- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Conference Proceedings Journal
(Biological Sciences, Chemical Sciences, Physical Sciences, Medicine, Engineering & Technology)
(Discontinued)
ISSN: 2210-2892 ― Volume 10, 2020
Applicability of Micro Inoculation Culture (MIC) for Rapid Monitoring of Total Coliform Contaminants in the Food Industry
Wipavadee Sangadkit1, Anat Deepatana2, Aluck Thipayarat3, *
Abstract
Unlike medical samples from clinics, samples associated with food products and the environment they come into contact with during their processing are characterized by low initial cell counts and large sample volumes. This calls for different strategies of handling, especially in low-resource settings and in less advanced food industrial laboratories. This paper compares three popular industrial methodologies; MPN method, Petrifilm™ by 3M, and the standard pour plate technique, relative to a modified surface spread technique using 96-well microtiter plates (MIC). The colony enumeration results obtained from each technique showed good agreement. The miniaturized rapid protocol efficiently managed a large number of samples using multichannel autopipettes and a high-throughput design utilizing 96-well microtiter plates. Useful colony counts were obtained within 12-16 h. The analytical efficacy of the miniaturized protocol surpassed those of the three conventional methods. The colony counts from ready-to-eat product samples showed comparable results to the pour plate technique, displaying good agreement with the universally-accepted standard. The feedback by QC staff from a local Thai food factory revealed good overall acceptance of the MIC method with respect to usability, protocol design and method efficiency. The proposed miniaturized technique gave highly consistent results of colony count numbers and good colony separation. This colony enumeration consistency suggests that the miniaturized rapid protocol can economically replace the slower more complex standard protocols as an in-house protocol for food processing environment swabs.
Article Information
Identifiers and Pagination:
Year: 2016Volume: 7
First Page: 126
Last Page: 133
Publisher Id: TOPROCJ-7-1-126
DOI: 10.2174/2210289201607010126
Article History:
Received Date: 29/6/2015Revision Received Date: 20/05/2016
Acceptance Date: 31/05/2016
Electronic publication date: 30/06/2016
Collection year: 2016
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution-Non-Commercial 4.0 International Public License (CC BY-NC 4.0) (https://creativecommons.org/licenses/by-nc/4.0/legalcode), which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Office of Education, Faculty of Engineering, Burapha University, 169 Long-Hard Bangsaen Road, Saen Sook Sub-district, Mueang District, Chonburi 2013 1, Thailand; Tel.: +66 2 4709246; Fax: +66 2 4709240; Email: athipaya@yahoo.com
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 29-6-2015 |
Original Manuscript | Applicability of Micro Inoculation Culture (MIC) for Rapid Monitoring of Total Coliform Contaminants in the Food Industry | |
1. INTRODUCTION
Sanitary assessment of water, workers, and processing environment, is of vital importance to the food industry. It could help prevent food products from pathogen contamination. The rapid and accurate detection of bacterial indicators for sanitation, Escherichia coli (E. coli) total coliforms (TC), is needed to provide emergency feedback for monitoring the processing line. There are several options available for the qualitative enumeration of E. coli and total coliform in foods. The most probable number (MPN) technique has been widely accepted and usually employed for routine investigation [1Oblinger, J.L.; Koburger, J.A. Understanding and teaching the most probable number technique. J. Milk Food Technol., 1975, 38, 540-545.-4Suwansonthichai, S.; Rengpipat, S. Enumeration of coliforms and Escherichia coli in frozen black tiger shrimp Penaeus monodon by conventional and rapid methods. Int. J. Food Microbiol., 2003, 81(2), 113-121.
[http://dx.doi.org/10.1016/S0168-1605(02)00190-3] [PMID: 12457585] ]. The MPN technique, however, has some obvious drawbacks. It is labor-intensive, expensive, and time-consuming [4Suwansonthichai, S.; Rengpipat, S. Enumeration of coliforms and Escherichia coli in frozen black tiger shrimp Penaeus monodon by conventional and rapid methods. Int. J. Food Microbiol., 2003, 81(2), 113-121.
[http://dx.doi.org/10.1016/S0168-1605(02)00190-3] [PMID: 12457585] , 5Bredie, W.L.; de Boer, E. Evaluation of the MPN, Anderson–Baird– Parker, Petrifilm E. coli and Fluorocult ECD method for enumeration of Escherichia coli in foods of animal origin. J. Food Microbiol., 1992, 161, 197-208.
