- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Paleontology Journal
(Discontinued)
ISSN: 1874-4257 ― Volume 5, 2014
Phylogenetic and Biogeographic Analysis of Ordovician Homalonotid Trilobites
Curtis R Congreve*, Bruce S Lieberman
Abstract
Cladistic parsimony analysis of the trilobite family Homalonotidae Chapman 1980 produced a hypothesis of relatedness for the group. The family consists of three monophyletic subfamilies, one containing Trimerus Green 1832, Platycoryphe Foerste 1919, and Brongniartella Reed 1918; one containing Plaesiacomia Hawle and Corda 1847 and Colpocoryphe Novák in Perer 1918; and one containing Eohomalonotus Reed 1918 and Calymenella Bergeron 1890. All genera are monophyletic, except Brongniartella, which is paraphyletic; as it was originally defined it “gives rise” to Trimerus and Platycoryphe.
A modified Brooks Parsimony Analysis using the phylogentic hypothesis illuminates patterns of biogeography, in particular, vicariance and geodispersal of homalonotids, during the late Ordovician. The analysis yields three major conclusions about homalonotid biogeography: homalonotids originated in Gondwana; Avalonia and Laurentia were close enough during the late Ordovician to exchange taxa, especially when sea level rose sufficiently; and long distance dispersal events occurred between Armorica and Florida, and also between Arabia and a joined Laurentia-Avalonia.
Article Information
Identifiers and Pagination:
Year: 2008Volume: 1
First Page: 24
Last Page: 32
Publisher Id: TOPALOJ-1-24
DOI: 10.2174/1874425700801010024
Article History:
Received Date: 8/4/2008Revision Received Date: 3/8/2008
Acceptance Date: 7/8/2008
Electronic publication date: 10/9/2008
Collection year: 2008
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Department of Geology and Natural History Museum and Biodiversity Research Center, University of Kansas; Lawrence, Kansas. USA; E-mail: oldjack327@yahoo.com
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 8-4-2008 |
Original Manuscript | Phylogenetic and Biogeographic Analysis of Ordovician Homalonotid Trilobites | |
INTRODUCTION
The Homalonotidae Chapman 1890 [1] is a distinctive group of relatively large Ordovician-Devonian trilobites. They are not especially diverse, although they are common in nearshore environments. However, because of their shovel-like cephalon and tendency towards effacement, they have received some interest among paleontologists in general and trilobite workers in particular (Fig. 1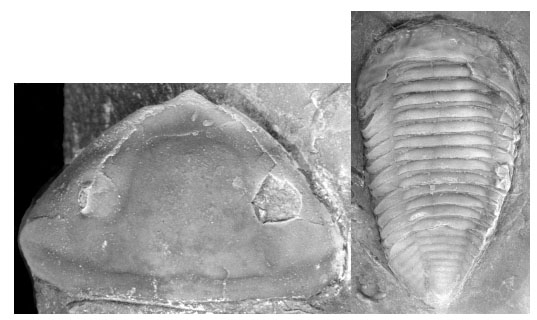 ). There have been debates about taxonomy of the Homalonotidae. These are caused in part by the group’s close evolutionary affinity to its sister taxon, Calymenidae Burmeister 1843 [2] (see Edgecombe [3] for a phylogeny of trilobite families to support this relationship). In particular, this has caused paleontologists to suggest different family-level assignments for some genera (see [4-9] for varying opinions on homalonotid classification). Also, the Ordovician homalonotids are rather distinct, such that there is a morphological discontinuity between these and the more derived Silurian and Devonian forms [7]. Here we revisit the issue of homalonotid taxonomy using a phylogenetic analysis. Our focus is primarily on Ordovician homalonotids since these are most critical from the perspective of reconstructing taxonomic patterns in the group because they are phylogenetically basal, and also this study may provide information on the number of taxa affected by the end Orodovician mass extinction. On the whole, our reconstructed phylogenetic patterns correspond most closely to Thomas’ [7] taxonomy of the family. Further, we use the phylogenetic hypothesis to reconstruct biogeographic patterns in the group by conducting a modified Brooks Parsimony Analysis [10-11]. The biogeographic analysis makes it possible to consider the role of biogeography in the end Orodovician mass extinction.
