- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Medical Informatics Journal
(Discontinued)
ISSN: 1874-4311 ― Volume 13, 2019
Digital Management of a Hysteroscopy Surgery Using Parts of the SNOMED Medical Model
Anastasios Kollias*, 1, Minas Paschopoulos2, Angelos Evangelou3, Marios Poulos1
Abstract
This work describes a hysteroscopy surgery management application that was designed based on the medical information standard SNOMED. We describe how the application fulfils the needs of this procedure and the way in which existing handwritten medical information is effectively transmitted to the application’s database.
Article Information
Identifiers and Pagination:
Year: 2012Volume: 6
First Page: 15
Last Page: 25
Publisher Id: TOMINFOJ-6-15
DOI: 10.2174/1874431101206010015
Article History:
Received Date: 15/2/2012Revision Received Date: 25/3/2012
Acceptance Date: 4/4/2012
Electronic publication date: 18/5/2012
Collection year: 2012
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Laboratory of Information Technology, Department of Archives and Library Sciences, Ionian University, Greece; Tel: +302661087429; Fax: +302661087433; E-mail: tkodka@ionio.gr
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 15-2-2012 |
Original Manuscript | Digital Management of a Hysteroscopy Surgery Using Parts of the SNOMED Medical Model | |
INTRODUCTION
In light of problems arising in the medical sector due to the increasing role of modern technology in our everyday lives, it is obvious that modern medical science needs to fully develop and exploit all forms of medical information originating from existing medical incidents. The main interface between different medical computer systems is served by the international medical standards which were formulated after years of studying and processing medical terminology.
SNOMED, the international standard for medical terminology, is a complete encoded system of medical information that is becoming the most recognized tool for exploiting medical knowledge. SNOMED achieves interoperability between different types of medical information systems [1 Lussier YA, Shagina L, Friedman C. Automating SNOMED coding using medical language understanding: a feasibility study In: Proc AMIA Symp. Philadelphia, PA: Hanley & Belfus 2001; pp. 418-22.] and also defines rules and processes in order to solve the problem of interoperability between computer systems based on different principles, as its content is not purely medical. Rapidly developing high-level technologies, and the need to solve multiple medical problems, are leading to the invention and application of new diagnostic and surgical procedures that require the authoring of computer applications capable of recording the highly specialized new data [2 Brook P, Health A. Standards and interoperability in healthcare information systems: current status, problems, and research Issues 2010.Moorhead, MN, USA. 2010. In: 5th Midwest Relevante Metadatenstandards Version 1.2.1 LABIMI/F 19 Deliverable Association for Information Systems Conference MWAIS; Available at: http://aisel.aisnet.org/mwais2010/18/]. One of these procedures is hysteroscopy, which constitutes the latest development in endoscopic surgery of the female genitalia. The study outlined here attempts to study and codify the results of this procedure.
Problems
The problems which were detected and which this study is attempting to solve are the following:
- Novel medical processes that are based on the application of new technologies contain unique characteristics and, as a result, cannot use existing computer control applications [3 Crichton C, Davies J, Gibbons J, Harris S, Tsui A, Brenton J. Metadata-driven software for clinical trials Proceedings of the 2009 ICSE Workshop on Software Engineering in Health Care Los Alamitos: IEEE 2009; pp: 1-11.]. When attempted, it causes problems for the functionality of medical procedures.
- The time required to develop and refine a specialized computer application capable of bridging the characteristics of a medical procedure with international medical standards for exploiting such information is disproportionately large to the initial development of the medical procedure itself [4 Kuhn K, Lenz R, Elstner T, Siegele H, Moll R. Experiences with a generator tool for building clinical application modules Methods Inf Med 2003; 42: 37-44.]. The result is the accumulation of a very large volume of medical information at medical organizations which either remains in primary electronic format or in handwritten documents.
- Each medical incident is unique for the attending physician, depending on the uniqueness of the patient, though digitization requires the “adaptation and placement” of incidents into groups with common elements and terms, aiming to exploit information stemming from the final result [5 Hillestad R, et al. Can electronic medical record systems transform health care? Potential health benefits, savings, and costs Health Aff 2005; 24: 1103.].
- In the case of surgery, the effort to digitize and record an incident with a computer application based on medical models may influence the medical development of an incident itself, eventually causing delays and dysfunctions that decrease the final reliability of the undertaking [6 Li Z, Xuan-Dong Y, Kai-Yang L. Digitization and DICOM standardization of X-ray image signal Wuhan Univ J Nat Sci 2004; 9: 463-7.].
