we were able in the next step to perform a proportions test to compare the mean scores of each group. The completeness of the specification by the expert physicians was significantly lower than the results of the knowledge engineers, (z=2.4, p=0.033) and the collaborative teams (z=2.86, p=0.004). There was no significant difference (p>0.05) in the completeness of the specification by the knowledge engineers and by the collaborative teams. Table 5 summarizes these results.
Results for Correctness
The overall mean correctness of the elements included within the specifications of the participant individuals and teams, was 94.11% (1872 correct elements from an overall number of 1989 elements tested). The knowledge engineers' mean correctness levels were 91.4%, 94.12% and 95.02%. The expert physicians' mean correctness levels were 90.05%, 91.4% and 91.4%. The collaborative teams' mean correctness levels were 98.19%, 98.64% and 96.83%.
When we compared the correctness scores among all the pairs of participants within the three groups of users, no significant differences were found (p>0.05). Therefore we were able in the next step to conduct proportion tests to compare the mean correctness scores of each group. Table 6 summarizes the result for the comparison of the correctness between the three types of users. The correctness of the specification by the collaborative teams, was significantly higher than the one produced by either the expert physicians (z=5.5, p<0.001) or by the knowledge engineers (z=3.93, p<0.001). There was no significant difference (p>0.05) between the correctness of the specifications produced by the knowledge engineers and by the physicians.
Results Regarding Time Measures
For each of the participants, we measured the time to complete the training session and the time to complete the specification and then calculated the total time. On average, the duration of the training session was 1 hour and 51 minutes and the duration of the specification was 3 hours and 51 minutes (the overall mean time for training and specification was 5 hours and 42 minutes). Table 7 summarizes the time needed for completing the tasks for each of the participants and the average times required by the three different types of users.
The effect of the group type on the training, specification and overall time was examined using three separate one-way ANOVAs. The time was the dependent variable and the group was the independent variable. The results show that the effect of the group was significant for the analysis of the overall time (F(2,6)=7.06, p=0.026) and insignificant for the analysis of the training and the specification times separately.
A post-hoc Duncan test showed that for the analysis of the overall time, the time needed by both knowledge engineers and the collaborative teams was significantly shorter than the time required by the physicians (p<0.05). However, there was no significant difference between the time needed by the knowledge engineers and by the collaborative teams.
It should be mentioned, however, that the physicians' sessions were typically composed of several shorter sessions, given their external time constraints. This limitation might well have had an effect on the time measurement results (see discussion).
Results for Usability
To evaluate the usability of the knowledge map interface, we asked each of the participants to fill out a standard SUS questionnaire after completing the specification task. The mean overall usability score was 80 ± 8.19 on a scale of 1 to 100. Table 8 shows the result for physicians and knowledge engineers. It should be noted that the SUS questionnaire was filled out on an individual basis not by teams.
To compare the usability scores of the two different types of users, we used a t-test, assuming equal variances. The mean score given to the interface by the knowledge engineers (85.83 ± 6.06) was significantly higher (t(10)=-3.52, p<0.01) than the mean score the physicians gave it (74.12 ± 5.4).
Subjective Results
Several more interesting insights emerged from observing and debriefing the participants while performing the specification and after completing the tasks. While performing the specification, the knowledge engineers used the interface in a more fluent fashion, perhaps due to the fact that the interface included various functions that are common in these kind of graphical interfaces. For example, in order to relocate a concept and layout it on the map, it is necessary to locate the mouse pointer on the edge of the concept’s shape and then drag and drop it to the new position. When locating the mouse on the center of the shape and clicking it, the concept changes its state for renaming instead of moving. Another example of a common graphical interface function is the option to use the “ctrl + mouse click” in order to select multiple shapes, and to move them all together. It is possible to assume that the experience of the engineers in using professional software, such as development environments, enabled them to easily learn and use the various functions of the knowledge map interface, which they may have found simpler to operate than the physicians did. Another interesting insight is the way the users organized the layout of the concepts in the map, during the specification and when finished. The physicians did not pay much attention to the organization of the map and used the function to hide other concepts which they did not need so that they would not bother them at that moment. The engineers, on the other, were more concerned in keeping the map organized. Fig. (9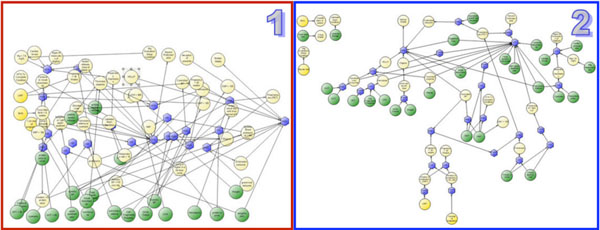 ) demonstrates the difference in the layout of two maps; the first was created by a physician and the second by an engineer.