[http://dx.doi.org/10.1016/0168-1605(92)90080-M] ]. To obtain the final colony counts, this technique requires up to 10 days. Industry commonly replaces the conventional MPN with commercial available media and protocols, for example Chromocult® Coliform agar (CCA), Fluorocult® LMX broth, and Petrifilm™E. coli count plates. Industry is constantly seeking more efficient methods with shorter detection times and more prompt and useful microbial information to monitor the routine manufacturing.
There have been attempts to develop rapid alternatives and shorten the detection time for E. coli in food and environmental samples [6Anderson, J.M.; Baird-Parker, A.C. A rapid and direct plate method for enumerating Escherichia coli biotype I in food. J. Appl. Bacteriol., 1975, 39(2), 111-117.
[http://dx.doi.org/10.1111/j.1365-2672.1975.tb00551.x] [PMID: 1104553] -9Firstenberg-Eden, R. Electrical Impedance Method for Determining Microbial Quality of Foods. In: Rapid Methods and Automation in Microbiology and Immunology; Hobermehl, K.O., Ed.; Springer-Verlag: Berlin, 1985.
[http://dx.doi.org/10.1007/978-3-642-69943-6_83] ]. Most rapid techniques based on enzyme-linked immunosorbent assay (ELISA) and DNA hybridization are fast and sensitive, but are not yet practical for routine screening of large numbers of industrial samples. Pre-enrichment is often required to obtain high enough cell concentrations (more than 4 log CFU/ml) for accurate detection [10Franco, W.; Hsu, W.Y.; Simonne, A.H. Survival of Salmonella and Staphylococcus aureus in mexican red salsa in a food service setting. J. Food Prot., 2010, 73(6), 1116-1120.
[PMID: 20537270] ]. Although the identification time is reduced, long incubation time made them as unattractive as the conventional methods. Alternatively, miniaturization of microbiological assessment through reduction of media and culture volumes has shown potential to decrease the detection time and provided better efficacy of microbial enumeration [11Fung, D.Y. Historical development of rapid methods and automation in microbiology. J. Rapid. Meth. Aut. Mic., 1992, 1, 1-14.
[http://dx.doi.org/10.1111/j.1745-4581.1992.tb00066.x] ].
Several authors have demonstrated that miniaturization methods facilitate detection and enumeration of target bacteria without altering sensitivity or specificity [12Khueankhancharoen, J.; Thipayarat, A. Application of modified drop plate technique (MDPT) and logistic model to optimize non-selective substrates for Salmonella typhi resuscitation. Asian. J. Food. Agro. Ind., 2011, 4(6), 349-358.-14Supanivatin, P.; Khueankhancharoen, J.; Saeaung, W.; Boonyaprapasorn, A.; Thipayarat, A. Industrial Implementation of Fast Total Plate Count Analysis Applying Micro Inoculation Culture on Frozen Ready-to-eat Food Products In: Proceeding of the International Conference for a Sustainable Greater; Mekong Subregion: Bangkok, Thailand, 2010. ]. A rapid method utilizing 24-well microplates successfully detected Listeria spp. in food samples [15Kang, D.H.; Fung, D.Y. Development and evaluation of a 24 well microtitre plate method for isolation of Listeria spp. or Listeria monocytogenes from foods. Lett. Appl. Microbiol., 1999, 28(4), 280-284.
[http://dx.doi.org/10.1046/j.1365-2672.1999.00532.x] [PMID: 10212440] ]. This technique was shown to be superior to the conventional streak plate or spread plate techniques when handling large volume industrial samples. Previous studies using multiple microwell techniques (both 24- and 96-well formats) for viable cell counts of various bacteria have proven practical for routine inspection of industrial food and environmental samples, because large numbers of samples can be handled effectively saving both money and analytical time [3Pavic, A.; Groves, P.J.; Bailey, G.; Cox, J.M. A validated miniaturized MPN method, based on ISO 6579:2002, for the enumeration of Salmonella from poultry matrices. J. Appl. Microbiol., 2010, 109(1), 25-34.
