- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Medical Informatics Journal
(Discontinued)
ISSN: 1874-4311 ― Volume 13, 2019
The Integrated Proactive Surveillance System for Prostate Cancer
Haibin Wang*, 1, Mahendra Yatawara 1, Shao-Chi Huang 1, Kevin Dudley 1, Christine Szekely 2, Stuart Holden 3, Steven Piantadosi 4
Abstract
In this paper, we present the design and implementation of the integrated proactive surveillance system for prostate cancer (PASS-PC). The integrated PASS-PC is a multi-institutional web-based system aimed at collecting a variety of data on prostate cancer patients in a standardized and efficient way. The integrated PASS-PC was commissioned by the Prostate Cancer Foundation (PCF) and built through the joint of efforts by a group of experts in medical oncology, genetics, pathology, nutrition, and cancer research informatics. Their main goal is facilitating the efficient and uniform collection of critical demographic, lifestyle, nutritional, dietary and clinical information to be used in developing new strategies in diagnosing, preventing and treating prostate cancer.
The integrated PASS-PC is designed based on common industry standards – a three tiered architecture and a Service- Oriented Architecture (SOA). It utilizes open source software and programming languages such as HTML, PHP, CSS, JQuery, Drupal and MySQL. We also use a commercial database management system – Oracle 11g. The integrated PASS-PC project uses a “confederation model” that encourages participation of any interested center, irrespective of its size or location. The integrated PASS-PC utilizes a standardized approach to data collection and reporting, and uses extensive validation procedures to prevent entering erroneous data. The integrated PASS-PC controlled vocabulary is harmonized with the National Cancer Institute (NCI) Thesaurus. Currently, two cancer centers in the USA are participating in the integrated PASS-PC project.
The final system has three main components: 1. National Prostate Surveillance Network (NPSN) website; 2. NPSN myConnect portal; 3. Proactive Surveillance System for Prostate Cancer (PASS-PC). PASS-PC is a cancer Biomedical Informatics Grid (caBIG) compatible product. The integrated PASS-PC provides a foundation for collaborative prostate cancer research. It has been built to meet the short term goal of gathering prostate cancer related data, but also with the prerequisites in place for future evolution into a cancer research informatics platform. In the future this will be vital for successful prostate cancer studies, care and treatment.
Article Information
Identifiers and Pagination:
Year: 2012Volume: 6
First Page: 1
Last Page: 8
Publisher Id: TOMINFOJ-6-1
DOI: 10.2174/1874431101206010001
Article History:
Received Date: 5/12/2011Revision Received Date: 31/1/2012
Acceptance Date: 16/2/2012
Electronic publication date: 2/3/2012
Collection year: 2012
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Research Informatics Core, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA, 90048, USA; Tel: (01)310-423-3315; Fax: (01)310-423-4020; E-mail: haibin.wang@cshs.org
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 5-12-2011 |
Original Manuscript | The Integrated Proactive Surveillance System for Prostate Cancer | |
INTRODUCTION
Prostate cancer is a cancer that forms in tissues of the prostate (a gland in the male reproductive system found below the bladder and in front of the rectum). It is the most common cancer affecting men in the United States. More than 200,000 new cases are expected to be diagnosed in 2011 [1Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics CA Cancer J Clin 2011; 61: 69-90.]. The majority of diagnosed men will not have disease that will result in prostate cancer specific mortality; however, nearly 30,000 men will die from prostate cancer this year. The introduction of serum prostate specific antigen (PSA) screening (around 1990) led to a transient increase in prostate cancer diagnosis. Furthermore, the pattern of initial presentation of patients shifted to men with low volume disease. The long natural history of this disease has been characterized [2Pound CR, Partin AW, Eisenberger MA, Chan DW, Pearson JD, Walsh PC. Natural history of progression after PSA elevation following radical prostatectomy JAMA 1999; 281(17 ): 1591-7.] and as a result has raised concerns that there may be excessive use of local intervention in men with low risk disease. This is accentuated by the recognized morbidity of the various forms of local therapy. In 2011, the initial report from the Veterans Administration sponsored PIVOT study [3American Urological Association, PIVOT study. Available from: http://clinicaltrials.gov/ct2/show/NCT00007644 [cited 2012 Jan 26];] demonstrated a lack of mortality benefit for men with low-risk prostate cancer who underwent surgical intervention. Conversely, in ongoing active surveillance series, it has been shown that approximately one third of men deemed appropriate for active surveillance show evidence of progression that merits consideration of intervention while the remainder either remain stable and eventually terminate follow-up or, due to excessive anxiety, elect to proceed with therapy despite a lack of evidence of progression [4Tosoian JJ, Trock BJ, Landis P, et al. Active surveillance program for prostate cancer: an update of the Johns Hopkins experience J Clin Oncol 2011; 29: 2185-90.].