) demonstrates the difference in the layout of two maps; the first was created by a physician and the second by an engineer.
5.3. . Experiment 3: Evaluation of the Guideline Exploration Module
This experiment was focused on the usability of the Gesher guideline exploration module. This module is used for exploring the hierarchical structure of clinical guidelines (see Fig. 8 ), during the specification process. One common scenario in using this module includes presenting the guideline to senior medical experts to validate the specification and/or extend the guideline. This module allows easy navigation through the hierarchical structure of the guideline and provides visualization of the procedural aspects. The generic structure used for the hierarchical representation of the procedural aspects of the guideline is based on DeGeL's meta-ontology and thus supports the display of guidelines from multiple ontologies.
), during the specification process. One common scenario in using this module includes presenting the guideline to senior medical experts to validate the specification and/or extend the guideline. This module allows easy navigation through the hierarchical structure of the guideline and provides visualization of the procedural aspects. The generic structure used for the hierarchical representation of the procedural aspects of the guideline is based on DeGeL's meta-ontology and thus supports the display of guidelines from multiple ontologies.
5.3.1. Experiment 3: Evaluation Methods
In this experiment we created a specification for one aspect of the congestive heart failure (CHF) guideline, managing the Coumadin treatment. We then asked several participants to use the exploration module. The evaluation was conducted with 12 participants: physicians, nurses and students. After being presented with a short demonstration of the tool, they were then asked to use the graphic interface and to answer four short questions regarding the guideline (e.g., what is the Coumadin dose in the loading stage for patients with low BMI levels). After completing the tasks, the participants completed the SUS questionnaire to evaluate the usability of the module.
5.3.2. Experiment 3: Results
The overall mean time for training the users was 4 minutes and 20 seconds. The mean time to answer each question was 1 minute and 40 seconds; the mean score for the users' responses to the questions was 85.6%; the mean usability score for the system was 71 (according the SUS scale).
6. SUMMARY AND DISCUSSION
In this study, we focused on developing and evaluating a computational framework to support the process of clinical-guideline specification. We presented a digital library and a suite of computer tools that we developed for creating and maintaining a repository of clinical guidelines that are represented in increasingly formal format.
In order to evaluate the tools we developed and to learn about the guideline specification process, we conducted two experiments and additionally implemented a less-formal practical assessment. The two experiments involved 15 participants who were using these tools to perform different tasks in the specification process. In these experiments we evaluated the ability of the knowledge specification framework to support the intended users in specifying the procedural and declarative aspects of the clinical guideline.
In the first experiment, the clinicians found the system functional and usable in supporting the structuring of the guideline's consensus and in specifying the structured text representation. The Gesher system was found reasonably functional and usable in supporting the specification of the structured consensus and the structured representation. It is particularly encouraging that the intern successfully completed the tasks at a level of completeness and correctness that was not inferior to the senior physician.
To better understand why only 50% of the knowledge elements that were marked-up originated in the source guideline text, it is important to note that in the current methodology that we used for the guideline specification, the expert physician created much of the knowledge when creating the consensus, and could not be pinned down to a specific text in the source guideline. In particular, much of that knowledge was implicit in the guideline and not made sufficiently explicit. The physician was relied upon to apply the guideline to complete the missing details. However, that result suggests a significant difficulty regarding attempts to use natural language processing (NLP) techniques to automatically structure guidelines; much of the necessary text to be understood simply does not exist.
The knowledge map interface was functional and usable in supporting the specification of the semi-formal representation. Regarding completeness and correctness, the quality of the resultant specifications was very high. A mean time of 5 hours and 42 minutes, including the one-time training session, for completing the task of specifying all concepts within a relatively complex guideline such as the PET, was very reasonable, and provides another indication as to the feasibility of performing this task using the knowledge map. The usability scores given to the interface were high as well. An interesting fact is that the collaborative teams and the knowledge engineers produced a specification within a considerably shorter time that was significantly more complete than the physicians. This fact may be possibly due to several factors: the experience of the knowledge engineer in completing such tasks in complex computer environments; the more technical and organized approach of the engineers in solving these types of problems; and the fact that the physicians, given their external time constraints, had to complete the task in multiple short sessions, which might have had a cognitive effect (at each session, they needed to remember their work from previous sessions and continue from this stage).