). There have been debates about taxonomy of the Homalonotidae. These are caused in part by the group’s close evolutionary affinity to its sister taxon, Calymenidae Burmeister 1843 [2] (see Edgecombe [3] for a phylogeny of trilobite families to support this relationship). In particular, this has caused paleontologists to suggest different family-level assignments for some genera (see [4-9] for varying opinions on homalonotid classification). Also, the Ordovician homalonotids are rather distinct, such that there is a morphological discontinuity between these and the more derived Silurian and Devonian forms [7]. Here we revisit the issue of homalonotid taxonomy using a phylogenetic analysis. Our focus is primarily on Ordovician homalonotids since these are most critical from the perspective of reconstructing taxonomic patterns in the group because they are phylogenetically basal, and also this study may provide information on the number of taxa affected by the end Orodovician mass extinction. On the whole, our reconstructed phylogenetic patterns correspond most closely to Thomas’ [7] taxonomy of the family. Further, we use the phylogenetic hypothesis to reconstruct biogeographic patterns in the group by conducting a modified Brooks Parsimony Analysis [10-11]. The biogeographic analysis makes it possible to consider the role of biogeography in the end Orodovician mass extinction.
Materials Analyzed
Specimens from the Yale Peabody Museum (YPM) YPM 7449A, 7449B, 33872, 33870, 204407, 204410, 6575, 204408, 204412, and 204411 and Harvard’s Museum of Comparative Zoology (MCZ) MCZ 190759, 190778, 190828, and 190832 were used in our analysis. For key references on homalono-tids, see [4, 7, 8, 12-17].
Methods
Morphological terminology follows [18].
Taxa Analyzed- Sixteen taxa were considered in this phylogenetic analysis.Neseuretus Hicks, 1873 [19] was used as the outgroup; it is widely considered to be a basal calymenid. For instance, see [7, 8, 20] though see [21-22] for a contrary viewpoint. The taxa analyzed in the ingroup had been originally assigned to Plaesiacomia Hawle and Corda, 1847 [23], Trimerus Green, 1832 [24], Platycoryphe Foerste, 1919 [25], Calymenella Bergeron, 1890 [26], Brongniartella Reed, 1918 [27], Eohomalonotus Reed, 1918 [27], and Colpocoryphe Novák in Perer, 1918 [28]. The hierarchical placement of several of these genera has been a matter of contention. Although traditionally placed with Homalonotidae, Henry [8] had argued that Colpocoryphe belonged in Calymenidae based on hypostomal structures that suggested the genus was closely related to Neseuretus. He also argued that Platycoryphe and Calymenella should be removed from Homalonotidae and placed in Calymenidae, primarily based on thoracic characters [9]. However, we include these three genera in Homalonotidae based on characters of the cephalon, glabella, and pygidium that we discuss more fully below.
Character Analysis- The characters used for this phylogenetic analysis come from the dorsal side of the mineralized exoskeleton. Hypostomal characters were not included because the hypostome is rarely preserved in homalonotids and for too many of the taxa analyzed incomplete information was available. The characters are listed below in approximate order from anterior to posterior position on the organism.
- anterior margin outline --- dorsal view (convex = 0 / not convex = 1)
- preglabellar field expansion (sag.) --- dorsal view (roughly twice length of LO [sag.] = 0 / roughly the length of L0 [sag.] = 1)
- ccephalic outline --- dorsal view (lanceolate = 0 [anterior margin width > width of L0 and lateral margin weakly convex] / subovate = 1 [anterior margin width > width of L0 and lateral margin strongly convex] / triangular = 2 [anterior margin width ≤ width of L0 and lateral margin weakly convex])
- glabellar furrows (encroaching sagittal axis of glabella = 0 / restricted to lateral margins or indistinct = 1)
- anterior margin of glabella --- dorsal view (not strongly convex = 0 / strongly convex = 1)
- inflation of anterior margin of cephalon --- dorsolateral view (inflated = 0 / not inflated = 1)
- ala distinctness --- dorsal view (distinct = 0 / indistinct or absent = 1). The ala is a semicircular lobe adjacent to the basal glabella outlined by a furrow of variable depth.