These data together underscore the need for understanding the natural history and biology of low-risk disease and the impact of the practice patterns of active surveillance on men with low risk disease. Several academic institutions have programs of active surveillance in which men with low-risk cancers have undergone intense observation. Low-risk cancers are typically those which 1) are small volume prostate cancers that cannot be felt on a prostate examination (digital rectal exam) and 2) lack aggressive histological morphology (microscopic appearance). These active surveillance routines have been institution specific; as such, a more comprehensive active surveillance approach is necessary. Active surveillance that is accompanied with biospecimen collection represents a key need in prostate cancer research and an evolution of this process has been coined “pro-active surveillance”.
Currently, there are many open source and commercial clinical data management systems available such as Research Electronic Data Capture (REDCap) [5Harris P, Taylor R, Thielke R, Payne J, Gonzalez N, Conde J. Research electronic data capture (REDCap) – A metadata-driven methodology and workflow process for providing translational research informatics support J Biomed Info 2009; 42: 377-81.], OpenClinica [6OpenClinica Available from: https://www.ope nclinica.com/ [cited 2011 July 20];], and Medidata [7Medidata Available from: http://www.mdsol. com/ [cited 2011 July 20];], etc. Here, we give a brief overview of REDCap and OpenClinica.
REDCap is a secure Web application for building and managing online surveys and databases. It employs a novel workflow methodology and the software solution is designed for rapid development and deployment of electronic data capture tools to support clinical and translational research [2Pound CR, Partin AW, Eisenberger MA, Chan DW, Pearson JD, Walsh PC. Natural history of progression after PSA elevation following radical prostatectomy JAMA 1999; 281(17 ): 1591-7.]. It is built using PHP and MySQL.
OpenClinica is powerful software for collecting and managing clinical trial data. It allows you to build your own studies, design electronic Case Report Forms (eCRFs), and conduct a full range of Electronic Data Capture (EDC) and Clinical Data Management (CDM) functions. It is a Web-based system and is built using J2EE and Oracle or PostgreSQL.
The integrated PASS-PC is a Web-based distributed, heterogeneous clinical data system developed to support the research study entitled “Active Surveillance of Prostate Cancer” for multi-center clinical sites.
The study has two objectives:
Primary Objective
To carefully observe men (active surveillance) with screened detected low risk prostate cancer and manage them without immediate curative intervention.
Secondary Objective
To explore urine and serum collected in order to develop and evaluate new and existing biomarkers for prostate cancer, evaluate biomarker changes, study gene expression profiles, and evaluate nuclear proteins.
Web-based data management systems offer great potential for facilitating the conduct of large scale or multi-center clinical studies [8Avidan A, Weissman C, Sprung CL. An internet web site as a data collection platform for multicenter research Anesth Analg 2005; 100(2
): 506-11.-11Zhang GQ, Siegler T, Saxman P, et al. VISAGE: a query interface for clinical research Proceedings of the 2010 AMIA clinical
research informatics summit, San Francisco. ]. Investigators and researchers working across multiple sites with varying infrastructure can access data and analytical tools in these systems on a real-time basis, minimizing the logistical challenges in multi-center collaboration, providing improved monitoring capability, and facilitating new mechanisms for producing high quality validated data [10Tran VA, Johnson N, Redline S, Zhang GQ. OnWARD: Ontology-driven web-based framework for multi-center clinical studies J Biomed Inform 2011.
]. The integrated PASS-PC is a Health Insurance Portability and Accountability Act (HIPAA) compliant and caBIG [12Eschenbach A, Buetow K. Cancer informatics vision: caBIGTM Caner Inform 2006; 2: 22-4.] compatible Web-based clinical data management system incorporating three main components:
- National Prostate Surveillance Network (NPSN) website – an informational website for proactive surveillance of prostate cancer;
- NPSN myConnect Portal – A secure patient registration web portal;
- PASS-PC – Secure study management portal for researchers and study coordinators.
