- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Systems Biology Journal
(Discontinued)
ISSN: 1876-3928 ― Volume 5, 2014
Doomsday Book, Vol. II
William F. Wade*
Abstract
For humans, power is in part control over their lives. History is a collection of past experiences that empower informed actions. A repertoire is the catalog of your things. A large diverse repertoire can be critical or it can be irrelevant. A great hammer is nice if the job involves a nail. You might have lots of wrenches, but not the one that bends the right way for the “tough nut”. The wrong wrench eventually gets the nut loose and off; the right wrench gets it done faster. The right wrench is needed to turn the tough nut. Specific antibodies (Abs) protect against a number of bacterial, viral, and protozoan infections. Protective, antigen-specific Abs are wrenches that can turn the tough nuts (protective antigens of pathogens). Having the right Abs in advance of an infection makes protection more likely. The immunome comprises all the Abs in the immune system’s tool box. Only a small subset of the immunome protects against certain infectious diseases. Perhaps the immunome needs “outside” instruction in how to maximize its protective Ab capacity in the shortest time possible to maximize vaccine-induced immunity.
Article Information
Identifiers and Pagination:
Year: 2009Volume: 2
First Page: 8
Last Page: 17
Publisher Id: TOSYSBJ-2-8
DOI: 10.2174/1876392800902010008
Article History:
Received Date: 3/2/2009Revision Received Date: 16/2/2009
Acceptance Date: 16/2/2009
Electronic publication date: 09/04/2009
Collection year: 2009
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http: //creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Department of Microbiology and Immunology, Dartmouth Medical School, Lebanon, NH 03756, USA; Tel: 603-650-6896; Fax: 603-650-6223; E-mail: william.f.wade@dartmouth.edu
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 3-2-2009 |
Original Manuscript | Doomsday Book, Vol. II | |
THE IMMUNOME
In 1085 A. D., the Domesday Book was commissioned by England’s William the Conqueror. The book was a complete listing of his kingdom’s property and its owners (www.domesdaybook.co.uk). A10th and 11th century Anglo-Saxon practice termed the “Danegeld” was a uniform tax to pay for the defense (tribute) of England against common enemies like the Danish Vikings. William codified his tax base (repertoire of funds), not for tribute but for defense by force of arms. The human host must be defended by force of arms against common enemies that take the form of viral, bacterial, and protozoan invaders. In 2009, A. D. we do not have a complete listing, a Domesday Book that defines all the protective vaccine targets for various invaders we face daily.
The immunome is to immunology what the genome is to genetics. Dr. Pederson defined the immunome as the totality of expressed Abs and antigen receptor (T cell receptor, TCR) genes present in all living humans [1]. Others expanded the definition to include all the genes associated with the immune response [2] or restrict the immunome to peptide epitopes bound to MHC Class I or II molecules that activate antigen-specific T cells by binding TCR [3]. Antibodies and TCRs are structurally unique proteins that recognize foreign antigens, non-self macromolecules that are structural or surface components of pathogens. Recognition of non-self is the first step in activating the immune system to advance suitable effectors to control or eliminate an infection.
A unifying concept implicit to the definitions of the immunome is immunity. Human B cells make Abs to protect the host against viral, bacterial, and protozoan infections. Humans developed their immune Ab repertoire over evolutionary time, but more importantly they can also evolve the Ab repertoire in real time (life of the host) in response to infection. For the purposes of this discussion, the total expressed immunome is limited to the collected gene segments of the immunoglobulin (Ig) loci which recombine to provide surface-expressed, antigen-specific Abs on B cells. B cell expressed Abs, that bind pathogen-associated antigens are usually activated to proliferate and eventually secrete antigen-specific Abs. Pathogen-specific Ab may or may not be protective. Protection depends on the target’s role in pathogenesis and the quality of the Abs.