- The usual strategy that is followed in the application of medical models while coding medical information is to use the medical standards entirely as well as independently to then bind medical terms [7 Young A. The creation of medical knowledge: some problems in interpretation Soc Sci Med Med Anthropol 1981; 15: 379-86.] with the incident’s data. However, this strategy is inferior in speed and functionality, since there are time-consuming intermediary calculations during this process of binding and the final user is not exempted from the requirement to have a full awareness of the function and structure of the entire model.
Aims and Objectives
The present study describes the methods used in the creation of a computer application to manage a hysteroscopy surgery based on the SNOMED medical model. Processes were defined and executed to transcribe the existing handwritten medical information of the surgery into a form that can be input into a database, thereby allowing it to be fully exploited. This practice is considered to be a research priority according to the resolutions of the European parliament [8Available at www.Hl7.org ].
During this effort, we attempted to construct and propose a process of coding hysteroscopy incidents. Up to now, hysteroscopy has not been interoperable with the SNOMED medical model [9Available at: www.cda.org ]. Also, the Clinical Document Architecture (CDA) [9Available at: www.cda.org ] has not yet adopted a relative architecture of harmonizing hysteroscopy to the medical model.
This work advanced in two stages. The first stage was to create the environment of the hysteroscopy medical process based on the established practice that is followed by doctors of this specialty. In the second stage, a mapping of the application fields to the elements of the SNOMED model was carried out using a mechanism for mapping XML schemas-SNOMED to object-relational database systems.
MATERIALS AND METHODS
The development of the application, together with the planning and implementation of transcribing the handwritten documents into the database, aimed to approach and interconnect two different concepts: the actual medical incident and its digitized form. The lack of existing information in electronic form was exploited: the format design, analysis and implementation were made flexibly. The logic of incorporation within the application was therefore selected in all the data input processes, segments of tables, record sets or individual codes belonging to the SNOMED model and the registration of primary information conforming to its database requirements.
This process, compared with the usual tactic that is followed in many similar cases (i.e., where the entire and independent tables of the model are used and then bound to medical terms depending on the data of the incident), outclasses them in speed and functionality. On one hand, time-consuming intermediary calculating binding procedures are avoided while on the other, the end user is not freed from the requirement to be fully aware of the operation and structure of the entire model. This results in trust and recognition by the doctor-users of the usefulness of models in medical informatics. The very flexible possibility of data output and input in the form of messages (Clinical Document Architecture: CDA) was added for registered medical incidents, which will lead to the creation of an information network for hysteroscopy and simultaneously will bridge the gap in communication and interoperability with other applications.
The methods that were followed in the design and implementation of the management application were determined by the fact that two basic conditions should be satisfied. The first was complete harmonization of all registered information with the requirements of the SNOMED medical standard; the second was that the operation of the application itself should by no means influence the medical process of hysteroscopy (rather, it should help by applying organization).
The SNOMED standard is not simply a dictionary of medical terms. It contains structures that define processes of data input, necessary as well as unnecessary property elements, essential term attributes, and non-term attributes that also determine the field values [10 Mochizuki M. Application methods of SNOP, SNOMED, ICD codes Mod Med Lab 2003; 31: 1409-0.]. Existing applications that use SNOMED for the registration of medical information [11 Elkin PL. Evaluation of the content coverage of SNOMED CT: ability of SNOMED clinical terms to represent clinical problem lists Mayo Clin Proc 2006; 81: 741-8.] are almost exclusively based on the fact that the operator has explicit knowledge of the structure and the function of the standard. These applications are controlled by the model in data input, since most of them use the organization of the model for user guidance. This helps because it determines techniques that contain the collective experience of many individuals that contributed to its construction and its continuous enrichment; however, it limits the possibilities for alternatives during data input. This was fundamentally problematic in the effort to apply the standard to a new medical process (hysteroscopy), since in no way should the medical procedure be altered from what the doctor had defined. It was therefore necessary to devise methods by which to adapt the standard to the specific medical process and not the procedure to the standard, something that initially appeared to be incompatible with the definition of the standard itself.