The integrated PASS-PC is based on a legacy stand-alone Microsoft Access database developed by John Hopkins University in 2006. After investigation of REDCap, OpenClinica, and Medidata, etc., we decided to design and implement an in-house system to 1) facilitate migration of data from the legacy Access database and 2) efficiently integrate with the NPSN myConnectdatabase.
METHODS
The integrated PASS-PC team elected to use a “confederation model” [13Sherman S, Shats O, Ketcham MA, et al. PCCR: Pancreatic cancer collaborative registry Cancer Informatics 2011; 10: 83-91.], as opposed to traditional data repository or network models that assume control of an individual center’s data. A confederation model assumes that each participating site retains all rights to the acquired data that can be used by other integrated PASS-PC participants only after obtaining required permissions and approved by its Institutional Review Board (IRB). It is essential to have a standardized approach to data collection and reporting for this model to be successful.
The integrated PASS-PC is designed and implemented based on common industry standards – a three tiered distributed architecture and a Service-Oriented Architecture (SOA).
Based on the legacy standalone MS Access database developed by John Hopkins, the integrated PASS-PC defines and establishes the criteria for standardization of collection forms and identified research questions that must be addressed. Baseline and followup questionnaires and a dietary food frequency questionnaire (FFQ) have been implemented in the integrated PASS-PC.
The integrated PASS-PC establishes a core data set of information to which all participating centers must contribute. The core data elements include the most common questions used in clinical, nutritional, and quality of life studies. Additionally, the core data set includes the following data elements: registering institution, staff member performing data entry, and patient identification code.
Patients will provide information on demographic, lifestyle, physical activity, dietary habits, family history, male and female relatives’ health, and medical history; whereas information on diagnostic studies, pathology/ staging, treatment, surgeries, and biospeciments can only be provided by research coordinator.
The integrated PASS-PC controlled vocabulary is harmonized with NCI Thesaurus. The data elements have been defined based on the NCI Cancer Data Standards Registry and Repository (caDSR) [14Cancer Data Standards Registry and Repository (caDSR) Available from: https: //cabig.nci.nih.gov/concepts /caDSR/ [cited 2011 July 20];].
First, the data model is built using the open source data modeling software – ArgoUML [15ArgoUML Available from: http://argouml.tigris.org/ [cited 2011 July 20];], and metadata is created based on NCI’s controlled vocabulary. Next, the Unified Modeling Language (UML) data model is loaded into the NCI’s caDSR production server. Finally, the data definition language (DDL) script for Oracle database is generated from the data model. The process flow for the system model design is shown in Fig. (1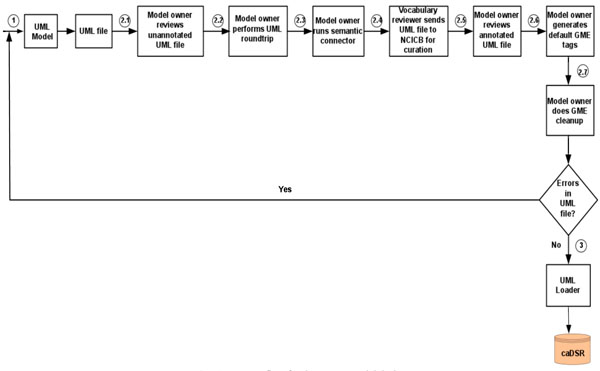 ).
).