- glabella convex on entire lateral margin --- dorsal view (present = 0 / absent = 1)
- glabella expands laterally in the medial section of L1 to form a bell shape --- dorsal view (absent=0/present=1)
- glabella posterior margin --- dorsal view (strongly convex = 0/ not strongly convex = 1)
- shape of posterior part of fixigenae --- dorsal view (subangular = 0 / rounded = 1)
- posterior fixigenal angle --- dorsal view, relative to transverse line (30-40º = 0 / >55º = 1)
- lateral processes on axial rings (present = 0 / absent = 1)
- cephalon lateral convexity --- lateral view (distinct = 0 / indistinct = 1)
- occipital ring (thickest medially, with anteriorly directed lateral wing-like processes = 0 / uniform thickness, posteriorly curved, with indistinct or absent lateral wing-like processes = 1 / uniform thickness or widest medially, but parallel to thoracic axis, with anteriorly directed lateral wing-like processes indistinct or absent = 2) *the specimen used to code Plaesiacomia exul did not possess a complete occipital ring so the coding for this taxa was accomplished by extrapolation, using what was left of the structure.
- glabellar furrows --- dorsal view (deep = 0 / shallow or absent = 1)
- pygidial axis shape --- dorsal view (funnel-shaped = 0 / ovate = 1)
- swollen tubercle on pygidial axial terminus --- dorsal view (present = 0 / absent = 1)
- posterior pygidial pleurae (distinct = 0 / indistinct = 1)
- pygidial outline --- dorsal view (conical = 0 / subconical = 1)
- number of pygidial axial furrows ( > 5 = 0 / ≤ 3 = 1)
- posterior pygidial margin --- dorsal view (convex = 0 / concave = 1)
- a coaptive pygidial groove, parallel to lateral pygidial margins that connects to anterior cephalic margin during enrollment --- dorsal view (absent = 0 / present = 1)
- pygidial lateral convexity --- dorsal view ( distinct = 0 / indistinct = 1)
- pygidial dorsal convexity --- lateral view (pronounced = 0 / not pronounced = 1)
- lateral expansion of the last axial segment of the pygidial axis --- lateral view (absent = 0 / present = 1)
Phylogenetic Analysis- The data (Table 1) were analyzed using PAUP 4.0 [29]. A branch and bound search was used to determine the most parsimonious tree for this data matrix. All multistate characters were treated as unordered. Bootstrap and Jackknife statistical tests, as well as a test of Bremer [30] support, were performed to assess the statistical strength of our results. The Bootstrap and Jackknife tests were performed using PAUP [29] and were analyzed heuristically with 1,000 replicates; five most parsimonious trees were sampled at each replication. A Bayesian analysis using MrBayes v.3.1.2 [31] was also performed on the data, with the nst=6 and rates=invgamma. This allows rates of change to vary between and within transformation series. The mcmc went through 10,000,000 generations, sampling every 1000 generations. All matrix data were compiled into Nexus files using Macclade v.4.08 [32] and Mesquite v.2.01 [33] and trees were generated using FigTree v.1.1.2 [34].
Specific Taxa Analyzed- Plaesiacomia exul (Whittington 1953) [35], P. vacuvertis Thomas 1977 [7], P. oehlerti (Kerforne 1900) [36], Colpocoryphearago (Rouault 1849) [37], C. roualti Henry 1970 [38], Calymenellaboisselli Bergeron 1890 [26], C. alcantarae Hammann & Henry 1978 [39], Brongniartellabisulcata (M’Coy 1851, ex Salter, MS.) [40], B.trentonensis (Simpson 1890) (YPM 7449A and 7449B, MCZ 190828 and 190832) [41], Trimerusdelphinocephalus (Green 1832) [24] (YPM 33872, 33870, 204407, 204410, 6575, 204408, 204412, and 204411), Eohomalonotussdzuyi Hammann & Henry 1978 [39], Platycoryphedyaulax Thomas 1977 [7], P. dentata Dean 1961 [17], P.christyi (Hall 1860) [42], and P. vulcani (Murchison 1839) [43] for a total of fifteen ingroup taxa. Neseuretusvaningeni Dean & Martin 1978 [18], was chosen as the outgroup for the analysis because it is a well-preserved, complete specimen of Neseuretus from the lower Ordovician of eastern Newfoundland.