B cells that can bind immunogenic (able to induce an immune response) antigens can differentiate in to long-lived, circulating memory B cells and sessile plasma cells that secrete immunogen-specific Ab (Fig.1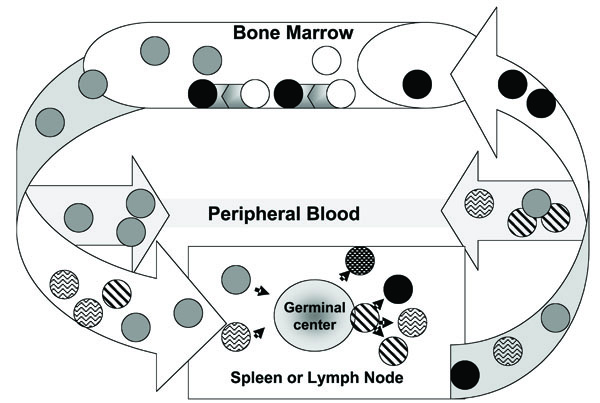 ). There are three major subsets of circulating human B cells that are differentiated based on surface markers: non-antigen selected, naïve B cell (CD19+, IgD+ CD27-), antigen-selected, not isotype-switched memory B cells (CD19+, IgD+ CD27+), and antigen-selected, isotype-switched (IgG+, IgA+, IgE+) memory B cells (CD19+, IgD- CD27+) [4, 5]. Naïve B cells contain the evolutionary record of the immunome; somatically mutated, memory B cells express the real time antigen-selected immunome. The evolutionarily immunome is encoded in the germ line DNA of the Ig loci. The real time, antigen-selected repertoire reflects somatically mutated germ line Ab sequences. The evolutionary immunome ensured the survival of the populations because it was selected to recognize a very large set of antigens. The memory immunome, which improves the germ line repertoire, results from individual responses to vaccinations or infections. The memory immunome repertoire includes B cells that respond to a previous encounter with a immunome-antigen set, faster, and with higher affinity Abs. If we knew the protective immunome (germ line or memory) we could design vaccines to ensure selection of only the “right” B cells. The protective immunome, Abs that prevent infectious disease could be used to define age and geographically appropriate vaccines for different infectious agents.
). There are three major subsets of circulating human B cells that are differentiated based on surface markers: non-antigen selected, naïve B cell (CD19+, IgD+ CD27-), antigen-selected, not isotype-switched memory B cells (CD19+, IgD+ CD27+), and antigen-selected, isotype-switched (IgG+, IgA+, IgE+) memory B cells (CD19+, IgD- CD27+) [4, 5]. Naïve B cells contain the evolutionary record of the immunome; somatically mutated, memory B cells express the real time antigen-selected immunome. The evolutionarily immunome is encoded in the germ line DNA of the Ig loci. The real time, antigen-selected repertoire reflects somatically mutated germ line Ab sequences. The evolutionary immunome ensured the survival of the populations because it was selected to recognize a very large set of antigens. The memory immunome, which improves the germ line repertoire, results from individual responses to vaccinations or infections. The memory immunome repertoire includes B cells that respond to a previous encounter with a immunome-antigen set, faster, and with higher affinity Abs. If we knew the protective immunome (germ line or memory) we could design vaccines to ensure selection of only the “right” B cells. The protective immunome, Abs that prevent infectious disease could be used to define age and geographically appropriate vaccines for different infectious agents.
INTRINSIC SIGNATURES EQUAL INFORMATION
Advances in informatics and computational biology have fostered the collection, the classification, and the understanding of several intrinsic biological signatures: the genome, the proteome, and the immunome. Data mining is an accepted means of investigation to make rapid progress in understanding and devising controls to manipulate complex systems. This accounting, along with a systems biology perspective, is shaping the way we think about how to conduct basic research to provide more effective, yet less expensive health care.
Intrinsic biological signatures develop over evolutionary time. They are maintained as encoded instructions that specify the correct responses to environmental challenges for species, proteins, and systems. Research based on the sequences of the human genome and of multiple pathogens (e.g., V. cholerae and most of the common viruses) has proven the value of knowing intrinsic signatures. The immune system is inducible and dynamic in its response to pathogens. The humoral arm of the immune system provides Abs that bind intrinsic signatures of macromolecules expressed or secreted by pathogenic organisms. To optimize humoral immunity with improved vaccine design and delivery, we must know the signature of the protection human immunome.
IMMUNOGLOBULIN STRUCTURAL INFORMATION IS ENCODED IN DISCONTINUOUS EXONS THAT ASSORT TO CONTRIBUTE 107 OR MORE SPECIFICITIES
Antibodies are found at the portals of entry used by pathogens to infect the host. Vaccination prepositions pathogen-specific Abs at these portals to prevent or limit the infection to local damage rather than systemic. The generic, monomeric Ab structure is composed of two heavy (H) chains and two light (L) chains (Fig.2 ). The H and L chain constant domains are connected to their respective V domain, the composite of which binds antigen. Constant domains are coupled to Ab tissue distribution (H chain), Ab effector function (H chain), and Ig-fold stabilization (H and L chain). During B cell development in the bone marrow, the Diversity (D) and Joining (J) exons are recombined in a semi-random with an upstream Variable (V) to form the VH. The L chains use V and J segments to form the VL.
). The H and L chain constant domains are connected to their respective V domain, the composite of which binds antigen. Constant domains are coupled to Ab tissue distribution (H chain), Ab effector function (H chain), and Ig-fold stabilization (H and L chain). During B cell development in the bone marrow, the Diversity (D) and Joining (J) exons are recombined in a semi-random with an upstream Variable (V) to form the VH. The L chains use V and J segments to form the VL.