Procedure Analysis
In an effort to arrive at an accommodating solution it was decided to follow the medical procedure as it is defined by medical practice [12 Hamou JE. Hysteroscopy and micropolhysteroscopy: text and atlas. Norwalk CT: Appleton & Lange 1994.] (Fig. 1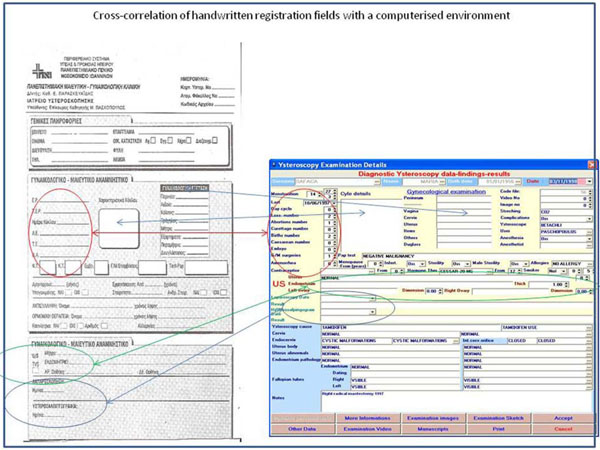 ). Though in the backstage and with methods that will be described below, the model was satisfied completely; only in exceptional cases (Fig. 2
). Though in the backstage and with methods that will be described below, the model was satisfied completely; only in exceptional cases (Fig. 2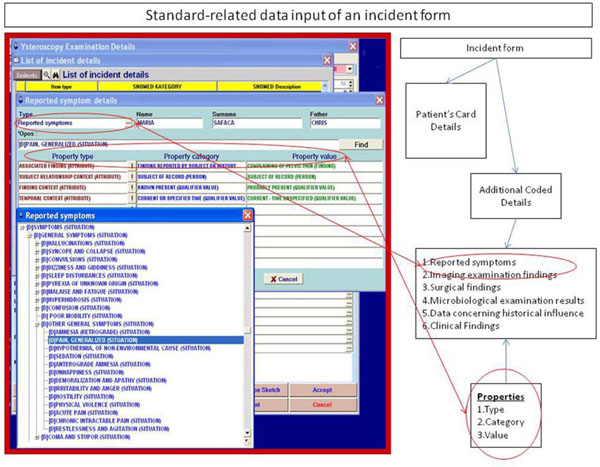 ) was any direct input made to the existing data. The steps followed are shown in Fig. (1
) was any direct input made to the existing data. The steps followed are shown in Fig. (1 ).
).
 |
Fig. (1) Cross-correlation of handwritten registration fields with a computerized environment |
 |
Fig. (2) Data input according to the rules of the standard. |
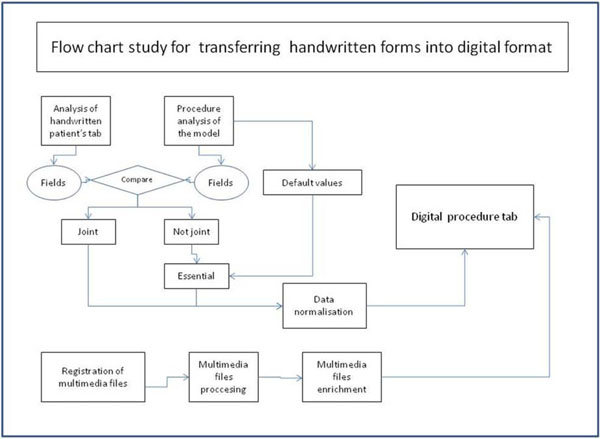 |
Fig. (3) Flow chart illustrating how handwritten forms were transferred into computerized format. |
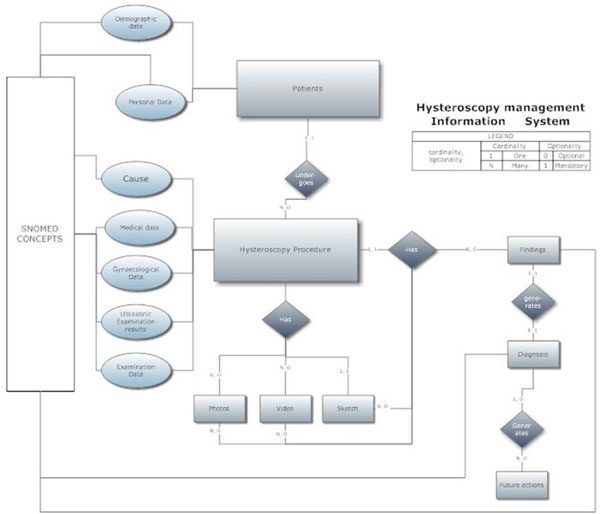 |
Fig. (4) Depiction of relational tables of the database in the Entity-Relation-Diagram (ERD). |
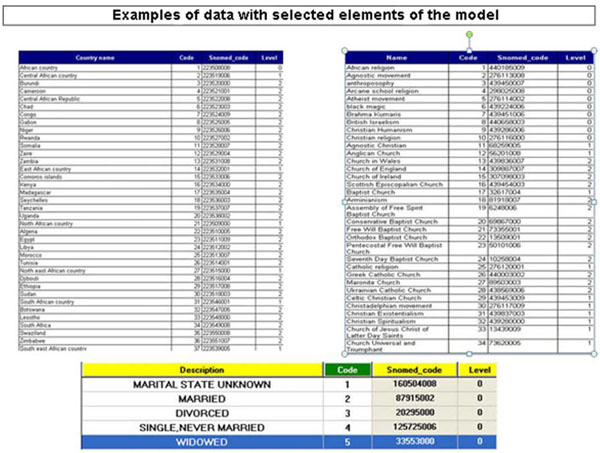 |
Fig. (5) Examples of data with selected elements of the model. |
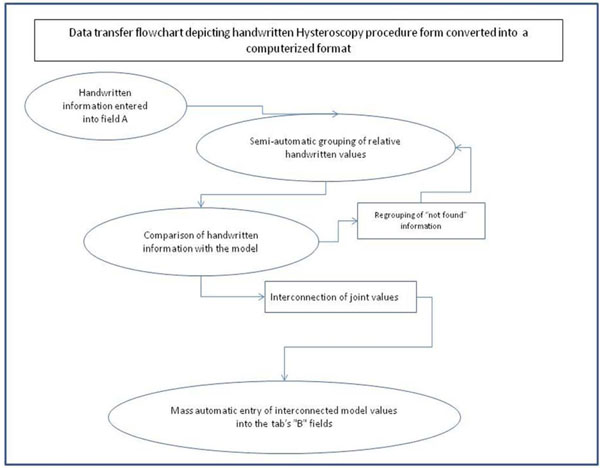 |
Fig. (6) Data transfer flowchart. |
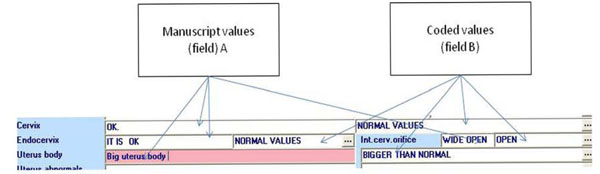 |
Fig. (7) Example of the “double field” use. |
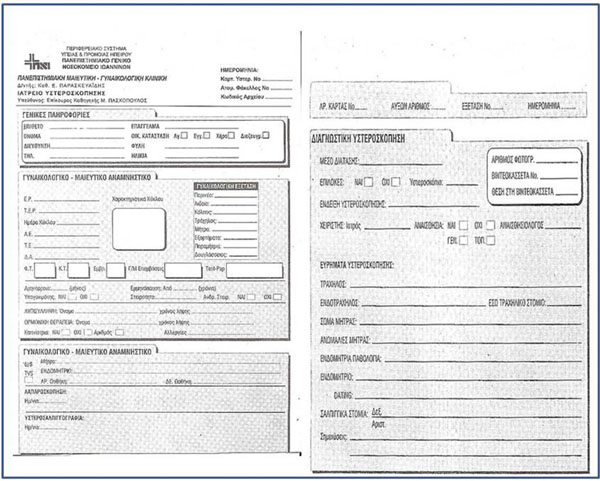 |
Fig. (8) Example of handwritten registration form. |
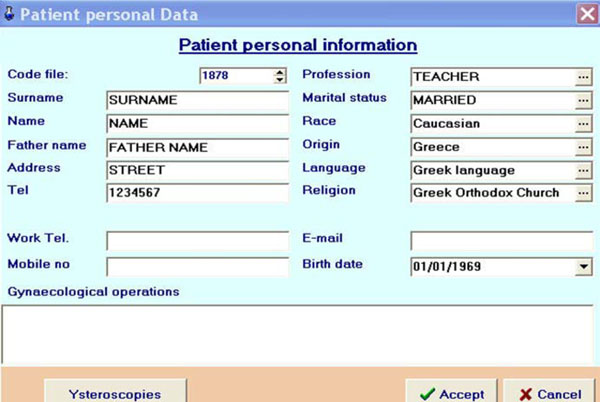 |
Fig. (9) Input of patient's personal information into the database. |
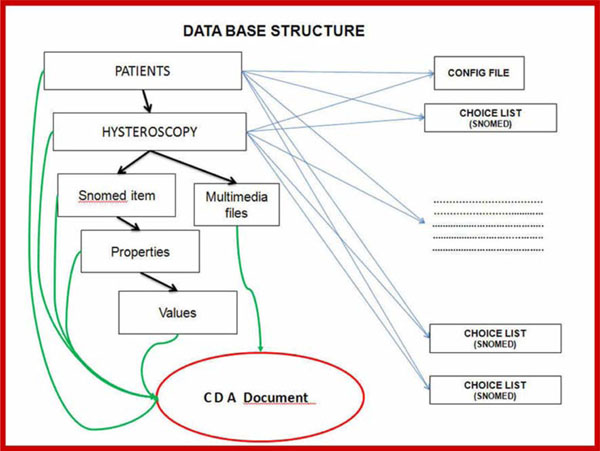 |
Fig. (10) Database Medical Document creation process. |
The proposed process includes six stages, which are schematically represented in Fig. (3 ):
):