[PMID: 20059618] , 16Chenu, J.W.; Pavic, A.; Cox, J.M. A novel miniaturized most probable number method for the enumeration of Campylobacter spp. from poultry-associated matrices. J. Microbiol. Methods, 2013, 93(1), 12-19.
[http://dx.doi.org/10.1016/j.mimet.2013.01.013] [PMID: 23384829] , 17Kim, S.; Fung, D.Y. Modified microtiter count method for viable cell counts from pure cultures and food model samples. Food Microbiol., 2005, 22, 595-599.
[http://dx.doi.org/10.1016/j.fm.2004.11.011] ]. In this paper, several common protocols to estimate coliforms and E. coli were evaluated. A novel miniaturization method was also proposed to reduce the cultivation volume of CCA and decrease the detection time on both pure culture and industrial samples.
2. MATERIALS AND METHODS
2.1. Preparation of Test Microorganism
Escherichia coli DMST 4609 was obtained from the Thailand Institute of Scientific and Technological Research, Bangkok, Thailand. One loopful of the stock culture was inoculated into tryptic soy broth (TSB, Difco Laboratories, Sparks, MD) at 37 °C for 24 h to reach the initial cell stock density of approximately 107 CFU/ml. To validate the proposed miniaturized method against the MPN, pour plate and Petrifilm™ techniques, the initial cell stock was diluted to 102, 103, 104, 105, and 106 CFU/ml.
2.2. Detection of E. coli and TC From Commercial Foods and Its Production Line Facilities
2.2.1. Most Probable Number Method
In the MPN method [18Weagant, S.D.; Grant, M.A. Bacteriological Analytical Mannual (BAM) Online: Chapter 4 Enumeration of Escherichia coli and the Coliform Bacteria. Available at: http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm064948.htm [2015, July 2].], 1 ml of a sample suspension at a proper dilution was pipetted into triple tubes containing lactose broth (Oxoid CM451) and these tubes were incubated at 37 °C for 24 h. Tubes with gas formation were gently agitated and a loopful of each suspension then transferred to tubes of EC medium (DIFCO 0314-01-0) that were subsequently incubated at 44.5 °C for 48 h to confirm test for E. coli. And then a loopful of suspension from each gassing EC medium was streaked onto eosin methylene blue agar (EMB) (Oxoid CM63) and incubated at 37 °C for 24 h to complete test for E. coli. Their numbers per 1 ml of sample were calculated from the MPN table.
2.2.2. Pour Plate Method
The pour plate cell count was performed using CCA. The sample suspension at 1 ml was pipetted onto an empty Petri dish and homogeneously mixed with melting CCA. The inoculated plate was incubated at 37 °C for 24 h. Each dilution was tested using duplicates of CCA plates. Coliform colonies appear pink red and E. coli colonies purple.
2.2.3. Petrifilm™E. coli/Coliform Method
Again, 1 ml of the same sample suspension was inoculated onto the surface of a Petrifilm™E. coli/coliform count (EC) plate. The cover film was reinstalled slowly on the plate which was then incubated at 35 oC for 24 h according to the manufacturer’s manual [19Coliforms and Escherichia coli Counts in Foods: Petrifilm™ E.coli / Coliform Count Plate Method. AOAC Official Method 991.14, 1991.]. Red colonies surrounded by trapped gas were coliforms and purple colonies with trapped gas were E. coli. Duplicate trials were performed per dilution.
2.2.4. Miniaturized Technique
Micro inoculation culture (MIC) plates were fabricated using CCA in a 96-well microtiter plate format (Nunc, Rochester, NY, USA). Each microwell contained up to 0.5 ml of CCA and the inoculation volume was fixed at 10 μl applied onto solidified CCA. The miniaturized microwells were incubated at 37 ºC and it took normally 12–15 h to detect bacterial colonies. Total coliforms and E. coli from all methods were reconfirmed by reisolation on EMB agar and IMViC testing [20Andrews, W. Manual of Food Quality Control; Food and Agriculture Organizations of the United Nation: USA, 1992. ]. Real samples from a local food exporter, including green papaya salad, fried rice, and cake topping as well as environmental swab samples from its production lines, were collected to evaluate coliforms and E. coli enumeration by each technique described above.