Each VH and VL domain has three complementary determining regions, (CDR 1-3) which are the principle points of contact between antigens and Abs. The third CDR, which is the most diverse, is the product of combinatorial assortment of either the VDJ or the VJ exons. Maximum diversity and thus antigen specificity is attributed to the VH CDR3. Experiments have proven that a limited number of VH and VL gene segments given somatic mutation of CDR3 (HV) can provide a seemingly comprehensive Ab repertoire [6, 7]. The CDR1 and CDR2 loops encoded in the VH and VL gene family members are often viewed as canonical with respect to length and shape. When folded as a mature Ab, the H and L V domains containing the CDR1 and CDR2 combined with CDR3 region generate promiscuous antigen binding structures [8]. The germ line H and L V region family members combine to form at least seven different folds that position the CDRs into “shapes” that are the starting conformations or the “complementarities” that Abs expressed by naïve B cells use to bind a pathogen’s immunome-antigen set [9, 10]. Initial vaccination or infection, results in selection of naive B cells expressing appropriately shaped germ line Abs. In contrast, antigen-selected memory B cells usually express “somatic mutated Ab” wherein the Ab encoding DNA is altered to improve fine specificity and increase the affinity. The modification of Ab’s antigen combining site optimizes Ab and immunogen interaction by reducing the number of Ab conformers (shapes) the antigen binds [11].
There are differences in recombination and subsequent selection results that in biases between the neonatal and adult Ab repertoires that likely results in protective immunome differences as we age [12-14]. VH3 and VH4 family members (the two largest VH families) account for 70% of the adult B cell repertoire [15, 16]. In B cell subtypes, VH6 family usage differs between neonates and adults [17]. The Ab repertoire can differ between CD5+ (expressed in neonates) and CD5- B cell subsets [15]. The diversity introduced by somatic mutation of IgM memory (innate memory) B cells differ with age. It takes between 2 and 5 years after birth to develop an adult repertoire of IgM memory B cells [18-20]. The less complex fetal and neonatal Ab repertoires are enriched for poly-reactive Abs of low affinity [21, 22]. Neonatal Abs generally have longer CDR3 regions that present flatter areas composed more often of small, polar amino acids (tyrosine, threonine, serine and proline) that promote promiscuous binding of antigens [23-25]. As B cells undergo affinity maturation, somatic mutation of the Fab, there is a trend to eliminate outlier CDR gene usage, to decrease the CDR3 length, and to modify the amino acid composition and hydrophobicity of the CDR3 region [26].
THE PROTECTIVE IMMUNOME
In the developing world, and to a lesser extent in the developed world, there is a pressing and largely unmet need for infectious disease (e.g., RSV, Cholera, and Malaria) vaccines. During infection or in response to vaccination, humans do not make all possible protective Abs with equal efficiency, nor are all selected Abs protective. In rotavirus infection, infant (≤ 2 years old) B cells which are predominantly CD5+ use VH3-23, while CD5-B cells use VH1 and VH4; VH1-46 was associated with the immunodominant response in CD5- B cells from children and adults [27]. An open question is do these differences reflect the need to select different Abs (degree of conformational flexibility) at different ages to be protected against rotavirus infection. Parasitic diseases such as Malaria or Schistosomiasis result in protective Ab responses that only develop after repeated exposure to parasite antigens [28, 29]. It is highly probably that identifying the protective immunome for parasitic diseases will aid vaccine target discovery so younger individuals can be immunized effectively without the need for repeated infections.
Childhood vaccines are delivered multiple times to guarantee adequate immunity. This is due to the need to select Abs shapes and then to sharpen them for efficacy. Another reason for multiple vaccination with the same immunogen is because of the limitations in inducing immunity imposed by developmental delays of immune system’s support cells and structural elements (marginal zones or bone marrow) required for function of certain B cell subsets [30].
Dr. Pederson’s immunome definition should be modified to focus on the protective immunome, the essential component of the total immunome. It is reasonable to think the protective immunome is a manageable set that can be experimentally determined. Somatic mutations of antigen-selected Abs represent the product of punctuate evolution of B cell fitness, whereby they now express an evolved, more protective Abs. Can we modify the antigen-induced real time evolution of the protective immunome? If we knew the neonatal protective immunome based on the subsets of functional B cells, we could design immunogens to preferentially expand antigen-specific, protective B cells, thus advancing a larger percentage of memory B cells and plasma cells for sustained humoral immunity and memory.