We also found that the collaborative physician-engineer teams produced a specification with a significantly higher correctness level than both physicians and knowledge engineers separately. We attribute this result to the fact that the correctness of the semi-formal specification consists of clinical elements, which obviously were more familiar to the physicians while the engineers more easily understood the logical and technical elements related to the interface and to the specification model. The fact that the knowledge engineers awarded the interface a better usability score to the interface is quite intuitive; the engineers are used to performing this sort of task and working with this kind of graphical user interface. That the knowledge engineers and the collaborative teams completed the task in a significantly shorter time than the expert physicians might be connected to the fact that the interface was more usable to the engineers and that the physician faced with various external constraints had to complete the task in multiple shorter sessions. However, our overall conclusion is that the knowledge map interface for specifying declarative knowledge within clinical guidelines enables both knowledge engineers and physicians to specify that knowledge with high levels of completeness and correctness within reasonable time frames.
In the third experiment we evaluated the interface for guideline exploration. The interface was found usable by participants (score of 71) after a very short training (average of 4.3 minutes). The participants used this module for answering questions regarding certain knowledge within the complex structure of the hierarchical guideline. The questions were answered in a very short time, less than two minutes.
Regarding the specification of the formal representation, we examined it via an informal though quite insightful project, the specification of a complex congestive heart failure (CHF) guideline. The specification of this guideline was performed by two knowledge engineers in collaboration with an experienced CHF nurse, and approved by expert physicians from the Maccabi health maintenance organization. The formal representation of the guideline that was generated by the Gesher system was also validated by an automatic application of that guideline to a database of real patient records, using the runtime application engine for simulating and validating (by domain experts) the recommendations.
In future research, once the soundness and completeness of the knowledge specified by clinical editors has been demonstrated, the consistency among specified knowledge components can be measured, including a comparison to an existing body of knowledge. In fact, in an additional study that we carried out but which is not within the scope of the current project, we developed a methodology for validating and verifying the declarative knowledge.
6.1. Limitations of the Study
The study has several limitations. Conducting the evaluation with additional clinical guidelines from other medical domains, published by different professional associations, would have enhanced our ability to generalize our conclusion. In the first experiment only two clinicians were used, due to the very detailed level of structuring that we required (more than 2000 knowledge elements for each specification).We chose to use the limited time we had with the participants for creating a full specification of the PET guideline at all presentation levels. The PET guideline is very complex and it was important for us to create a full specification guideline in order to assess the feasibility and the efforts required for such a task. The number of participants, although sufficient for the overall evaluation, might be larger in future experiments and could strengthen the significance of the results when comparing different types of users (e.g., knowledge engineers versus. expert physicians). However, the second and third experiments focused on smaller tasks, but included more users. Overall, 27 users participated in the three tasks. In addition, although informal discussions with the participants of the study highlighted the usefulness of the knowledge-map constructs, we did not evaluate formally in a controlled manner each of its components. Such a study might be a suitable subject for further research.
6.2. Advantages of the Study
Over the past two decades, various groups have been conducting research to apply computerized methods to clinical guidelines. In this study we developed a framework for guideline specification in an attempt to address several of the unresolved issues mentioned in the background section, such as lack of an integrated guideline-specification methodology, use of a central guideline repository, and support for specification of both declarative and procedural knowledge. Our approach emphasizes the importance of a central digital library for sharing and reusing the clinical guidelines in a distributed environment. Providing the ability to share the guidelines allowed us to better support a collaborative specification process by expert physicians and knowledge engineers who are often located in remote locations. We also suggested an overall methodology for gradual specification that involves these different types of users. Since we aimed for representing the guidelines in a format that allows automatic application for tasks such as decision support, the methodology was designed to achieve a formal level of representation, in a gradual process. Similar to other groups developing tools to achieve formal specification, the methodology emphasizes the formal model (i.e. "model-centric") and begins with specifying a formal consensus for the guideline by the domain experts. Linking between the formal guidelines to the free-text sources allows us to easily maintain the formal guidelines and strengthens the validity of their recommendations. Consequently we used the intermediate structured-text representation and included references to the source guidelines in a manner similar to the "document-centric" approaches. The methodology also included the semi-formal specification that supports a semi-automatic application. To validate the overall process we evaluated the methodology and the framework in three experiments. These experiments included 27 participants and resulted in high levels of validity for the specification process. The framework was also given high usability scores by the users (and even higher by knowledge engineers), an encouraging fact that strengthens our belief in the feasibility of representing formal clinical guidelines and implementing our overall vision for automating guideline-based care.
ACKNOWLEDGEMENTS
This research was supported in part by NIH award LM-06806.
