- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Analysis of Genetic Variation of Two NADC30-like Strains of Porcine Reproductive and Respiratory Syndrome Virus in China
Zhi Zhou, Jiajun Wu, Shuo Zhang, Bo Hou, Tao Han, Jing Wang, Qi Xu, Dongyue Wang, Yinyi Liu, Shengpeng Xin, Xinyan Zhai*
Abstract
Background:
NADC30-like strains of porcine reproductive and respiratory syndrome virus first appeared in Chinese swine herds in 2012.
Objective and Method:
To explore the possible genetic diversity of these strains, we sequenced and analyzed the complete genomes of two NADC30-like strains. These isolates shared 95.4% homology with NADC30.
Result:
The two strains displayed a discontinuous deletion of 131 amino acids in NSP2, mutations of amino acids in GP3 and GP5, and a 3-nucleotide deletion in the 3′ untranslated region. Phylogenetic analysis showed that the two isolates formed a new branch and clustered in a subgroup with NADC30 isolates from North America.
Conclusion:
We conclude that the above two NADC30-like strains may have been introduced from North America to China, where they acquired new genetic diversity.
Article Information
Identifiers and Pagination:
Year: 2017Volume: 11
Issue: Suppl-1, M7
First Page: 90
Last Page: 97
Publisher Id: TOVJ-11-90
DOI: 10.2174/1874357901711010090
Article History:
Received Date: 06/10/2016Revision Received Date: 01/11/2016
Acceptance Date: 06/02/2017
Electronic publication date: 30/06/2017
Collection year: 2017
open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution 4.0 International Public License (CC-BY 4.0), a copy of which is available at: https://creativecommons.org/licenses/by/4.0/legalcode. This license permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
* Address correspondence to this author at the China Animal Disease Control Center, No. 20 Maizidian Street, Chaoyang District, Beijing, 100125, P. R. China; Tel: +86-10-59198898; Fax: +86-10-59198899; E-mail:Zhaixy2010@sina.cn
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 06-10-2016 |
Original Manuscript | Analysis of Genetic Variation of Two NADC30-like Strains of Porcine Reproductive and Respiratory Syndrome Virus in China | |
INTRODUCTION
Porcine reproductive and respiratory syndrome (PRRS) is one of the most important infectious diseases in swine. This disease leads to reproductive failure in sows and respiratory disease in pigs of all ages [1Wensvoort G, Terpstra C. et al. Mystery swine disease in The Netherlands, the isolation of Lelystad virusVet Q 1991;13(3):121-130., 2Meulenberg JJ, Hulst MM, de Meijer EJ, et al. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology 1993; 192: 62-72.
[http://dx.doi.org/10.1006/viro.1993.1008] ]. PRRS virus (PRRSV), which belongs to the Arteriviridae family in the order Nidovirales, is an enveloped virus with a single-stranded positive-sense RNA genome [1Wensvoort G, Terpstra C. et al. Mystery swine disease in The Netherlands, the isolation of Lelystad virusVet Q 1991;13(3):121-130.-3Allende R, Lewis TL, Lu Z, et al. North American and European porcine reproductive and respiratory syndrome viruses differ in non-structuralprotein coding regions. J Gen Virol 1999; 80: 307-15.
[http://dx.doi.org/10.1099/0022-1317-80-2-307] ]. The whole PRRSV genome is ~15 kb and contains at least 10 overlapping open reading frames (ORFs): ORF1a and ORF1b encode viral nonstructural proteins, whereas ORF2a, ORF2b, and ORF3–7 encode viral structural proteins [4Firth AE, Zevenhoven-Dobbe JC, Wills NM, et al. Discovery of a small arterivirus gene that overlaps the GP5 coding sequence and is important for virus production. J Gen Virol 2011; 92(Pt5): 1097-106.
[http://dx.doi.org/10.1099/vir.0.029264-0] -7Snijder EJ, Meulenberg JJ. The molecular biology of arteriviruses. J Gen Virol 1998; 79: 961-79.
[http://dx.doi.org/10.1099/0022-1317-79-5-961] ].
PRRSV is divided into European (Type 1) and North American (Type 2) genotypes, which share only 60% homology at the nucleotide level [7Snijder EJ, Meulenberg JJ. The molecular biology of arteriviruses. J Gen Virol 1998; 79: 961-79.
[http://dx.doi.org/10.1099/0022-1317-79-5-961] , 8Oleksiewicz MB, Bøtner A, Madsen KG, et al. Sensitive detection and typing of porcine reproductive and respiratory syndrome virus by RT-PCR amplification of whole viral genes. Vet Microbiol 1998; 64(1): 7-22.
[http://dx.doi.org/10.1016/S0378-1135(98)00254-5] ]. Although PRRSV isolates are always identified in their respective geographic regions, coexistence of the two types has been found in Europe, North America, and Asia. Such coexistence has had a major impact on PRRS diagnosis and management [9Madsen KG, Hansen CM, Madsen ES, et al. Sequence analysis of porcine reproductive and respiratory syndrome virus of the American type collected from Danish swine herds. Arch Virol 1998; 143(9): 1683-700.
[http://dx.doi.org/10.1007/s007050050409] -11Balka G. Genetic diversity of porcine reproductive and respiratory syndrome virus strains circulating in Hungarian swine herds. Vet Microbiol 2008; 127: 128-35.
[http://dx.doi.org/10.1016/j.vetmic.2007.08.001] ]. In China, emergence of PRRSV Types 1 and 2 has been reported previously [12Guo B, Chen Z, Liu W, et al. Isolation and identification of porcine reproductive and respiratory syndrome (PRRS) virus from fetuses suspicious of PRRS. Chin J Anim Poult Infect Dis 1996; 17: 1-4., 13Chen NH, Cao Z, Yu XL, et al. Emergence of novel European genotype porcine reproductive and respiratory syndrome virus in mainland China. J Gen Virol 2011; 92: 880-92.