LIMITED SPACE SUGGESTS WE SHOULD OPTIMIZE THE PATH TO PLASMA CELL SURVIVAL
We don’t know the exact numbers or composition of the expressed human VH and VL repertoire, but estimates of the human Ab repertoire diversity are 0.5-1.0 x 107 and higher [6, 31]. The uncertainty is due to the “fudge factor” immunologists use to calculate the influence of non-combinatorial diversity on the generation of a unique Ab. Immunogen-selected B cells (naïve or memory) develop into plasmablasts and plasma cells, the later of which needs to gain access to survival niches in the bone marrow, lamina propria (gut), and inflamed tissues to sustain circulating and mucosal Ab [30]. Plasma cells, whether or not they secrete protective Abs, occupy limited space in the “survival niches”. Stromal cells maintain the limited number of B cell survival niches that new developing B cells, old plasma cells, and new plasma cells compete for. Regardless of the exact number, the potential to make a very large number of plasma cells, able to secrete Abs specific for multiple relevant and irrelevant B cell epitopes [32] suggests the number of plasma cells is large. It is not known how new plasma cells replace old plasma cells, but it is obvious that given the large number of potential plasma cell and the limited number of survival niches, that it is important to preferentially ensconce useful plasma cells. If we create age-appropriate immunogens that efficiently super-select protective Ab expressing B cells, we will reduce the competition for survival niches and optimize vaccine-induced immunity.
THE MISSING PIECE AND THE PATTERN WE NEED TO WEAVE
There is an outstanding issue that currently limits weaving information together to develop approaches or methods to engineer better vaccines. We know the germ line sequences of the Ig H and L chain loci. We know the genome sequence of multiple pathogens and much about potential protective Ab targets for those pathogens. We know a good deal about B cell biology, especially the pathways involved in affinity maturation of B cells. We know where to look for protective Ab producing B cells. We do not know the complete expressed human immunome. In specific, we don’t know the expressed, protective immunome present in different aged humans at steady state (germ line) or after immunogen exposure. In fairness, the search for defined parts of the immunome for specific diseases is being pursued by virologists, bacteriologist, parasitologists, and immunologists. The results to date support the concept presented here, but the scope of the current efforts will not define the total immunome or the protective immunome at the level that needs to be known. The power of a larger n (more recombinant Ab libraries and more epitope screening) from an expansive and more inclusive survey will increase the likelihood of finding rare, age-related protective immunome signatures.
Theoretically it is possible to discover the Ab repertoire that defines all the protective immunogens for the foremost human pathogens. There is a limited set of pathogens (not including serotypes) that account for the majority of human morbidity and death. The Center for Disease Control lists some 40-50 infectious agents that are major causes of human disease. Simple viruses might have two surface proteins each with three protective B cell epitopes. Complex viruses, bacteria, and protozoa have more surface expressed immunogenic targets. The number of protective epitopes for more complex pathogens might be an order of magnitude higher. Given these approximations, 1-3 x103 is a conservative estimate of the number of protective target epitopes. If the actual number is 2 logs higher, 105, and the total Ab immunome is 108-9, then protective B cell epitope embody between 0.01 - 0.1% of the total immunome. The number of plasma cell niches in the bone marrow is thought to be around 109 [33]. Competition to ensure the protective plasma cells are available for serum memory (circulating Abs and memory B cells) is complicated by selection of the right B cells from a large population of irrelevant specificities. Competition for survival niches occurs against a background of B cell activation and differentiation to gut flora immunogens, non-protective immunogens of pathogens, and allergens.
Evolution of the immune system was based on its interactions with pathogens and established the baseline immunome (Ig loci). This immunome, likely did not have to respond to many closely spaced infections (life-threatening) so the competition for survival niches during evolution of the immune system was probably not limiting. The vaccination protocols we have established, selects and expands the baseline immunome into a more evolved protective immunome within a short period of time. The question is can intelligent immunogen design for new vaccines increase the percentage of protective plasma cells successfully competing for survival niches?
FV (FRACTION VARIABLE) LIBRARIES - AN ESSENTIAL TOOL FOR EX VIVO ISOLATION AND SELECTION OF THE IMMUNOME
Recombinant VH and VL Ab libraries can be made in the form of scFv (single chain, HV and LV chains tethered by a linker) or Fab (fraction antibody binding), a more stable version of a scFv with VH and VL-proximal constant domains. scFv and Fab libraries have been made from peripheral blood and germinal center B cells [34]. B cells provide mRNA to generate full length VH and VL cDNAs for cloning into the pIII coat protein gene of a ssDNA phage or phagemid. This results in surface expression of an antigen binding structure at the phage’s tip [35]. Phagemids are the preferred cloning system for efficient generation of highly complex (large, 109-11) libraries containing multiple high affinity clones [36]. Libraries can be stored long term and used for repeated isolation of multiple antigen-specific reagents.
Recombinant phages of interest are selected for binding to identified substrates, followed by washing unbound phage away from bound. Several rounds of selection and amplification can result in multiple clones of antigen-specific reagents. New protocols have been devised that allow discrimination of the conformational state of surface proteins which will be important for selecting the complete protective immunome [37]. There are often immunodominant Ab responses to particular epitopes on specific immunogens. Immunodominant Ab responses can comprise a larger proportion of the memory repertoire. In order to find the complete set of protective Abs it is necessary to manipulate the screening process. These rare, non-immunodominant but protective and perhaps germ line sequence reagents can be selected by “blocked panning” which removes or prevents selection of scFv of Fab reagents that bind the immunodominant antigen. An initial screen of pathogen-associated targets and their validation as protective targets would eventually reduce the complexity of the library for a more in-depth screen. Second and third generation arrays that express non-immuno-dominant or rare B cell epitopes will improve isolation of reagents that represent protective VH and VL that are not highly represented in the B cell repertoire of immune individuals.