2.2.5. Preference Testing
Preference surveys evaluated the panelist’s likes and dislikes of the proposed techniques. All ten staff members from the quality control department who had experienced using the conventional agar culture protocol were participated in the survey. The questionnaires were formulated to determine the intrinsic characteristics of employee satisfaction using a nine-point hedonic scale from 1 to 9. The score meaning can be interpreted as follows: 9 = like extremely, 8 = like very much, 7 = like moderately, 6 = like slightly, 5 = neither like nor dislike, 4 = borderline of acceptability, 3 = dislike moderately, 2 = dislike very much, and 1 dislike extremely [21Hasan, A.; Belgin, S.; Serdar, C. Assessment of the microbiological quality of meals sampled at the meal serving units of a military hospital in Ankara, Turkey. Food Contr., 2004, 15(5), 379-384.
[http://dx.doi.org/10.1016/S0956-7135(03)00101-4] , 22Juyun, L.H. A review of methods and theory. Food Qual. Prefer., 2011, 22, 733-747.]. The scores were the average value from the ten panelists and the standard deviations were included in the report to show the range of variability of responses.
2.2.6. Financial Comparison
The cost of material and operating time from the two techniques were compared. The volume of media (CCA) and the amount of disposable materials (glass Petri dishes and 96 micro-deep wells) were monitored. Pavic et al. [3Pavic, A.; Groves, P.J.; Bailey, G.; Cox, J.M. A validated miniaturized MPN method, based on ISO 6579:2002, for the enumeration of Salmonella from poultry matrices. J. Appl. Microbiol., 2010, 109(1), 25-34.
[PMID: 20059618] ] and Chenu et al. [16Chenu, J.W.; Pavic, A.; Cox, J.M. A novel miniaturized most probable number method for the enumeration of Campylobacter spp. from poultry-associated matrices. J. Microbiol. Methods, 2013, 93(1), 12-19.
[http://dx.doi.org/10.1016/j.mimet.2013.01.013] [PMID: 23384829] ] recommended the method for calculating the operating cost with slight modification. The times to analyze 36 samples by ISO 4833-1:2013 and the microscale technique were measured.
2.2.7. Statistical Analysis
TC counts obtained were transformed to log CFU/ml. All data were analyzed at p<0.05 for significant values by ANOVA. Paired t-test was used to test the significant differences between the means of two sets of data and to assess the agreement.
3. RESULTS AND DISCUSSION
3.1. Comparison of MIC to Other Commercial and Standard E. coli Enumeration Techniques
The use of the MIC technique has several advantages over other commercial and standard protocols in terms of compactness, short detection time and less medium usage (Table 1). The miniaturized microwells allowed multiple dilutions of food and environment samples to adequately cover the normally encountered range of industrial contamination. In industrial situations, there are often cases of under-diluted samples since most factories are cost-conscious and CCA is rather costly for many environmental swab samples. The compactness of the 96-well microtiter plate (5 mm width, 10 mm length and 1.5 mm thickness) accommodates 96 samples, equivalent to 96 Petri dishes using the pour plate protocol. Also the 96-well format facilitates the use of commercial multi-channel auto pipettes, either 8- or 12-channel, and diminishes the sample dilution and liquid transferring loads of lab workers. Our interviews with QC&QA personnel carrying out the MIC protocol in a food factory setting have generally been positive since our first MIC implementations. This is particularly true, if these factory microbiologists had been previously following mostly ISO protocols using the pour plate technique as their regular routine.
The only obvious disadvantage of the MIC protocol was the smaller inoculum volume. The 50.28 mm2 agar surface was able to accept optimally 10-20 μl of sample inoculums. Higher inoculum volume resulted in smearing on the agar surface or less well-defined colonies after incubation. The two orders of magnitude smaller sample volume compared to the other protocols results in a minimum detection limit of 2 log CFU/ml.