Results
Analysis results and comparison between phylogenetic methods- The parsimony analysis yielded the single most parsimonious tree with a length of 54, a CI of 0.5185, and an RI of 0.7615 (Fig. 2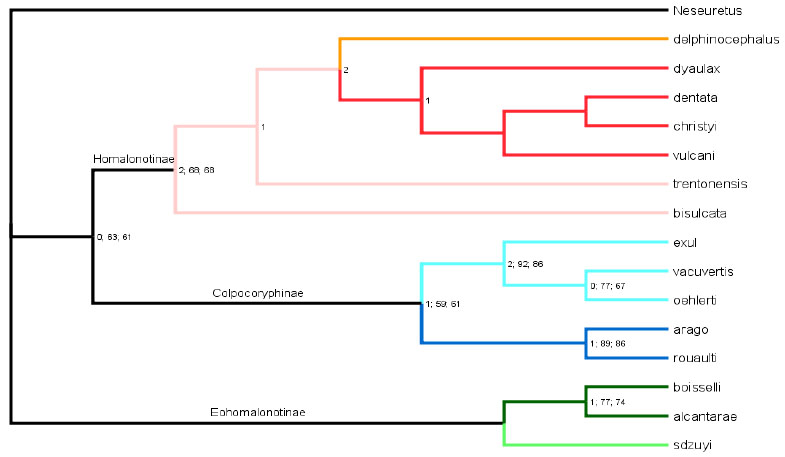 ). The Bayesian analysis also yielded a tree, although none of the posterior probabilities were significant with 95% confidence (Fig. 3
). The Bayesian analysis also yielded a tree, although none of the posterior probabilities were significant with 95% confidence (Fig. 3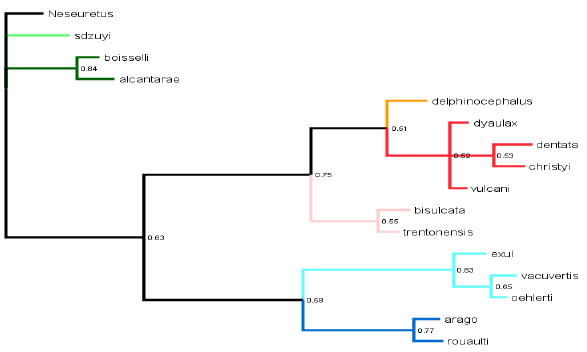 ). The nodes with the highest posterior probabilities in the Bayesian analysis also had the highest Jackknife and Bootstrap values in the parsimony analysis. High Bremer support values, however, did not strongly correlate with high posterior probabilities; for instance, the node that defines a monophyletic group with Trimerus and Platycoryphe has a Bremer support value of 2, but a posterior probability of only 51%. Focusing on the topologies of both trees, the relationships implied by the parsimony tree basically concur with those implied from the Bayesian derived tree, with two exceptions. In particular, the Bayesian analysis predicted Brongniartella was monophyletic, while the parsimony analysis indicated Brongniartella was paraphyletic (in essence “giving rise” to both Trimerus and Platycoryphe). Further, the parsimony analysis indicated that Eohomalonotus grouped with Calymenella, while the Bayesian analysis placed both taxa in a polytomy. For the purposes of taxonomy and biogeography, we will be using the tree generated from the parsimony analysis as our phylogenetic hypothesis. The Bayesian tree can be treated as another means of gauging support for different aspects of the tree, in addition to the Jackknife/Bootstrap and Bremer support methods.