Two types of libraries can be constructed, non-immune and immune. Non-immune libraries contain the baseline immunome that is selected and expanded by unknown immunogens. It is not known if different populations with unique diets and environmental encounters have the same basic immunome. B cell biology suggests that these differences in antigen exposure, if translated to differences in the expressed immunome, might predispose individuals to autoimmunity or moderate their vaccine responses. Immune libraries, which are usually less complex, are derived from immunized or infected hosts. Immune libraries have an improved capacity to bind the pathogen’s immunome-antigen set, protective or non-protective. Primary and secondary immune libraries from sequential immunization or infection with the same immunogen set reflect the intrinsic immunome and the memory immunome, respectively.
DO ScFv OR Fab RECOMBINANT LIBRARIES REPRESENT THE BEST PLACES TO SELECT THE BEST WRENCHES?
Some have noted differences in scFv and Fab recombinant libraries compared to mAbs derived from single B cells [38]. Random re-association of VH and VL cDNA not normally found in “real” B cells can occur in scFv or Fab libraries. However, random assortment of H and L chains in immature B cells is part of the normal molecular biology of Ab assembly. The percentage of irrelevant “non-normal” pairing of VH or VL recombinant libraries compared to normal pairing found in single B cells has not been determined. If there is a significant number of miss-paired HV and HL in individual scFv or Fab, their inclusion in the immunome as it accrues can be managed later by statistical analysis. ScFv or Fab reagents with VH and VL chain associations that do not occur in vivo might be an advantage for finding unique serologic reagents to discover non-dominate immunogens or to produce reagents for passive Ab therapy. Others have noted limited VH and VL diversity in scFv compared to mAbs specific for the same antigen, but not necessarily the same B cell epitope [37]. The length of some CDR3 of scFv is reportedly shorter that those in mAb of similar specificity [6, 15].
These differences do not diminish the value scFv or Fab libraries have already provided and that they could provide in the classification of the immunome. Phage recombinant libraries have identified targets for immuno-cancer therapy, unique reagents for diagnostic tests, and reagents to identify vaccine development or passive Ab therapy [39, 40]. Data indicate the microarray identification of Abs parallels that of conventional ELISA-based identification [41]. Recombinant phage technology is capable of being the major means to capture the protective and total immunome.
SORTING THE WHEAT FROM THE CHAFF
Methods are in place to collect and read the human immunome. Technology is available to represent the protein components of various pathogens in a multi-array format for efficient and accurate screening [42, 43]. A recent report indicated that over 1000 scFv clones bound to a single protein, yet only 40% inhibited binding to its receptor [32]. For a viral protein it would be important to know which of the 40% binders were the most protective or specific for less mutable B cell epitopes.
The effector function of Abs is as important as the specificity of the Ab. After B cell collection and generation of recombinant libraries it will be necessary to link the scFv or Fab reagents panned by the immunogens to the protective nature of the selected reagents. Library cloning vectors have been designed to be fascicle enough for reengineering the VH and VL into a functional Ab which will be required for some tests of protection. The means (virus neutralization, complement killing etc) to do this will depend on the disease model in current use. The modal sequence of protective Abs and the extremes with respect to Ab affinity will be further characterized to note changes in the protective immunome with age and time between infection or vaccination.
We should begin the identification of the protective immunome with IgG subclasses, but eventually should define the protective efficacy of IgM and sIgA for pathogen specific immunome-antigen sets. This is important as the multi-valiancy of IgM and sIgA might alter the protective repertoire, especially in young children wherein lower affinity solutions still function due to increased avidity afforded by increased valence of the Abs.
It is not uncommon to generate thousands of scFv clones and recover complete sequence from greater than 80% of them [32]. Computer programs (Phred and Phrap) exist to capture the phage sequence data, verify its accuracy, and process it for later use [44, 45]. Human Ig data bases and analysis programs (BLASTN) are available to compare VH and VL chain family usage and CDR3 lengths/polymor-phism with age and geographical location of the B cell donor [46-48]. New programs to parse sequences to correlate the age of the donor, type of B cell used to make the library, the endemic pathogen burden, and other variables will need to be developed. The collection of the immunome data set will prompt improved methods to streamline the collection and analysis of the information. These new data sets will need to be easily searchable and available to scientists for vaccine design and development.