- The data used in the process were analyzed (Table 1). The fields and the order of data input were researched before the corresponding terms were located in the medical model. Wherever it was judged necessary for fields that had not been forecasted in advance, it was planned for them to be added into the application given default values without user intervention.
- The model's tables and the relationships and properties of the medical terms used were analyzed in order to ascertain to which point they coincided with the handwritten procedures.
- When the model required particular term properties that were not present on the handwritten form, this was to be completed by a mechanism independent to the data entry procedures.
- The possible values for every field were located, connecting them to the input values from the handwritten forms with even more terms from the model's dictionary. Thus, special lists in table form were created, from which values could be selected by the user. The latter was rendered possible after almost all values from handwritten forms were input and partially revised according to an appropriate regrouping of handwritten forms containing “relative” data. The value being input was to be selected from a user-defined list, and not from one containing the exact term given by the model, although the value that was being input into the table was the standard model value (Fig. 3
 ).
).
- Techniques for storing and sharing multimedia files were incorporated in such a way that there would be no need for the operator to have specialized materials or knowledge (Fig. 3
 ).
).
- Finally, the input environment was changed to the form used in handwritten documents, with only minor modifications made in cases where it was obviously difficult to apply the model (Fig. 3
 ). Natural language data entry techniques were used with parallel binding to the model's terminology.
). Natural language data entry techniques were used with parallel binding to the model's terminology.
Application Design