Considering the sample residue in transferring glass pipettes, the inherent analytical errors using the standard methods never permit the colony enumeration accuracy to single digit CFU counts. There is a counting guideline between 30 and 300 CFU per plate, which is never practical in industrial applications, since it is rare that under normal circumstances lab personnel have any real idea of the degree of contamination a priori. Hence, there is always some inherent error of at least 1 log CFU/ml for all other routine methods included in this study. To validate the use of MIC compared to the standard techniques, pure culture was first used to prepare the standards of known cell densities. Many authors have performed the same validation procedure in search of a rapid, easy and accurate way to assess industrial viable cell counts and replace the current microbial analytical routines shown in Fig. (1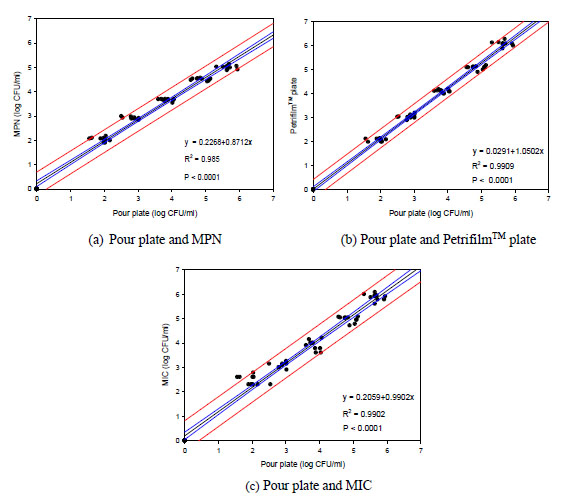 ).
).
To validate the MIC method against other standard protocols, an evaluation of different methods was conducted to contrast the performance of the pour plate technique to MPN, Petrifilm™ EC plate and MIC techniques. Other investigators have done similar evaluations to test newly-developed methodology [16Chenu, J.W.; Pavic, A.; Cox, J.M. A novel miniaturized most probable number method for the enumeration of Campylobacter spp. from poultry-associated matrices. J. Microbiol. Methods, 2013, 93(1), 12-19.
[http://dx.doi.org/10.1016/j.mimet.2013.01.013] [PMID: 23384829] , 23Finney, M.; Smullen, J.; Foster, H.A.; Brokx, S.; Storey, D.M. Evaluation of Chromocult coliform agar for the detection and enumeration of Enterobacteriaceae from faecal samples from healthy subjects. J. Microbiol. Methods, 2003, 54(3), 353-358.
[http://dx.doi.org/10.1016/S0167-7012(03)00068-X] [PMID: 12842481] ]. In Fig. (1 ), all regression lines showed slopes close to 1. The correlation coefficient between any of these methods ranging from 0.985 to 0.9902 suggested good agreement between the data and linear approximation from 0 to 6 log CFU/ml. Multiple analysis of variance showed that there were no differences between the methods at the 5% confidence level. However, the log CFU readings from the MIC technique were less reliable due to its higher lower bound limitation (2 log CFU/ml). At this low cell concentration, other techniques allowing more sample volume were more accurate. Only MPN returned slightly lower cell counts on average. Essentially all techniques resulted in practically the same readings providing the sample inoculums were prepared with proper dilution for colony evaluation. The short detection time, user-friendliness, and high throughput nature by MIC technique made this protocol practical for cell enumeration in food industry, especially with resource-poor settings. Our previous studies also demonstrated the versatility and practicality of the similar MIC format in enumerating other types of bacterial colonies (i.e., Listeria monocytogenes, Salmonella spp.) [24Liamkaew, R.; Thipayarat, A.; Saranak, J. Listeria detection in microscale solid state inoculation with minimal selective agents. Food Contr., 2014, 43, 183-192.
), all regression lines showed slopes close to 1. The correlation coefficient between any of these methods ranging from 0.985 to 0.9902 suggested good agreement between the data and linear approximation from 0 to 6 log CFU/ml. Multiple analysis of variance showed that there were no differences between the methods at the 5% confidence level. However, the log CFU readings from the MIC technique were less reliable due to its higher lower bound limitation (2 log CFU/ml). At this low cell concentration, other techniques allowing more sample volume were more accurate. Only MPN returned slightly lower cell counts on average. Essentially all techniques resulted in practically the same readings providing the sample inoculums were prepared with proper dilution for colony evaluation. The short detection time, user-friendliness, and high throughput nature by MIC technique made this protocol practical for cell enumeration in food industry, especially with resource-poor settings. Our previous studies also demonstrated the versatility and practicality of the similar MIC format in enumerating other types of bacterial colonies (i.e., Listeria monocytogenes, Salmonella spp.) [24Liamkaew, R.; Thipayarat, A.; Saranak, J. Listeria detection in microscale solid state inoculation with minimal selective agents. Food Contr., 2014, 43, 183-192.