 |
Fig. (1) Process flow for the system model design. The algorithmic steps for the process flow are described as: Begin Step 1: Build the object and data models using UML modeling tool such as ArgoUML or Enterprise Architect [16Enterprise Architect Available from: http://www.sparxsystems.com/ [cited 2011 July 20];]. Step 2: There are several separate steps in running Semantic Integration Workbench (SIW): Step 2.1 (by Model Owner): Review unannotated XML Metadata Interchange (XMI) or UML file built in Step 1. Step 2.2 (by Model Owner): Perform XMI or UML roundtrip. Step 2.3 (by Model Owner): Run semantic connector. If no errors such as invalid data types and “unbounded array” in model found in steps 2.1-2.3, then proceed to step 2.4; otherwise, go to step 1 to correct the errors and repeat steps 2.1-2.3. Step 2.4 (by Vocabulary Reviewer): Send XMI or UML file via email to the NCICB to curate the file. Step 2.5 (by Model Owner): Review annotated XMI or UML file. Step 2.6 (by Model Owner): Generate default Global Model Exchange (GME) tags. Step 2.7 (by Model Owner): GME cleanup. If no errors found in steps 2.4-2.7, then proceed to step 3; otherwise, go to step 1 to correct the errors and repeat steps 2.1-2.7. Step 3: Run UML Loader by the NCICB to load the approved annotated XMI or UML file into the caDSR. End. |
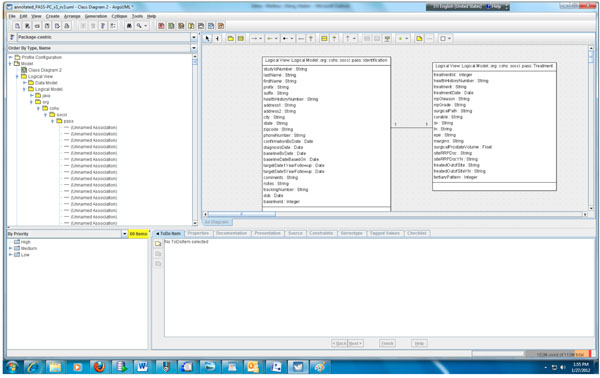 |
Fig. (2) Partial object model of the PASS-PC. |
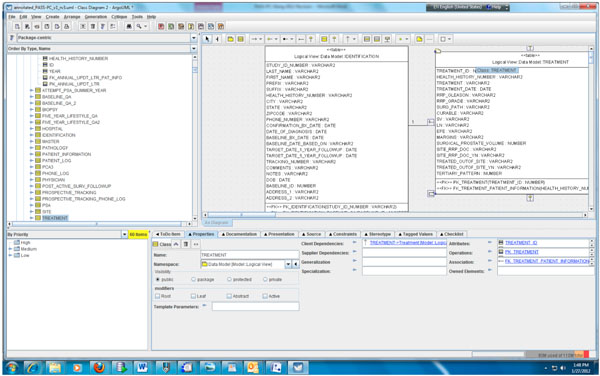 |
Fig. (3) Partial data model of the PASS-PC. |
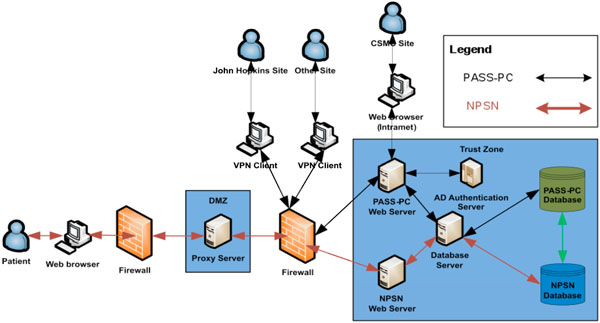 |
Fig. (4) The high level system architecture of the integrated PASS-PC. |
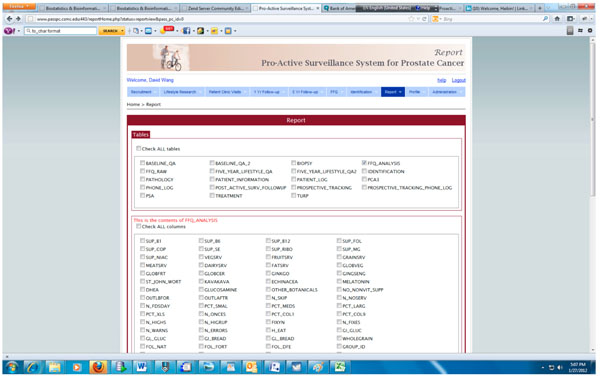 |
Fig. (7) Query interface for generating report in PASS-PC. |
According to step 1, we first need to create the object and data models for the underlying PASS-PC database. Here, we use ArgoUML 0.28, a free open source application, to create the models. The output of this procedure is a UML (XML format) file. Screenshots of the partial object and data models in ArgoUML are shown in Figs. (2 , 3
, 3 ), respectively.
), respectively.
The PASS-PC web application is developed using HTML, PHP, CSS and JQuery with an Oracle 11g backend database. The Apache web server and Oracle database server are running on the Red Hat Linux.
The NPSN website is developed using PHP and the Drupal content management framework, and runs on the Red Hat Linux Apache web server.
The patient’s registration web portal (myConnect) has a login interface on the NPSN website and utilizes a MySQL backend database.