We first studied and analyzed the model and final export to the database tables of the application of all data and relationships connected to hysteroscopy (see Appendix). Because the model did not determine the process that ought to be followed [11 Elkin PL. Evaluation of the content coverage of SNOMED CT:
ability of SNOMED clinical terms to represent clinical problem
lists Mayo Clin Proc 2006; 81: 741-8.], this design was based on established medical practices [13 Neis KJ, Brandner P, Hepp H. Hysteroscopy: textbook and atlas. NY: G. Thieme Verlag 1994.]. Three stages were included in the outline. In Stage A, the incident data were recorded (Personal Data, Medical data, Gynaecological Data and Ultrasonic Examination Results) upon which the "Cause" of the examination was justified. In Stage B, the examination data were recorded (Examination Data, Photos, Video, Sketch and Findings). Finally, in Stage C, the final diagnosis and decision (Future Actions) were recorded. These stages are presented in the Entity-Relation-Diagram (ERD) shown in Fig. (4 ).
).
This stage offered both the usability sought by the physician [13 Neis KJ, Brandner P, Hepp H. Hysteroscopy: textbook and atlas. NY: G. Thieme Verlag 1994.] as well as the flexibility that was provided by the reduced encoding of the model and its data (Fig. 4 ). The creation of independent tables with elements from the model was judged essential because the application required only components of the model's elements and not the complete tables. Thus, the choice of components from the model’s basic tables that were related to hysteroscopy, and their incorporation into the application's files, added speed and accuracy to the processing of information (Fig. 5
). The creation of independent tables with elements from the model was judged essential because the application required only components of the model's elements and not the complete tables. Thus, the choice of components from the model’s basic tables that were related to hysteroscopy, and their incorporation into the application's files, added speed and accuracy to the processing of information (Fig. 5 ).