B CELL SAMPLING - ARGUMENTS FOR SPECIFIC B CELL SUBSETS OR THE TOTAL B CELL REPERTOIRE
An important question that needs to be addressed is whether all B cells (circulating, those in secondary lymphoid organs, bone marrow or lamina propria) or only circulating B cells (peripheral blood) and perhaps only a subset of those B cells should be isolated and used to generate recombinant libraries. Specialized B cell such as CD5+ and marginal zone B cells exist. These tissues-associated B cells have skewed repertoires that encoded “innate” Abs that are mostly IgM, poly-reactive, and the Ig loci in germ line configuration [49, 50]. In tissues and organs, resident B cells, plasmablasts, and short or long-lived plasma cells represent successful clones that have been selected and maintained for humoral immunity. All of the different B cell types will be important to collect so that the entire protective immunome is collected. The source of B cells for generating recombinant libraries will depend on the question being asked and the state of development of the immunome data base.
Blood born, memory B cells are easily isolated at times closely coupled to recent infections or vaccinations. Peripheral blood, B cells (CD27- and CD27+) of neonates and young children will need to be evaluated as both represent baseline comparisons for the adult Ab repertoire. Memory B cells (CD27+) from peripheral blood of the young, middle aged, and old will be used to generate scFv or Fab libraries. The utility of knowing the total immunome of naive B cells (CD27-) will be important, but acquiring the memory B cell (CD27+) repertoire selected by immunization or infection will be more useful for vaccine design and thus should have priority.
The time point for B cell sampling for library production is a variable to consider as the number of immunogen-specific memory B cells in the blood will vary somewhat post antigen encounter. In general, memory B cells and plasmablasts leave the secondary lymphoid organs soon (4-7days) after antigen encounter. Plasmablasts rapidly try to enter their survival niches; memory B cells circulate in the blood for months. Even if B cells are collected at different time (days to weeks) post antigen encounter, a large donor cohort will provide for statistically significant coverage of the immunome expressed by memory B cells.
MICROARRAY ANALYSES, THE CRITICAL STEP LINKING THE SELECTED IMMUNOME TO PROTECTIVE IMMUNOGENS
Microarray platforms are high through-put formats that have dramatically changed the way we determine if one biologic interacts with another. The ability to print (accessible but immobilized) peptides, proteins, small molecules, or nucleic acids in a micro-format available to interact with a search medium allows the rapid identification of unique biologic signatures. This technology has revealed much about different aspects of biology from how genes are expressed in response to environmental stressors or cell differentiation to whether allergic patient’s IgE binds a specific allergen component [51, 52].
The most robust format for microarray analyses is based on nucleic acid, whereby cDNA mini genes representing the genome or a subset of the genome of a particular organism are printed in arrays which are interrogated with mRNA from the organism of interest. These types of analyses can indicate how the host responds to different stressors or activators. Proteins and peptides have also been used in the microarray format with the “what binds what” end point in mind. Peptide arrays are easy to produce and manipulate, but suffer from not being full-length immunogens and thus their use as targets to screen recombinant Ab libraries is limited to linear B cell epitope discovery.
Zhu and co-workers generated a microarray containing most of the proteins encoded by S. cerrevisiae [53]. Rolfs and co-workers demonstrated the potential of high through-put “functional” proteomics screening using the complete set of clones expressing the open reading frame of individual genes of V. cholerae [54]. While doable, the expression of the complete proteome of most pathogens (bacteria or protozoan) is currently impractical and perhaps not necessary. Native, pathogen-expressed surface structures representing conformational epitopes have been used to screen recombinant libraries (Plasmodium sp, HIV, RSV) [55-58]. There are already multiple, identified viral surface proteins and many bacterial surface proteins available for screening libraries. Other targets can be identified serologically and by targeting sequence analysis to predict bacterial membrane proteins. The set of surface structures of protozoa and metazoans is larger, but much is known for problematic pathogens such as Plasmodium that could direct selection of the protective immunome [58]. To begin the screening of the protective immunome, academic laboratories can take the lead in developing the libraries of expressed surface structures of the top 3 viral, bacterial, and protozoan pathogens.
THE SCOPE
The human genome project was met with idealism and skepticism; now most scientists agree the efforts were worthwhile. The full utility of knowing the human genome and that of various pathogens will be advanced by knowing the protective immunome. All the technologies and the infrastructure required to pursue and define the human immunome are established. The number of existing scFv and Fab libraries attest to the efficiency of the VH and VL primer set for cDNA synthesis and library construction. Academic and pharmaceutical laboratories throughout the world should serve as the centers for collecting and processing peripheral blood B cells. In more rural or underdeveloped areas, simple kits can be used to extract mRNA for the libraries obviating cold chain issues and thus enhance coverage. It will be important to annotate the source of the B cells as completely as possible with respect to geographical location, age, and vaccination and infection history. Algorithms exist for comparison of new sequences to the VH and VL data bases from existing pathogen-specific scFv, Fab, and mAbs (single B cell source) [59, 60].