The MySQL and Oracle databases are located on the two different physical Red Hat Linux database servers at CSMC’s Enterprise Information Services (EIS) data center. These two databases exchange stored patient health information (PHI) in an encrypted format via Web Service call. In current project’s phase I stage, there is only unidirectional information flow – from myConnect’s MySQL database to PASS-PC’s Oracle database via Web Service on hourly basis. In project’s phase II stage, there will be bidirectional information flow between myConnect’s MySQL database and PASS-PC’s Oracle database via Web Service.
The high level system architecture is shown in Fig. (4 ).
).
Fig. (5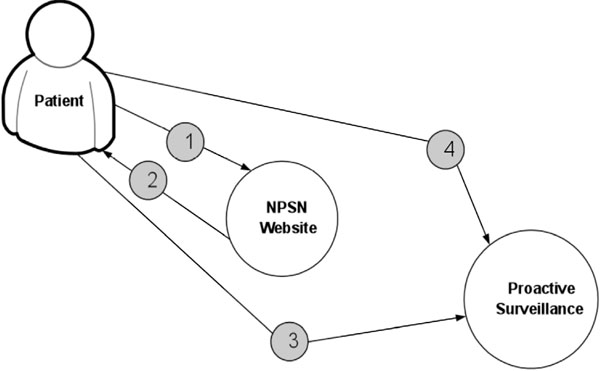 ) displays the procedure for determining patient’s eligibility for the study. Fig. (6
) displays the procedure for determining patient’s eligibility for the study. Fig. (6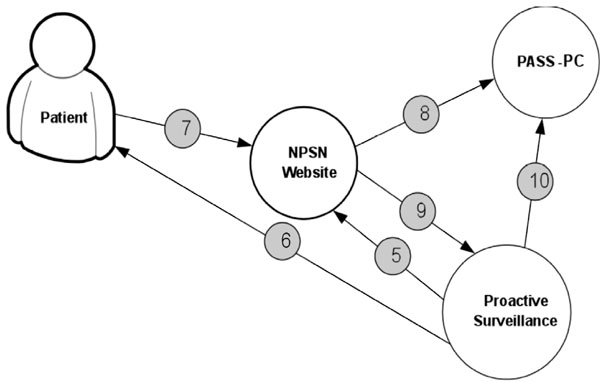 ) shows the procedure of PASS-PC connectivity to NPSN and myConnect web portals.
) shows the procedure of PASS-PC connectivity to NPSN and myConnect web portals.
Cedars-Sinai Medical Center is the Data Coordinating Center (DCC) for this Active Surveillance study. Multiple clinic sites will be able to simultaneously access and store PHI in the integrated PASS-PC. As such, security is the first priority in designing the system in order to meet HIPAA compliance.
In keeping with the CSMC’s EIS standards, the web server is SSL encrypted and all browsers accessing the server will use https. This applies to both internal and external users.
User logins to databases associated with integrated PASS-PC is via 2-factor authentication utilizing username, password and control-ID.
Authenticated users will be presented with different web interfaces based on their respective user role such as Program Administrator, Research Coordinator orViewer which is assigned by a system administrator at the time of user registration. A program administrator can view, edit and add PHI records in the PASS-PC database. He/she can also add new users to its site. A program administrator can generate different types of reports based on various database queries and export reports in a format compatible with common statistical software packages such as SAS. A research coordinator can view, edit and add PHI records in the PASS-PC database. He/she can also generate reports based on various database queries and export reports in a format compatible with common statistical software packages such as SAS. A viewer can only view PHI records and generate reports.
All users belong to a clinical site and are restricted to accessing PHI associated with that site.
All PHI is stored in an encrypted format in both PASS-PC and myConnect databases.
Passwords are also encrypted and are set at a minimum of 12 characters and must include one number, one upper case letter, one lower case letter and one special character. All users will be forced to change their passwords on initial login and every six months thereafter.
All users will be automatically logged out after 10 minutes of inactivity.
RESULTS
The integrated PASS-PC is implemented as a cancer research informatics data repository to support the proactive surveillance for prostate cancer study. Fig. (7 ) presents the screenshot of the query interface for generating various reports which can be downloaded and imported into standard statistics software.
) presents the screenshot of the query interface for generating various reports which can be downloaded and imported into standard statistics software.