).
Study and analysis of the currently used handwritten hysteroscopy form and adaptation of its elements were made not from the form itself but were accomplished according to the SNOMED coding. In order to achieve this, the binding of each tab field to those of the model's was studied, and a connection between them was achieved. Strictly following the model, these were added where necessary in the application’s background, not in the foreground, so as not to influence the existing data input process (Table 1). Wherever possible, this was achieved with the use of predetermined values from the model given to fields (e.g., sex) that needed to be created and were customizable.
Next, the problem of regrouping and coding the already registered values in existing fields was studied and analyzed (Fig. 6 ). The main problems were found in fields such as "Indication for examination" ("Hysteroscopy cause”) or “Pathology intra-uterine” (“Endometrial pathology”), as the values of these fields could not be standardized. To manage this problem, two fields (Fig. 7
). The main problems were found in fields such as "Indication for examination" ("Hysteroscopy cause”) or “Pathology intra-uterine” (“Endometrial pathology”), as the values of these fields could not be standardized. To manage this problem, two fields (Fig. 7 ) were created: one for the doctor's past description and another which is a list from the model and which will be completed in the future.
) were created: one for the doctor's past description and another which is a list from the model and which will be completed in the future.
The possibility of exporting and importing medical documents in the CDA format based on the SNOMED model was investigated and materialized. To protect personal data a special scheme was created as a subset of the described elements of the method to succinctly describe a hysteroscopy medical incident (Document Type Definition).
Construction and Implementation
Construction
The proposed process was realized using the work environment of the Delphi programming language, with the support of the open source library of the Flashfiler database. The construction of the application proceeded as follows:
- Creation of the database, tables, fields and their relationships with each other. It must be noted that in order to define the relationships between the tables, both the needs of the application as well as the prerogatives set by the model were taken into account.
- Selected elements from the analysis of the procedure were transferred from the model's tables to the database's table.
- The application's work environment was constructed following the rules that were established during the analysis of the procedure.
- 3000 handwritten hysteroscopy examination cards (Fig. 8
 ) were digitized and incorporated as images into the database; their information was subsequently entered into the application. The handwritten content of the critical fields was analyzed, grouped according to similar values and, with the aid of the doctor, were redefined and matched to elements of the model. The application was enriched with multimedia elements in the form of images and videos at the resolution of the sketch made by the doctor during the examination.
) were digitized and incorporated as images into the database; their information was subsequently entered into the application. The handwritten content of the critical fields was analyzed, grouped according to similar values and, with the aid of the doctor, were redefined and matched to elements of the model. The application was enriched with multimedia elements in the form of images and videos at the resolution of the sketch made by the doctor during the examination.
- A segment of code that bypassed the given registration form and followed the model's structure was added to the application (Fig. 2
 ), in order to cover exceptional cases which had not been foreseen in the handwritten form.
), in order to cover exceptional cases which had not been foreseen in the handwritten form.
Implementation
For the materialization, a volume of registered cases of hysteroscopy surgeries was obtained from the Gynaecological Clinic of the Faculty of Medicine at the University Hospital of Ioannina. The registration process included the following stages [14 Paschopoulos M, Paraskevaidis E, Stefanidis K, Kofinas G, Lolis D. Vaginoscopic approach to outpatient hysteroscopy J Am Assoc Gynecol Laparosc 1997; 4: 465-7.]:
- The compulsory details were input into the patient’s file (Fig. 9
 ).
).
- The hysteroscopy incidents of each patient were recorded. In cases where the field content could not be bound directly to the model's elements, they were input as free text (in field A; see Fig. 7
 ). In this way, until the data from the existing handwritten forms were entered, the database was being enriched with the handwritten descriptions. When the process was complete, in collaboration with the doctor, these entries were regrouped and bound to the values of the options list belonging to the second field (Table 2). At application start-up in real-time, a value is only entered in field B (Fig. 6
). In this way, until the data from the existing handwritten forms were entered, the database was being enriched with the handwritten descriptions. When the process was complete, in collaboration with the doctor, these entries were regrouped and bound to the values of the options list belonging to the second field (Table 2). At application start-up in real-time, a value is only entered in field B (Fig. 6 ) from the updated option lists that is directly interconnected with the codes of the model.