The key to using the information in the human immunome extents past identifying selected Ab sequences. The full utility of the protective human immunome will only be realized if the selected, protective Ab repertoire is linked to the selector. There are multiple examples that have identified parts of the protective immunome for Mycobacterium, Plasmodium, and several viruses [56, 57, 61-63]. Many industrial and academic laboratories have established microarray systems (peptide, proteins of specific pathogens or carbohydrate) that could be used immediately. Once libraries are generated and characterized they would be kept at several NIH-funded repositories that would be responsible for the continued collection and distribution to scientists in established vaccine development groups. Focusing on several principal pathogens (RSV, Rotavirus, Salmonella, V. cholerae, HIV, or Plasmodium spp.) should speed the accrual of a pathogen-specific protective immunome. The broader question of how these sequences are distributed in the world-wide population will require more time for completion, but will be informative.
USES OF THE PROTECTIVE IMMUNOME
There are short and long term uses for the information contained within the human protective immunome and the total immunome. The most proximal use is vaccine development, especially for young children. Understanding how the protective immunome differs with age will be important for determining age appropriate immunogens. Young children require multiple immunizations partly because protective maternal Abs (transplacental or milk/colostrums) hampers the induction of the neonate’s immune system. Maternal anti-RSV G Abs and anti-RSV F Abs are protective, but restrict neonatal anti-G Ab development yet not the anti-F Ab response [64]. If we knew the protective anti-RSV immunome, we might discover that children have germ line solutions for protective anti-G protein Abs, while adults have a very effective anti-F Ab response resulting from somatic mutation. Based on this type of information, mothers would be immunized with RSV F protein to passively protect the infant while the infant’s anti-G Ab response is promulgated without silencing by maternal anti-G Ab.
A long term goal would be to determine the protective immunome of plasma cells, marginal zone B cells (functional only after 2-5 years), and CD5+ B cells (found in the young and old). The later two B cell types are important for certain infections based on their natural Ab repertoire. Defining the protective and total immunome will tell us whether different B cell subsets have repertoires skewed by living in different antigen environments. The compilation of the protective immunome might uncover a fundamental need to redesign vaccines that are age and geographically aligned. Other future uses derivative of the protective immunome might include a better understanding of how cross-reactivity of Abs participate in, or lead to autoimmune disease. The immunome might reveal if the Ab response to environmental antigens or pathogens regulates autoimmunity in a negative or positive manner. Harwanegg and Hiller reviewed the progress of microarray screening to define anti-IgE response to allergens [65]. They predicted that understanding the patterns of allergen-specific IgE or progress in allergen desensitization therapy (IgE to IgG) will significantly improve diagnosis and treatment. There are corollaries for the protective immunome analysis. Reduced vaccination costs are possible if pathogen-specific protective immunogen, microarrays determine a second booster for diphtheria, tetanus and pertussis is not required. The time for measles or mumps booster vaccines could be known and provided based on the need registered by knowing the individual protective immunome. In the future microarray chips with the top 100 protective immunogens will be able to clearly stage degrees of immunity to specific pathogens leading to comprehensive individualized diagnostics that correlate with the identified protective immunome.
INTELLIGENT IMMUNOGEN DESIGN FOR REAL TIME EVOLUTION OF THE GERM LINE REPERTOIRE
The thermodynamic analyses of Ab conformations, either bound or unbound with antigen has led to rules for Ab and antigen interactions that should be exploited for vaccine design. An initial theory for Ab/antigen binding was the induced fit concept whereby antigen binding one of several germ line Ab conformations resulted in the antigen’s embrace by the Ab [11, 66]. The lock and key model of Ab/antigen interaction predicts a more rigid Ab structure that antigen precisely fit into with minimal conformational change required for optimal binding.
As in most biological systems, both stories seem to be largely true and represent the evolution of the Ab response over time. The germ line, non-antigen selected B cell repertoire is thought to have an ensemble of “conformers” that it can access within femto- or nanoseconds to fit antigens and thus start the initial humoral response [11]. The entropy of the germ line Ab repertoire is high, allowing more antigens to sample one binding site thus maximizing the availability of binding sites for related or even different antigen which can select and activate more naïve B cells. The evolutionary advantage for this design is an easy and initial broad selection of B cells that can be shaped by somatic mutation of their Abs that will bind the original antigen better. For protein-specific Abs, entry of B cells into germinal centers for somatic mutation of the HV or LV and the frame work regions alters the Fab and reduces the number of conformers Abs access. Somatic mutation of antigen-selected Abs can change the angle and rigidity of the antigen binding site (paratope) resulting in lock and key model binding.