The integrated PASS-PC uses a two-factor authentication methodology to prevent unauthorized access to the system. User of the system needs to provide the correct username and password to pass the first layer of authentication, and then he/she has to provide the correct control id to pass the second layer of authentication. The integrated PASS-PC maintains an audit trail of all data entries and user activities to protect the authenticity, integrity and confidentiality of all data entries.
An entry is inserted into USER_AUDIT table whenever the following actions occur:
- User logs in to the PASS-PC database
- User logs out of the PASS-PC database
Variables included in entry of USER_AUDIT table are presented in Table 1.
An entry is inserted into OPERATION_AUDIT table whenever the following actions occur:
- User views a specific record
- User edits a specific record
- User creates a new record
Variables included in entry of OPERATION_AUDIT table are presented in Table 2.
Currently, there are about 800 prior patients in John Hopkins site needed to be re-consented to transfer their legacy data into the integrated PASS-PC. Currently, Cedars-Sinai Medical Center and John Hopkins Hospital are two participating sites. As a cancer research informatics computing platform for proactive surveillance for prostate cancer study, the integrated PASS-PC would lead to a better understanding and treatment of men with low risk prostate cancer.
The integrated PASS-PC is a multi-disciplinary project. Its success is based on the collaborative efforts of multiple individuals and teams with expertise in medical oncology, genetics, pathology, prostate cancer, nutrition, and information technology.
The NPSN website and myConnect web portal can be accessed at: http://www.npsn.net:443.
The PASS-PC can be accessed by participating clinic sites at: http://www.passpc.csmc.edu:443.
DISCUSSION
The integrated PASS-PC project, as a unique web-based, caBIG compatible and multi-center clinical data management system, has high reliability and extensibility. The integrated PASS-PC offers a number of benefits to its users, including: i) standardized data elements, vocabulary and forms for data collection; ii) high quality data validation for data entry; iii) highest level network and data security by firewall, vpn, two factor authentication, data encryption and audit trail; iv) generating various reports in spreadsheet format based on dynamic queries of data elements.
PCCR [13Sherman S, Shats O, Ketcham MA, et al. PCCR: Pancreatic cancer collaborative registry Cancer Informatics 2011; 10: 83-91.], a pancreatic cancer collaborative registry, is a multi-institutional Web-based system aimed to collect a variety of data on pancreatic cancer patients and high-risk subjects in a standard and efficient way. PCCR also utilized a “confederation model” as its architecture. Similar to the integrated PASS-PC, PCCR also adopted standardized data element, controlled vocabulary and forms for data collection. PCCR’s controlled vocabulary is in harmonization with NCI Thesaurus, as the same resource used by the integrated PASS-PC. Both systems follow the NCI caBIG compatible standards. The differences between the integrated PASS-PC and PCCR are: i) the integrated PASS-PC is developed in HTML, PHP, CSS, JQuery, Drupal, MySQL and Oracle 11g. PCCR is developed in Java/JSP and Oracle 10g; ii) the integrated PASS-PC consists of three components: NPSN website, NPSN myConnect portal and PASS-PC. It is based on the three-tier and service-oriented architecture. NPSN myConnect portal communicates with the PASS-PC through web service calls on an hourly basis. PCCR is a centralized registry.
BCCR [17Sherman S, Shats O, Fleissner E, et al. Multicenter breast cancer collaborative registry Cancer Inform 2011; 10: 217-6.], a breast cancer collaborative registry, is a multicenter Web-based system aimed to collect and manage a variety of data on breast cancer patients and breast cancer survivors. BCCR is developed by following the same methodology as PCCR, so it is similar to the integrated PASS-PC except that the differences we listed above between the integrated PASS-PC and PCCR.
Kong MY, et al. [18Kong MY, Dahlke C, Xiang Q, Qian Y, Karp D, Scheuermann RH. Toward an ontology-based framework for clinical research databases J Biomed Info 2011; 44: 48-58.] described an ontology-based framework for clinical research databases. The Ontology-Based eXtensible data model (OBX) was developed to serve as a framework for clinical research data in the Immunology Database and Analysis Portal (ImmPort). Similar to the integrated PASS-PC, OBX is a relatively simple conceptual model. The difference between two systems is that the integrated PASS-PC is a specialized system for proactive surveillance for prostate cancer study; OBX is a general data model for an immunology database and analysis portal.