). The nodes with the highest posterior probabilities in the Bayesian analysis also had the highest Jackknife and Bootstrap values in the parsimony analysis. High Bremer support values, however, did not strongly correlate with high posterior probabilities; for instance, the node that defines a monophyletic group with Trimerus and Platycoryphe has a Bremer support value of 2, but a posterior probability of only 51%. Focusing on the topologies of both trees, the relationships implied by the parsimony tree basically concur with those implied from the Bayesian derived tree, with two exceptions. In particular, the Bayesian analysis predicted Brongniartella was monophyletic, while the parsimony analysis indicated Brongniartella was paraphyletic (in essence “giving rise” to both Trimerus and Platycoryphe). Further, the parsimony analysis indicated that Eohomalonotus grouped with Calymenella, while the Bayesian analysis placed both taxa in a polytomy. For the purposes of taxonomy and biogeography, we will be using the tree generated from the parsimony analysis as our phylogenetic hypothesis. The Bayesian tree can be treated as another means of gauging support for different aspects of the tree, in addition to the Jackknife/Bootstrap and Bremer support methods.
 |
Fig. (2) Cladogram of the results from the parsimony analysis. Tree graphics generated using FigTree v.1.1.2 [34]. The stems that connect to an end member species have been color coded based on the genus they were traditionally assigned to, where Platycoryphe is red, Trimerus is orange, Brongniartella is pink, Plaesiacomia is light blue, Colpocoryphe is dark blue, Calymenella is dark green, and Eohamalonotus is light green. The values at the nodes are the results from the statistical tests. The first number is the Bremer Support value, the second is the Bootstrap value, and the third is the Jackknife value. Trees for the Bootstrap and Jackknife analyzes were generated using 50% majority rule consensus. |
We chose to include members of the genus Colpocoryphe in our analysis despite Henry’s [8] claim that the genus belongs to the Calymenidae based on hypostomal characters. We found that Colpocoryphe grouped with the ingroup and close to Plaesiacomia, which challenges aspects of Henry’s [8] hypothesis; however, we were unable to include hypostomal characters given their typically poor and incomplete state of preservation. In order to test how strongly the presence of Colpocoryphe affected the tree topology, all members of the genus were removed and the data matrix was analyzed again. The absence of Colpocoryphe had no affect on the topology. Henry [8] also argued Calymenella was a calymenid. Again, our phylogenetic results do not support this contention, but to test the effect including this taxon had on our result, we removed Calymenella from the analysis: the overall topology did not change.
Systematic Paleontology- According to our analysis Calymenella, Colpocoryphe, Plaesiacomia and Platycoryphe are monophyletic. Therefore, we do not redefine these taxa. Brongniartella as traditionally conceived is paraphyletic. Since bisulcata is the type species, we suggest that it be placed in a monotypic genus Brongniartella. Using the convention established by Wiley [44], we place trentonensis in “Brongniartella”, with the quote marks denoting the group’s paraphyly. (We are hesitant to create a monotypic genus for trentonensis simply because we have not included every known taxa of “Brongniartella” and thus do not know the entire structure of this paraphyletic group.) It was impossible to determine if Eohomalonotus or Trimerus as traditionally conceived were monophyletic since we only included one species of each of these taxa, and our primary emphasis was on Ordovician and Early Silurian exponents of the homalonotids.
The data suggests three larger monophyletic groups (subfamilies) within the Homalonotidae: one consisting of Trimerus-“Brongniartella”-Platycoryphe; another consisting of Colpocoryphe-Plaesiacomia; and the third consisting of Eohomalonotus-Calymenella. These subfamilies on the whole match those Thomas [7] identified. In particular, Thomas [7] grouped Trimerus, Brongniartella, and Platycoryphe within the Homalonotinae; he grouped Colpocoryphe and Plaesiacomia within the Colpocoryphinae Hupé, 1955 [45]; and he grouped Calymenella and Eohomalonotus within the Eohomalonotinae Hupé, 1953 [22]. Since our data supports Thomas’s [7] revision of these subfamilies, no new redefinition of these groups is required.
Genus BRONGNIARTELLA Reed 1918
TYPE SPECIES: Homalonotus bisulcata M’Coy 1851, ex Salter, MS [40].
DISCUSSION
Since the genus Brongniartella has been shown to be paraphyletic, we redefine the genus into a monotypic genus that includes only its type species, bisulcata, and refer the other species considered to the paraphyletic “Brongniartella”. For an in-depth diagnosis of Brongniartella bisulcata, refer to Dean [18].