Dr. Arie Koyfman, Dr. Adit Dubi-Sobol, Dr. Gay Bar, and Dr. Aviram Weiss were extremely helpful in the evaluation of all framework tools.
REFERENCES
| [1] | Grimshaw JM, Russel I T. Effect of clinical guidelines on medical practice: A systematic review of rigorous evaluations Lancet 1993; 342: 1317-22. |
| [2] | Ruben A, Laura P, Marie DW, Darrell JG, Neil RP. Clinical Information Technologies and Inpatient Outcomes A Multiple Hospital Study Arch Intern Med 2009; 169(2): 108-4. |
| [3] | Patkar V, Hurt C, Steele R, et al. Evidence-based guidelines and decision support services: a discussion and evaluation in triple assessment of suspected breast cancer Br J Cancer 2006; 95(11): 1490-6. |
| [4] | Quaglini S, Cavallini A, Gerzeli S, Micieli G. Economic benefit from clinical practice guideline compliance in strok patient management Health Policy 2004; 69: 305-15. |
| [5] | Shahar Y, Young O, Shalom E, et al. A hybrid, multiple-ontology framework for specification and retrieval of clinical guidelines J Biomed Inform 2004; 37(5): 325-44. |
| [6] | Shalom E, Shahar Y, Taieb MM, et al. A quantitative evaluation of a methodology for collaborative specification of clinical guidelines at multiple representation levels J Biomed Inform 2008; 41(6): 889-903. |
| [7] | Shalom E, Shahar Y, Taieb MM, et al. Ability of expert physicians to structure clinical guidelines: reality versus perception J Eval Clin Pract 2009 Dec; 15(6): 1043-53. |
| [8] | Peleg M, Tu S, Bury J, et al. Comparing computer-interpretable guideline models: a case-study approach J Am Med Inform Assoc 2003; 10(1): 52-68. |
| [9] | De Clercq P, Blom J, Korsten HHM , Hasman A. Approaches for creating computer-interpretable guidelines that facilitate decision support Artif Intell Med 2004; 31(1): 1-27. |
| [10] | Soloway E, Bachant J, Jensen K. Assessing the maintainability of XCON-in-RIME: coping with the problems of a very large rule-base AAAI-87 Proceedings 1987; 824-9. |
| [11] | Stefik M. Introduction to Knowledge Systems. San Fransisco, California: Morgan Kaufmann 1995; pp. 626-35. |
| [12] | Svatek V, Ruzicka M. Step-by-step mark-up of medical guideline documents Int J Med Inform 2003; 70: 329-5. |
| [13] | Shortliffe EH, Scott A, Bischoff MB, Campbell AB, Melle WV, Jacobs CD. ONOCIN: an expert system for oncology protocol management Proceedings of the 7th international Joint conference on artificial intelligence 1981. |
| [14] | Tu SW, Kahn MG, Musen MA, Ferguson JC, Shortliffe EH, Fagan LM. Episodic Skeletal-plan refinement on temporal data Commun ACM 1989; 32: 1439-55. |
| [15] | Musen MA, Tu SW, Das AK, Shahar Y. EON: A component-based approach to automation of protocol-directed therapy J Am Med Inform Assoc 1996; 3(6): 367-88. |
| [16] | Ohno-Machado L, Gennari JH, Murphy SN, et al. The guideline interchange format: a model for representing guidelines J Am Med Inform Assoc 1998; 5(4): 57-72. |
| [17] | Peleg M, Boxwala A, Bernstam E, Tu SW, Greenes RA, Shortliffe EH. Sharable representation of clinical guidelines in GLIF: Relationship to the Arden syntax J Biomed Inform 2001; 34: 170-81. |
| [18] | Boxwala AA, Peleg M, Tu S, et al. GLIF3: a representation format for sharable computer-interpretable clinical practice guidelines J Biomed Inform 2004; 37(3): 47-61. |
| [19] | Fox J, Johns N, Rahmanzadeh A. Disseminating medical Knowledge: the PROforma approach Artif Intell Med 1998; 14(1): 157-81. |
| [20] | Fox J, Patkar V, Thomson R. Decision support for health care: the PROforma evidence base Inform Prim Care 2006; 14(1): 49-54. |
| [21] | Shahar Y, Miksch S, Johnson P. The Asgaard project: A task-specific framework for the application and critiquing of time-oriented clinical guidelines Artif Intell Med 1998; 14(1-2): 29-51. |
| [22] | Miksch S. Plan management in the medical domain AI Commun 1999; 12(4): 209-35. |
| [23] | Seyfang A, Miksch S, Marcos M. Combining diagnosis and treatment using asbru Int J Med Inform 2002; 68(1): 49-57. |