In the market, REDCap, OpenClinica and Medidata are good clinical data management systems. But these systems are not suitable for this project because we need to migrate the legacy data stored in the MS Access database and the in-house software provides greater flexibility, control, extensibility and range of import/export options.
The integrated PASS-PC has a user-friendly graphical user interface (GUI) to help access the patient health information in the database. The security is first priority in designing and implementing the system. The current system also has some limitations. For example, it does not support the storage and retrieval of biospecimen images, and it does not integrate with other EMR systems currently running at Cedars-Sinai Medical Center.
CONCLUSIONS
The integrated PASS-PC project utilizes open source software and industry standards-based technologies in order to design, develop and deploy an extensible and integrative clinical data management platform. The design of the system follows the NCI’s caBIG paradigm to facilitate the integration among heterogeneous data systems and information sharing among these data systems.
The design principles of the integrated PASS-PC are generalized and should be informative to analogous efforts and programs.
Currently, the project is in phase I stage -unidirectional information flow from NPSN myConnect database to the PASS-PC database. Phase II is in the planning stage to implement bidirectional information flow between myConnect database and the PASS-PC database.
ACKNOWLEDGEMENTS
We thank the Prostate Cancer Foundation (PCF) for their donation in support of this project. We also thank Drs. Spencer Soohoo and Bob Varney for their support of providing the computing infrastructures in CSMC’s EIS Data Center.
REFERENCES
| [1] | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics CA Cancer J Clin 2011; 61: 69-90. |
| [2] | Pound CR, Partin AW, Eisenberger MA, Chan DW, Pearson JD, Walsh PC. Natural history of progression after PSA elevation following radical prostatectomy JAMA 1999; 281(17 ): 1591-7. |
| [3] | American Urological Association, PIVOT study. Available from: http://clinicaltrials.gov/ct2/show/NCT00007644 [cited 2012 Jan 26]; |
| [4] | Tosoian JJ, Trock BJ, Landis P, et al. Active surveillance program for prostate cancer: an update of the Johns Hopkins experience J Clin Oncol 2011; 29: 2185-90. |
| [5] | Harris P, Taylor R, Thielke R, Payne J, Gonzalez N, Conde J. Research electronic data capture (REDCap) – A metadata-driven methodology and workflow process for providing translational research informatics support J Biomed Info 2009; 42: 377-81. |
| [6] | OpenClinica Available from: https://www.ope nclinica.com/ [cited 2011 July 20]; |
| [7] | Medidata Available from: http://www.mdsol. com/ [cited 2011 July 20]; |
| [8] | Avidan A, Weissman C, Sprung CL. An internet web site as a data collection platform for multicenter research Anesth Analg 2005; 100(2 ): 506-11. |
| [9] | Cooper CJ, Cooper SP, del Junco DJ, Shipp EM, Whitworth R, Cooper SR. Web-based data collection: detailed methods of a questionnaire and data gathering tool Epidemiol Perspect Innov 2006; 3: 1. |
| [10] | Tran VA, Johnson N, Redline S, Zhang GQ. OnWARD: Ontology-driven web-based framework for multi-center clinical studies J Biomed Inform 2011. |
| [11] | Zhang GQ, Siegler T, Saxman P, et al. VISAGE: a query interface for clinical research Proceedings of the 2010 AMIA clinical research informatics summit, San Francisco. |
| [12] | Eschenbach A, Buetow K. Cancer informatics vision: caBIGTM Caner Inform 2006; 2: 22-4. |
| [13] | Sherman S, Shats O, Ketcham MA, et al. PCCR: Pancreatic cancer collaborative registry Cancer Informatics 2011; 10: 83-91. |
| [14] | Cancer Data Standards Registry and Repository (caDSR) Available from: https: //cabig.nci.nih.gov/concepts /caDSR/ [cited 2011 July 20]; |
| [15] | ArgoUML Available from: http://argouml.tigris.org/ [cited 2011 July 20]; |
| [16] | Enterprise Architect Available from: http://www.sparxsystems.com/ [cited 2011 July 20]; |
| [17] | Sherman S, Shats O, Fleissner E, et al. Multicenter breast cancer collaborative registry Cancer Inform 2011; 10: 217-6. |
| [18] | Kong MY, Dahlke C, Xiang Q, Qian Y, Karp D, Scheuermann RH. Toward an ontology-based framework for clinical research databases J Biomed Info 2011; 44: 48-58. |