High affinity Ab conformers for a specific immunogen fit the naïve immunogen, but can fit altered immungens better [67, 68]. The question becomes, can protective B cell epitopes be designed to maximize the selection of naïve, intermediate, and memory B cells based on the preferred conformers of those Abs. The complexity of the paratope for the early repertoire is such that immunogen binding (B cell selection) can take place in different conformer space, but also within the subspace of a conformer i.e., parts of the binding site rather than the binding site in total [69]. This flexibility for where antigen binds might allow selection of B cells (based on VH and VL shape and sequence) that are more “receptive” to mutations in the frame work or antigen binding residues with immunization. The result may be fewer intermediates to the final lock and key solution and thus less time and fewer immunizations for immunity.
Perhaps vaccine immunogens need to be of different designs depending on whether they are inducing the naïve or memory Ab repertoire. It is conceivable that a large but defined set of immunogens that are optimal for traversing different energy landscapers (Ab conformers) can be identified by small molecule screens in microarray format. Saturation mutagenesis of the protective B cell epitopes of various proteins of pathogens could be used to determine if some forms of the altered antigens select the naïve repertoire more efficiently. These non-native immunogens can be used to generate “new” vaccine to specifically target the immunome in young children or neonates. The “pre-native antigen” expansion before the interaction with native motifs would enhance the peripheral pool of protective B cells. It might be a particular advantage to set the immunome in motion at a young age so future antigen-specific B cells can enter the memory pool or the plasma cell pool thus providing more responsive B cells for subsequent booster immunizations.
The immunome project will require leadership at the government and industrial levels to focus the effort of the world’s scientific community. Special funding from the NIH to form partnerships between academic and pharmaceutical laboratories should be linked to preferential licensing of future vaccine that would incentivize the search for solutions that put the right Ab and the right immunogen together to solve the immunity puzzle.
THE DOOMSDAY BOOK, VOL. II - AN ACCOUNTING - A MEANS TO AN END
Defining the total expressed human immunome or just the protective immunome will be of the same scope, if not larger, than the human genome project. Classification of the protective immunome will begin to relate the real-time evolution of Abs with their efficacy. Identifying the protective immunome for particular pathogens will be useful for vaccine development, enabling “reverse engineering” to determine the best immunogen design to induce the protective immunome in an efficient, effective manner [70]. Energy savings through efficiency of use or new technology are the watch words of human progress. Optimized efficiency is a major step towards energy savings. The efficiency of the immune system is good in the context of survival of the species. It is reasonably fast (4-7 days) especially if the number of infectious agent encounters are low and the pathogens are not highly virulent. Currently, for young individuals who have more immediate and defined immunity needs, we require the immune system to respond early to multiple vaccinations to a larger antigen set.
The English people during the time of William the Conqueror likened the information, the invariant truth in the Domesday Book to that associated with Doomsday or the Last Judgment of Christians when the complete accounting of individual deeds will be reviewed by their God. The vision of the protective immunome needs to be communicated to, and understood by those empowered to make decisions about public health policy. If they are convinced of the value of knowing the protective immunome and sponsoring its acquisition, we can write the second volume of the Doomsday Book that based on its completeness will spell doom for intractable pathogens waging war against our immune system.
REFERENCES
Endorsements
Table of Contents
- THE IMMUNOME
- INTRINSIC SIGNATURES EQUAL INFORMATION
- IMMUNOGLOBULIN STRUCTURAL INFORMATION IS ENCODED IN DISCONTINUOUS EXONS THAT ASSORT TO CONTRIBUTE 107 OR MORE SPECIFICITIES
- THE PROTECTIVE IMMUNOME
- LIMITED SPACE SUGGESTS WE SHOULD OPTIMIZE THE PATH TO PLASMA CELL SURVIVAL
- THE MISSING PIECE AND THE PATTERN WE NEED TO WEAVE
- FV (FRACTION VARIABLE) LIBRARIES - AN ESSENTIAL TOOL FOR EX VIVO ISOLATION AND SELECTION OF THE IMMUNOME
- DO ScFv OR Fab RECOMBINANT LIBRARIES REPRESENT THE BEST PLACES TO SELECT THE BEST WRENCHES?
- SORTING THE WHEAT FROM THE CHAFF
- B CELL SAMPLING - ARGUMENTS FOR SPECIFIC B CELL SUBSETS OR THE TOTAL B CELL REPERTOIRE
- MICROARRAY ANALYSES, THE CRITICAL STEP LINKING THE SELECTED IMMUNOME TO PROTECTIVE IMMUNOGENS
- THE SCOPE
- USES OF THE PROTECTIVE IMMUNOME
- INTELLIGENT IMMUNOGEN DESIGN FOR REAL TIME EVOLUTION OF THE GERM LINE REPERTOIRE
- THE DOOMSDAY BOOK, VOL. II - AN ACCOUNTING - A MEANS TO AN END


 , Memory B cells,
, Memory B cells,
 , Somatically mutated B cell,
, Somatically mutated B cell,
 , Plasmablast,
, Plasmablast,
 , and Plasma cell,●.
, and Plasma cell,●.