) from the updated option lists that is directly interconnected with the codes of the model.
- A model of registration for "Total Incident" coverage was defined. The model contains all of the incident's medical details, as described by the model's encoding and filing procedures, in such a way as to facilitate the retrieval of reports and statistics and simultaneously communicate between the database and other computer systems via an input-output messaging system. Additionally, an internal communication file system with the same features was established in order to make briefing between surgeries that for various reasons were not interconnected. This facilitated the creation of a unified database between different surgeries capable of using the application (Fig. 10
 ).
).
Quantitative Comparison
This study is based on the development of medical management software that describes a medical incident using an automatically derived SNOMED medical standard. In contrast, many browsers [15Available at: http://www.nlm.nih.gov/research/umls/Snomed/snomed_browsers.html ] (e.g., CliniClue, Federation Health Browser, MedTalk, Minnow, Mycroft, NCI Terminology Browser, OpenKnoME 5.4d, ClaW Workbench, SNOB, SnoFlake, Snow Owl, Virginia Tech Browser, etc.) of that standard have been developed using an inverse philosophy. In other words, these browsers describe the medical incident via hierarchically sequenced SNOMED terminology.
It is known that the measure of SNOMED’s complexity is linked to the amount of steps needed in order to complete a medical incident description. The measure of this complexity can be described by the simplicity ratio
For the proposed study, E(X) = 1 because in this method the concept is referred with medical incident uniquely, thus
DISCUSSION
The application was based on certain requirements that were set either by the established medical procedure [17 Siegler AM, Lindemann HJ. Hysteroscopy: principles and practice. Philadelphia: JB Lippincott 1984.] or by the specific model structure. The main requirements were:
- It must be possible for either a doctor or other researcher to easily apply queries to the application's database [18 Paschopoulos M, Lolis ED, Alamanos Y, Koliopoulos G, Paraskevaidis E. Vaginoscopic hysteroscopy and transvaginal sonography in the evaluation of patients with abnormal uterine bleeding J Am Assoc Gynecol Laparosc 2001; 8: 506-10.]. Therefore, particular care was given to the database's design and to the creation of form and query applications.
- The application must be versatile and inclusive. This requirement led to the incorporation of segments of code that materialized data acceptance by other installations and a future prediction was made to interconnect remote surgeries over the internet, as far as the safety of personal data was ensured.
The complete operation of an original application for the registration of medical incidents of diagnostic hysteroscopy was designed and realised in the present study. The data are coded in accordance with the international medical model SNOMED, thus giving it the capability of functioning as a knowledge base through its coded content, and of interacting with other applications. Its main advantage is that while it offers the possibility of complete organization for medical incidents of a diagnostic hysteroscopy [19 Kontostolis E, Lolis ED, Koliopoulos G, Alamanos Y, Paraskevaidis E. The use of transvaginal sonography and vaginoscopic hysteroscopy in women on tamoxifen JSLSJ 2001; 5: 211-4.], there are no delays in its real-time operation. Additionally, it directly aids the future researcher by facilitating the analysis of incidents, since it uses elements of the model.
Problems can arise from the application of modern information technology techniques to medical processes. A multitude of these problems were confirmed, analyzed and solved. The problems were resolved using a “middle-of-the-road” approach, taking into consideration the huge value of encoded medical and primary information, while respecting the uncontradictable priority of the medical process.
Appendix
Example of Data Entry from SNOMED into the Database Application
procedure tform1.import_concepts;
var t,t1:textfile;
s:string;
ss1:string;
ii,posa:integer;
begin
assignfile(t,'c:\sna\concepts.txt');
reset(t);
posa:=0;
concepts.active:=true;
concepts.disablecontrols;
while not eof(t) do
begin
readln(t,s);
inc(posa);
if (posa mod 1000 =0) then
begin
form1.edit1.text:=inttostr(posa);
concepts.enablecontrols;
edit1.refresh;
concepts.disablecontrols;
end;
ii:=strtoint(ExtractWordw(1,s,''+#9));
with concepts do
begin
insert;
fieldbyname('conceptid').asinteger:=ii;
SS1:=gr(ExtractWordw(2,s,''+#9));
fieldbyname('Fullyspecifiedname').asstring:=ss1;
SS1:=gr(ExtractWordw(3,s,''+#9));
fieldbyname('snomedid').asstring:=ss1;
SS1:=gr(ExtractWordw(4,s,''+#9));
if ss1='Y' then fieldbyname('field1').asboolean:=true
else fieldbyname('field1').asboolean:=false;
SS1:=ExtractWordw(5,s,''+#9);
if ss1='Y' then
fieldbyname('isprimitive').asboolean:=true
else fieldbyname('isprimitive').asboolean:=false;
post;
end;
end;
closefile(T)
end.