Biogeography Analysis: Methods- We used our phylogeny to perform a biogeographic analysis using a modified version of Brooks parsimony analysis (BPA). This method is described in detail in [10, 11, 47], although some brief discussion is provided here, and has been used successfully to investigate biogeographic patterns in a variety of groups, including trilobites, e.g. [10, 11, 48-53]. Modified BPA makes it possible to detect patterns of geodispersal and vicariance. First, we created an area cladogram by replacing the names of the end member taxa with the geographic areas in which these taxa were found (Fig. 4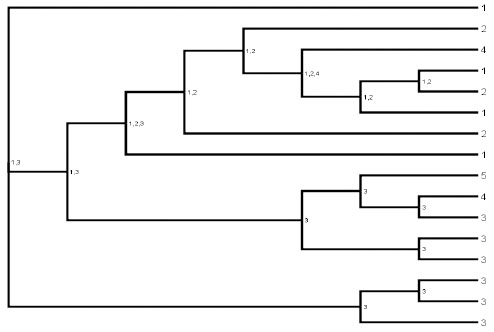 ). The areas used in the analysis were Avalonia (Newfoundland and Great Britain), Eastern Laurentia (the United States), Armorica (France and Spain), Arabia (Saudi Arabia), and Florida (Fig. 5
). The areas used in the analysis were Avalonia (Newfoundland and Great Britain), Eastern Laurentia (the United States), Armorica (France and Spain), Arabia (Saudi Arabia), and Florida (Fig. 5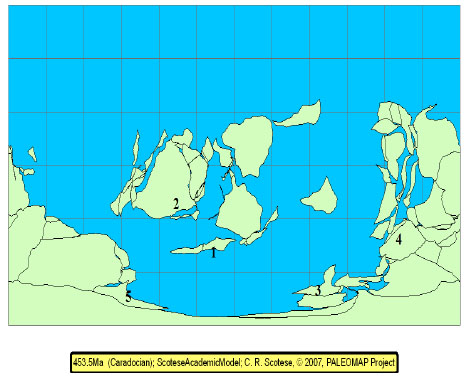 ) These areas were defined on the basis of geological evidence and because they contain large numbers of endemic taxa; in effect this follows the area descriptions and designations of [54-58].Next, the geographic locations for the ancestral nodes of the area cladogram were optimized using a modified version of the Fitch [59] parsimony algorithm. Then, the area cladogram was used to generate two matrices, one to code for patterns of vicariance and the other to code for patterns of geodispersal. The former provides information about the relative time that barriers formed, isolating regions and their respective biotas; the latter provides information about the relative time that barriers fell, allowing biotas to congruently expand their range [10, 11, 47]. Each matrix was then analyzed using an exhaustive search on PAUP 4.0 [29]. The results are presented in Fig. (6
) These areas were defined on the basis of geological evidence and because they contain large numbers of endemic taxa; in effect this follows the area descriptions and designations of [54-58].Next, the geographic locations for the ancestral nodes of the area cladogram were optimized using a modified version of the Fitch [59] parsimony algorithm. Then, the area cladogram was used to generate two matrices, one to code for patterns of vicariance and the other to code for patterns of geodispersal. The former provides information about the relative time that barriers formed, isolating regions and their respective biotas; the latter provides information about the relative time that barriers fell, allowing biotas to congruently expand their range [10, 11, 47]. Each matrix was then analyzed using an exhaustive search on PAUP 4.0 [29]. The results are presented in Fig. (6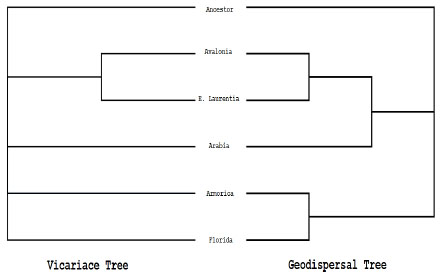 ). All matrix data was compiled into Nexus files using Mesquite v.2.01 [33] and trees were generated using FigTree v.1.1.2 [34].