[http://dx.doi.org/10.1016/j.foodcont.2014.03.007] , 25Khueankhancharoen, J.; Thipayarat, A.; Saranak, J. Optimized microscale detection of amino acid decarboxylase for rapid screening of Salmonella in the selective enrichment step. Food Contr., 2016. (in press).]. Therefore, the miniaturized colony counting technique is certainly an economical and adequate replacement for the standard plating technique for industrial colony enumeration of E. coli.
3.2. Application of Cell Enumeration For Food Samples and Environmental Swabs
A statistical comparison of colony counts generated by the MIC and ISO-approved pour plate techniques was performed to compare the techniques in an industrial setting. The colony counts of the two E. coli/coliform enumeration strategies were the same as would be utilized by QC staffs and were applied to real samples, including samples of some ready-to-eat products (Table 2) and swabs of food preparation environments from production lines (Table 3) previously rejected using Petrifilm™ kits. The use of the pour plate technique was to confirm the results with the PetrifilmTM and validate the use of the MIC techniques to that ISO standard protocol. The resulting number of colony forming units (CFU) generated in each sample by the two methods were compared using t-tests. There were no statistical differences between the MIC and the pour plate technique. For most samples, the cell counts of MIC and pour plate results agreed. All colonies found were pink in color suggesting all contaminated samples were mostly contaminated by coliforms with a few possibilies of atypical E. coli strains [26Manafi, M. New developments in chromogenic and fluorogenic culture media. Int. J. Food Microbiol., 2000, 60(2-3), 205-218.
[http://dx.doi.org/10.1016/S0168-1605(00)00312-3] [PMID: 11016610] ].
The proposed MIC technique facilitates routine measurement of processing and equipment cleanliness and accommodates frequent and large numbers of swab samples. The samples shown in Table 3 were a small fraction of the total number of samples actually tested. Most of the swab results were “not-detected” (ND) samples and only few samples were contaminated by coliforms. Generally the MIC technique produced higher CFU counts than did the pour plate technique Tables 2 and 3. A practical cleanliness evaluation of a processing environment such as provided by the MIC technique, calls for fast and accurate estimation of E.coli/coliform numbers. The pour plate counterpart is slower and less efficient in evaluating production facility hygiene and often increased evaluation time can cause serious monetary loss to food manufacturers. In theory, the frequency of production line swabs must be adequate to ensure good hygienic practice. However, at the present most food factories avoid more frequent swab sampling merely because it increases the production cost.
To decrease the cost associated with increased sampling frequency the cultivation time of E. coli/coliform was minimized to streamline the protocol and, hence, reduce the analytical cost. Also the feedback from industrial lab personnel trained to use the MIC was collected following their first experience using this technique. Table 4 provides an overview of how QC personnel from a local food factory perceived the MIC method compared with the pour plate technique. From all of the questions we asked, easy cleaning, less labor-intensity and rapid time to detection were among the highlighted perceived benefits of the MIC. It should be emphasized that the rapid detection significantly reduced the overall assay time compared to the conventional method. The high throughput provided quick screening of large industrial sample volumes and facilitated industrial batch release of product yielding negative contamination samples.
Although there is no significant statistical difference between the MIC technique and the standard pour plate technique in terms of detecting contamination, the authors feel there are several advantages to be gained from using the MIC technique. Monetary comparison of the consumption of all disposables, utilities and media by the MIC technique and pour plate method for E.coli/coliform detection in 36 food samples is presented in Table 5. The total cost of supplies including preparation, media, disposable material, and utility was US$6.12 for the MIC method and US$36 for the pour plate technique. Total saving for material cost was US$29.88/36 samples or 83%.