| [24] | Johnson PD, Tu SW, Booth N, Sugden B, Purves IN. Using scenarios in chronic disease management guidelines for primary care Proc AMIA Annu Symp 2000; 389-93. |
| [25] | Purves IN, Sugden B, Booth N, Sowerby M. The PRODIGY project-the iterative development of the release one model Proc AMIA Annu Symp 1999; 359-63. |
| [26] | Quaglini S, Dazzi L, Stefanelli M, Fassino C, Tondini C. Supporting tools for guideline development and dissemination Artif Intell Med 1998; 14(1-2): 19-37. |
| [27] | Quaglini S, Stefaneli M, Lanzola G, Caporusso V, Panzarasa S. Flexible guideline-based patient careflow systems Artif Intell Med 2001; 22: 65-80. |
| [28] | Terenziani P , Montani S , Bottrighi A , Torchio M , Molino G , Correndo G. The GLARE approach to clinical guidelines: main features Stud Health Technol Inform 2004; 101: 162-6. |
| [29] | Ram P, Berg D , Tu S , et al. Executing clinical practice guidelines using the SAGE execution engine Medinfo 2004; 11: 251-5. |
| [30] | De Clercq PA, Blom JA, Hasman A, Korsten HHM. Design and implementation of a framework to support the development of clinical guidelines Int J Med Inform 2001; 64(2- 3 ): 285-318. |
| [31] | Skonetzki S, Gausepohl H, Haak M, Knaebel S, Linderkamp O, Wetter T. HELEN, a modular framework for representing and implementing clinical practice guidelines Methods Info Med 2004; 43(4): 413-26. |
| [32] | Kawamoto K, Lobach DF. Design, implementation, use, and preliminary evaluation of SEBASTIAN, a standards-based web service for clinical decision support Proc AMIA Annu Symp 2005; 380-4. |
| [33] | Gordon C, Herbert I, Johnson P, Nicklin P, Reeves P. Telematics for clinical guidelines: a conceptual modelling approach Stud Health Technol Inform 1997; 43(Pt A): 314-8. |
| [34] | Shiffman RN, Michel G, Essaihi A, Thornquist E. Bridging the guideline implementation gap: a systematic, document-centered approach to guideline implementation J Am Med Inform Assoc 2004; 11(5 ): 418-26. |
| [35] | Hagerty CG, Chang J, Pickens DS, Kulikowski CA, Sonnenberg FA. Semi-automated encoding of guidelines Medinfo 2004; 1625. |
| [36] | Votruba P, Miksch S, Seyfang A, Kosara R. Tracing the formalization steps of textual guidelines Stud Health Technol Inform 2004; 101: 172-6. |
| [37] | Seyfang A, Miksch S, Conde CP, Wittenberg J, Marcos M, Rosenbrand K. A many-headed bridge between informal and formal guideline representations Proc Artif Intell Med (AIME) 2005. |
| [38] | Moskovitch R, Martins SB, Behiri E, Weiss A, Shahar Y. Comparative Evaluation of Full-text, Concept-based, and Context-sensitive Search J Am Med Inform Assoc 2007; 14(2): 164-74. |
| [39] | Moskovitch R, Shahar Y. Vaidurya: A multiple-ontology, concept-based, context-sensitive clinical-guideline search engine J Biomed Inform 2009; 42(1): 11-21. |
| [40] | Young O, Shahar Y, Lunenfeld E, et al. Runtime application of hybrid-Asbru clinical guidelines J Biomed Inform 2007; 40: 507-26. |
| [41] | Michael N, Munindar P. Service-Oriented Computing: Key Concepts and Principles IEEE Internet Comput 2005; 09(1): 75-81. |
| [42] | Wright A, Sittig F D. SANDS: An Architecture for Clinical Decision Support in a National Health Information Network Proc AMIA Annu Symp 2007; 816-20. |
| [43] | Middleton B. The clinical decision support consortium Stud Health Technol Inform 2009; 150: 26-30. |
| [44] | American College of Obstetricians and Gynecologists. Diagnosis and management of preeclampsia and eclampsia. Washington (DC) 2002; p. 33. |
| [45] | Brooke J. SUS-A quick and dirty usability scale, http://www.usability.serco.com/trump/documents/Suschapt.doc 1996. |
Endorsements
Browse Contents
Table of Contents
- INTRODUCTION
- OBJECTIVES
- BACKGROUND
- METHODS
- EVALUATION
- Experiment 1: Evaluation of the Interface for the Specification of Procedural Aspects of Clinical Guidelines
- SUMMARY AND DISCUSSION