). All matrix data was compiled into Nexus files using Mesquite v.2.01 [33] and trees were generated using FigTree v.1.1.2 [34].
 |
Fig. (4) Area cladogram. Tree graphics generated using FigTree v.1.1.2 [34]. The numbers code for the locations in which the taxa were found, where 1 = Avalonia, 2 = E. Laurentia, 3 = Armorica, 4 = Arabia, and 5 = Florida. The numbers at the nodes are the optimized locations of the ancestral taxa. |
 |
Fig. (5) Map of the late Ordovician (Caradoc) world generated with ArcView 9.2 and PaleoGIS [46]. The biogeographic areas used in this analysis are numbered 1 = Avalonia, 2 = E. Laurentia, 3 = Armorica, 4 = Arabia, and 5 = Florida. |
 |
Fig. (6) On the right the most parsimonious geo-dispersal tree and on the left the strict consensus of four most parsimonious vicariance trees. |
Results of the biogeographic analysis- The geodispersal analysis yielded the single most parsimonious tree of 37 steps. The tree suggests the most recent barriers to fall were those between E. Laurentia and Avalonia and those between Florida and Armorica. The next most recent barriers to fall were those between a combined E. Laurentia-Avalonia and Arabia. Finally, the oldest barriers were those between E. Laurentia-Avalonia-Arabia and Florida-Armorica. The vicariance analysis yielded four most parsimonious trees of 47 steps. A strict consensus of these four trees has only one resolved node: Avalonia and E. Laurentia, suggesting some vicariance between trilobites from these respective regions.
We also used the test of Hillis [60], the g1 statistic, to see whether the results from our analysis differ from those produced using random data. Our results differ from those generated using random data at the .01 level. Bootstrap, Jackknife, and Bremer support values were calculated for both trees. In the geodispersal tree, the node uniting Avalonia and Laurentia was most robust, with Bremer, Bootstrap, and Jackknife values of 2, 91%, and 87% respectively. In the vicariance tree, the node uniting Avalonia and Laurentia had Bremer, Bootstrap, and Jackknife values of 3, 95%, and 92% respectively.
Interpretation of biogeographic results and discussion- The close relationship between E. Laurentia and Avalonia is replicated in both the vicariance and geodispersal trees (Fig. 4 ). This suggests that the processes producing vicariance and geodispersal between these areas were similar, implicating cyclical processes, likely sea-level rise and fall, played an important role in generating the biogeographic patterns [10, 11, 47, 49]. In effect, this result largely matches paleomagnetic and tectonic evidence which indicates that Avalonia rifted from Gondwana during the early-mid Ordovician and began drifting towards Laurentia and Baltica; during the early Silurian, Avalonia and Baltica joined together to form Balonia; and this in turn collided with Laurentia during the Taconic orogeny [55, 58, 61-63]. Probably by the late Ordovician the Iapetus Ocean was effectively closed [61]. Our data suggest that either Laurentia and Avalonia were geographically close enough to each other during the late Ordovician to directly exchange taxa when sea level rose sufficiently, or they were indirectly exchanging taxa, with Baltica acting as an intermediary.
). This suggests that the processes producing vicariance and geodispersal between these areas were similar, implicating cyclical processes, likely sea-level rise and fall, played an important role in generating the biogeographic patterns [10, 11, 47, 49]. In effect, this result largely matches paleomagnetic and tectonic evidence which indicates that Avalonia rifted from Gondwana during the early-mid Ordovician and began drifting towards Laurentia and Baltica; during the early Silurian, Avalonia and Baltica joined together to form Balonia; and this in turn collided with Laurentia during the Taconic orogeny [55, 58, 61-63]. Probably by the late Ordovician the Iapetus Ocean was effectively closed [61]. Our data suggest that either Laurentia and Avalonia were geographically close enough to each other during the late Ordovician to directly exchange taxa when sea level rose sufficiently, or they were indirectly exchanging taxa, with Baltica acting as an intermediary.
Our results, in particular, the geodispersal tree (Fig. 6 ), also indicate a close biogeographic relationship between Avalonia-Laurentia and Arabia. When the patterns implied by the vicariance and geodispersal trees differ, as is the case with this aspect of the biogeographic results, it could be due to a tectonic collision or a chance long distance dispersal event [10, 11, 47, 49]. Given that there is no substantial tectonic evidence linking these regions, the dispersal between Avalonia-Laurentia and Arabia was unlikely to have been facilitated by a tectonic event, and instead may have been due to chance long distance dispersal between these regions. This dispersal could have been facilitated by a planktonic larval stage, however homalonotids are presumed to have had benthic larvae [64]. Dispersal also could have been facilitated by chains of island arcs that allowed organisms to island-hop to Gondwana.