CONCLUSION
An alternative protocol (i.e., MIC technique) was proposed and validated with the common industrial practices (MPN, pour plate, and Petrifilm™ EC protocols) to detect and enumerate E. coli/coliform contamination. The MIC enabled rapid and accurate readings of E. coli/coliform colonies to those of conventional methods tested using the pure cultures as well as food and environmental samples commonly found in resource-poor industrial settings. This new less costly technique will enable a producer to sample product more frequently without increasing the quality control budget. Together with the integration of already existing technology for handling liquid samples, like multichannel autopipettes and 96-well microtiter plates, the use of MIC technique not only helps reduce the expense of analytical cost and expensive CCA, but also it can shorten incubation time to 12-15 h when comparing with the conventional method and enable the analysis of high volume microbial enumeration samples. This MIC technique was well received by lab operators and technicians from a local medium-size food factory, especially in comparison with their past experiences using the pour plate routines. Hence, the MIC technique is a more practical protocol to enumerate the E. coli and coliforms counts for finished food samples and production lines samples in industrial settings. Moreover this method can be applied for detecting other foodborne pathogens from clinical samples.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
ACKNOWLEDGEMENTS
This work was financially supported by a Research Grant from Burapha University through National Research Council of Thailand (Grant No. 114/2558) and a graduate scholarship from the Thailand Research Fund through the Royal Golden Jubilee Ph.D. Program (Grant No. PHD/0216/2552) to Wipavadee Sangadkit and Asst. Prof. Dr. Aluck Thipayarat. We thank Professors Gustav A. Engbretson, Brenda G. Engbretson and Kenneth W. Foster for helpful discussion and critical reading of the manuscript. We also thank our industrial partnership with Buono (Thailand) Co., Ltd. who provided the food samples tested.
REFERENCES
| [1] | Oblinger, J.L.; Koburger, J.A. Understanding and teaching the most probable number technique. J. Milk Food Technol., 1975, 38, 540-545. |
| [2] | Oblinger, J.L.; Koburger, J.A. The Most Probable Number Technique. In: Compendium of Methods for the Microbiological Examination of Food; 2nd ed.; American Public Health Association: USA. 1984. |
| [3] | Pavic, A.; Groves, P.J.; Bailey, G.; Cox, J.M. A validated miniaturized MPN method, based on ISO 6579:2002, for the enumeration of Salmonella from poultry matrices. J. Appl. Microbiol., 2010, 109(1), 25-34. [PMID: 20059618] |
| [4] | Suwansonthichai, S.; Rengpipat, S. Enumeration of coliforms and Escherichia coli in frozen black tiger shrimp Penaeus monodon by conventional and rapid methods. Int. J. Food Microbiol., 2003, 81(2), 113-121. [http://dx.doi.org/10.1016/S0168-1605(02)00190-3] [PMID: 12457585] |
| [5] | Bredie, W.L.; de Boer, E. Evaluation of the MPN, Anderson–Baird– Parker, Petrifilm E. coli and Fluorocult ECD method for enumeration of Escherichia coli in foods of animal origin. J. Food Microbiol., 1992, 161, 197-208. [http://dx.doi.org/10.1016/0168-1605(92)90080-M] |
| [6] | Anderson, J.M.; Baird-Parker, A.C. A rapid and direct plate method for enumerating Escherichia coli biotype I in food. J. Appl. Bacteriol., 1975, 39(2), 111-117. [http://dx.doi.org/10.1111/j.1365-2672.1975.tb00551.x] [PMID: 1104553] |
| [7] | Anonymous, Microbiological Methods for the Dairy Industry; Australian Standard 1095, Standard Association of Australia: Sydney, Australia, 1987. |
| [8] | Feng, P.C.; Hartman, P.A. Fluorogenic assays for immediate confirmation of Escherichia coli. Appl. Environ. Microbiol., 1982, 43(6), 1320-1329. [PMID: 7049088] |
| [9] | Firstenberg-Eden, R. Electrical Impedance Method for Determining Microbial Quality of Foods. In: Rapid Methods and Automation in Microbiology and Immunology; Hobermehl, K.O., Ed.; Springer-Verlag: Berlin, 1985. [http://dx.doi.org/10.1007/978-3-642-69943-6_83] |