ACKNOWLEDGEMENTS
Declared none.
CONFLICT OF INTEREST
Declared none.
REFERENCES
| [1] | Lussier YA, Shagina L, Friedman C. Automating SNOMED coding using medical language understanding: a feasibility study In: Proc AMIA Symp. Philadelphia, PA: Hanley & Belfus 2001; pp. 418-22. |
| [2] | Brook P, Health A. Standards and interoperability in healthcare information systems: current status, problems, and research Issues 2010.Moorhead, MN, USA. 2010. In: 5th Midwest Relevante Metadatenstandards Version 1.2.1 LABIMI/F 19 Deliverable Association for Information Systems Conference MWAIS; Available at: http://aisel.aisnet.org/mwais2010/18/ |
| [3] | Crichton C, Davies J, Gibbons J, Harris S, Tsui A, Brenton J. Metadata-driven software for clinical trials Proceedings of the 2009 ICSE Workshop on Software Engineering in Health Care Los Alamitos: IEEE 2009; pp: 1-11. |
| [4] | Kuhn K, Lenz R, Elstner T, Siegele H, Moll R. Experiences with a generator tool for building clinical application modules Methods Inf Med 2003; 42: 37-44. |
| [5] | Hillestad R, et al. Can electronic medical record systems transform health care? Potential health benefits, savings, and costs Health Aff 2005; 24: 1103. |
| [6] | Li Z, Xuan-Dong Y, Kai-Yang L. Digitization and DICOM standardization of X-ray image signal Wuhan Univ J Nat Sci 2004; 9: 463-7. |
| [7] | Young A. The creation of medical knowledge: some problems in interpretation Soc Sci Med Med Anthropol 1981; 15: 379-86. |
| [8] | Available at www.Hl7.org |
| [9] | Available at: www.cda.org |
| [10] | Mochizuki M. Application methods of SNOP, SNOMED, ICD codes Mod Med Lab 2003; 31: 1409-0. |
| [11] | Elkin PL. Evaluation of the content coverage of SNOMED CT: ability of SNOMED clinical terms to represent clinical problem lists Mayo Clin Proc 2006; 81: 741-8. |
| [12] | Hamou JE. Hysteroscopy and micropolhysteroscopy: text and atlas. Norwalk CT: Appleton & Lange 1994. |
| [13] | Neis KJ, Brandner P, Hepp H. Hysteroscopy: textbook and atlas. NY: G. Thieme Verlag 1994. |
| [14] | Paschopoulos M, Paraskevaidis E, Stefanidis K, Kofinas G, Lolis D. Vaginoscopic approach to outpatient hysteroscopy J Am Assoc Gynecol Laparosc 1997; 4: 465-7. |
| [15] | Available at: http://www.nlm.nih.gov/research/umls/Snomed/snomed_browsers.html |
| [16] | Wei D, Wang Y, Perl Y, Xu J, Halper M, Spackman KA. Complexity measures to track the evolution of a SNOMED hierarchy AMIA Annu Symp Proc 2008; 2008: 778-82. |
| [17] | Siegler AM, Lindemann HJ. Hysteroscopy: principles and practice. Philadelphia: JB Lippincott 1984. |
| [18] | Paschopoulos M, Lolis ED, Alamanos Y, Koliopoulos G, Paraskevaidis E. Vaginoscopic hysteroscopy and transvaginal sonography in the evaluation of patients with abnormal uterine bleeding J Am Assoc Gynecol Laparosc 2001; 8: 506-10. |
| [19] | Kontostolis E, Lolis ED, Koliopoulos G, Alamanos Y, Paraskevaidis E. The use of transvaginal sonography and vaginoscopic hysteroscopy in women on tamoxifen JSLSJ 2001; 5: 211-4. |