), also indicate a close biogeographic relationship between Avalonia-Laurentia and Arabia. When the patterns implied by the vicariance and geodispersal trees differ, as is the case with this aspect of the biogeographic results, it could be due to a tectonic collision or a chance long distance dispersal event [10, 11, 47, 49]. Given that there is no substantial tectonic evidence linking these regions, the dispersal between Avalonia-Laurentia and Arabia was unlikely to have been facilitated by a tectonic event, and instead may have been due to chance long distance dispersal between these regions. This dispersal could have been facilitated by a planktonic larval stage, however homalonotids are presumed to have had benthic larvae [64]. Dispersal also could have been facilitated by chains of island arcs that allowed organisms to island-hop to Gondwana.
The geodispersal tree also shows a grouping of Armorica and Florida (Fig. 6 ). During the late Ordovician, paleomagnetic and tectonic evidence suggests that Armorica had rifted away from the main continent of Gondwana [58, 61]. It is possible the rifted Armorica could have moved close enough to Florida to exchange taxa during this time period. However, since the vicariance tree does not record this rifting event, we cannot be sure if the separation of Armorica from Gondwana had the primary affect on the biogeographic patterns of homalonotids at the time, or instead these patterns were due to chance long distance dispersal. Furthermore, again there is no strong tectonic evidence to support a collision between Florida and Armorica.
). During the late Ordovician, paleomagnetic and tectonic evidence suggests that Armorica had rifted away from the main continent of Gondwana [58, 61]. It is possible the rifted Armorica could have moved close enough to Florida to exchange taxa during this time period. However, since the vicariance tree does not record this rifting event, we cannot be sure if the separation of Armorica from Gondwana had the primary affect on the biogeographic patterns of homalonotids at the time, or instead these patterns were due to chance long distance dispersal. Furthermore, again there is no strong tectonic evidence to support a collision between Florida and Armorica.
Our area cladogram (Fig. 4 ) also indicates that the homalonotids most likely originated in Gondwana, during a time when Avalonia was still connected to the main continent. This is because the area of the ancestral node of all homalonotids consists of a united Avalonia and Armorica. If we track patterns of biogeographic change up the tree, it appears that Avalonia then rifted from Gondwana, carrying with it a homalonotid fauna that diversified in Avalonia and later dispersed from Avalonia into Laurentia. Our data indicates Laurentian homalonotids have a close evolutionary relationship with Avalonian forms. Indeed, all Laurentian and Avalonian homalonotids group in a single subfamily (Figs. 2
) also indicates that the homalonotids most likely originated in Gondwana, during a time when Avalonia was still connected to the main continent. This is because the area of the ancestral node of all homalonotids consists of a united Avalonia and Armorica. If we track patterns of biogeographic change up the tree, it appears that Avalonia then rifted from Gondwana, carrying with it a homalonotid fauna that diversified in Avalonia and later dispersed from Avalonia into Laurentia. Our data indicates Laurentian homalonotids have a close evolutionary relationship with Avalonian forms. Indeed, all Laurentian and Avalonian homalonotids group in a single subfamily (Figs. 2 , 4
, 4 ). Ultimately, the movement of homalonotids into Laurentia appears to have had an important effect on macroevolutionary patterns in the group, as the group underwent substantial subsequent diversification after it entered that region.
). Ultimately, the movement of homalonotids into Laurentia appears to have had an important effect on macroevolutionary patterns in the group, as the group underwent substantial subsequent diversification after it entered that region.
ACKNOWLEDGEMENTS
We would like to thank Susan Butts from the YPM and Jessica Cundiff and Fred Collier from the MCZ for providing access to study material that was vital for the completion of this study, and also for loaning material used for study. We would also like to thank two anonymous reviewers and the editors for comments on an earlier version of this manuscript. Financial support for this research was provided by NSF DEB-0716162 and NASA Astrobiology NNG04G M41G.