| [10] | Franco, W.; Hsu, W.Y.; Simonne, A.H. Survival of Salmonella and Staphylococcus aureus in mexican red salsa in a food service setting. J. Food Prot., 2010, 73(6), 1116-1120. [PMID: 20537270] |
| [11] | Fung, D.Y. Historical development of rapid methods and automation in microbiology. J. Rapid. Meth. Aut. Mic., 1992, 1, 1-14. [http://dx.doi.org/10.1111/j.1745-4581.1992.tb00066.x] |
| [12] | Khueankhancharoen, J.; Thipayarat, A. Application of modified drop plate technique (MDPT) and logistic model to optimize non-selective substrates for Salmonella typhi resuscitation. Asian. J. Food. Agro. Ind., 2011, 4(6), 349-358. |
| [13] | Sangadkit, W.; Rattanabumrung, O.; Supanivatin, P.; Thipayarat, A. Practical coliforms and Escherichia coli detection and enumeration for industrial food samples using low – cost digital microscopy. Procedia Eng., 2012, 32, 126-133. [http://dx.doi.org/10.1016/j.proeng.2012.01.1246] |
| [14] | Supanivatin, P.; Khueankhancharoen, J.; Saeaung, W.; Boonyaprapasorn, A.; Thipayarat, A. Industrial Implementation of Fast Total Plate Count Analysis Applying Micro Inoculation Culture on Frozen Ready-to-eat Food Products In: Proceeding of the International Conference for a Sustainable Greater; Mekong Subregion: Bangkok, Thailand, 2010. |
| [15] | Kang, D.H.; Fung, D.Y. Development and evaluation of a 24 well microtitre plate method for isolation of Listeria spp. or Listeria monocytogenes from foods. Lett. Appl. Microbiol., 1999, 28(4), 280-284. [http://dx.doi.org/10.1046/j.1365-2672.1999.00532.x] [PMID: 10212440] |
| [16] | Chenu, J.W.; Pavic, A.; Cox, J.M. A novel miniaturized most probable number method for the enumeration of Campylobacter spp. from poultry-associated matrices. J. Microbiol. Methods, 2013, 93(1), 12-19. [http://dx.doi.org/10.1016/j.mimet.2013.01.013] [PMID: 23384829] |
| [17] | Kim, S.; Fung, D.Y. Modified microtiter count method for viable cell counts from pure cultures and food model samples. Food Microbiol., 2005, 22, 595-599. [http://dx.doi.org/10.1016/j.fm.2004.11.011] |
| [18] | Weagant, S.D.; Grant, M.A. Bacteriological Analytical Mannual (BAM) Online: Chapter 4 Enumeration of Escherichia coli and the Coliform Bacteria. Available at: http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm064948.htm [2015, July 2]. |
| [19] | Coliforms and Escherichia coli Counts in Foods: Petrifilm™ E.coli / Coliform Count Plate Method. AOAC Official Method 991.14, 1991. |
| [20] | Andrews, W. Manual of Food Quality Control; Food and Agriculture Organizations of the United Nation: USA, 1992. |
| [21] | Hasan, A.; Belgin, S.; Serdar, C. Assessment of the microbiological quality of meals sampled at the meal serving units of a military hospital in Ankara, Turkey. Food Contr., 2004, 15(5), 379-384. [http://dx.doi.org/10.1016/S0956-7135(03)00101-4] |
| [22] | Juyun, L.H. A review of methods and theory. Food Qual. Prefer., 2011, 22, 733-747. |
| [23] | Finney, M.; Smullen, J.; Foster, H.A.; Brokx, S.; Storey, D.M. Evaluation of Chromocult coliform agar for the detection and enumeration of Enterobacteriaceae from faecal samples from healthy subjects. J. Microbiol. Methods, 2003, 54(3), 353-358. [http://dx.doi.org/10.1016/S0167-7012(03)00068-X] [PMID: 12842481] |
| [24] | Liamkaew, R.; Thipayarat, A.; Saranak, J. Listeria detection in microscale solid state inoculation with minimal selective agents. Food Contr., 2014, 43, 183-192. [http://dx.doi.org/10.1016/j.foodcont.2014.03.007] |
| [25] | Khueankhancharoen, J.; Thipayarat, A.; Saranak, J. Optimized microscale detection of amino acid decarboxylase for rapid screening of Salmonella in the selective enrichment step. Food Contr., 2016. (in press). |
| [26] | Manafi, M. New developments in chromogenic and fluorogenic culture media. Int. J. Food Microbiol., 2000, 60(2-3), 205-218. [http://dx.doi.org/10.1016/S0168-1605(00)00312-3] [PMID: 11016610] |