[http://dx.doi.org/10.1099/vir.0.027995-0] ], although the most prevalent isolates in China are Type 2 strains.
Since 2012, NADC30-like PRRSV has been reported in Chinese swine herds and has caused huge economic losses [14Zhou L, Wang ZC, Ding YP, et al. NADC30-like strain of porcine reproductive and respiratory syndrome virus, china. Emerg Infect Dis 2015; 21(12): 2256-7.
[http://dx.doi.org/10.3201/eid2112.150360] ]. To explore the epidemic status and genetic diversity of NADC30-like PRRSV in China, we isolated two NADC30-like strains, sequenced their complete genomes, and compared their genetic relationships with previously examined NADC30 and NADC30-like PRRSV strains. We analyzed gene sequences of the 5′-untranslated region (UTR), 3′-UTR, hypervariable nonstructural protein 2 (Nsp2) gene, and most of the major viral structural proteins encoded by ORF3 and ORF5 of the two PRRSV isolates.
MATERIALS AND METHODS
Ethics Statement
This study was carried out at private farms. Animals were sampled with the permission of the farm’s managers. The animal field trial in this study was approved by the Animal Care and Ethics Committee of China Animal Disease Control Center. Conventional animal welfare regulations and standards were taken into account.
Virus Isolation
Twenty-six clinical samples of lungs, lymph nodes, livers, kidneys, and sera were collected from different farms in China. Samples were screened by a conventional RT-PCR assay for both genotypes of PRRSV [13Chen NH, Cao Z, Yu XL, et al. Emergence of novel European genotype porcine reproductive and respiratory syndrome virus in mainland China. J Gen Virol 2011; 92: 880-92.
[http://dx.doi.org/10.1099/vir.0.027995-0] ]. Samples that tested positive by PCR were further tested by real-time RT-PCR for differentiation of PRRSV Types 1 and 2 [13Chen NH, Cao Z, Yu XL, et al. Emergence of novel European genotype porcine reproductive and respiratory syndrome virus in mainland China. J Gen Virol 2011; 92: 880-92.
[http://dx.doi.org/10.1099/vir.0.027995-0] , 15Kleiboeker SB, Schommer SK, Lee SM, et al. Simultaneous detection of North American and European porcine reproductive and respiratory syndrome virus using real-time quantitative reverse transcriptase-PCR. Vet Diagn Invest 2005; 17: 165-70.
[http://dx.doi.org/10.1177/104063870501700211] ]. Positive tissues were homogenized with DMEM, frozen/thawed three times, and centrifuged at 10,000 × g for 15 min. Supernatant was passed through a 0.22-μm filter and inoculated on primary porcine alveolar macrophages (PAMs) and Marc-145 cells for virus isolation, as previously described [16Kim HS, Kwang J, Yoon IJ, et al. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch Virol 1993; 133: 477-83.
[http://dx.doi.org/10.1007/BF01313785] , 17Zeman D, Neiger R, Yaeger M, et al. Laboratory investigation of PRRS virus infection in three swine herds. J Vet Diagn Invest 1993; 5(4): 522-8.
[http://dx.doi.org/10.1177/104063879300500404] ]. Inoculated cells were maintained at 37 °C in a 5% CO2 atmosphere and monitored daily for cytopathic effects.
RNA Extraction and Genome Sequencing
Total RNA was extracted from cell cultures by using an RNeasy Mini kit (Qiagen, Germany) according to the manufacturer’s instructions. Whole genomes were sequenced as described previously [18Han W, Wu JJ, Deng XY, et al. Molecular mutations associated with the in vitro passage of virulent porcine reproductive and respiratory syndrome virus. Virus Genes 2009; 38: 276-84.
[http://dx.doi.org/10.1007/s11262-008-0322-1] ]. Briefly, 16 overlapping fragments covering the whole viral genome were amplified by RT-PCR using the corresponding primers. The PCR products were purified with the E.Z.N.A. Gel Extraction Kit (OMEGA, USA) and cloned into pEASY-T1 (Transgen, China). Recombinant plasmids were sequenced with an ABI Automatic DNA Sequencer (Invitrogen, China) and spliced artificially. Gene fragments were sequenced in triplicate and assembled with ContigExpress in Vector NTI Advance 11 [19Lu G, Moriyama EN. Vector NTI, a balanced all-in-one sequence analysis suite. Brief Bioinform 2004; 5(4): 378-88.
[http://dx.doi.org/10.1093/bib/5.4.378] ].
Multiple Sequence Alignment and Phylogenetic Analysis
Multiple alignments were performed for each ORF separately using the represented available whole genome sequences for Type 2 PRRSV, including VR-2332, CH-1a, JXA1, MN184A, NADC30, HENAN-XINX, JI580, HEN-HB, and CHsx1401. Using the MEGA4.0 program [20Tamura K, Dudley J, Nei M, et al. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 2007; 24(8): 1596-9.
[http://dx.doi.org/10.1093/molbev/msm092] ], phylogenetic trees were constructed from aligned nucleotide sequences by the neighbor-joining method. Trees were subjected to bootstrap analysis with 1,000 replicates to determine the reliability percentages at each internal node of the tree. Whole genome sequences of all PRRSVs mentioned in this study are available in GenBank (Table 1).
RESULTS
Virus Isolation and Complete Genome Sequence Analysis of Two NADC30-like Isolates
Following sample inoculation on primary PAM and Marc-145 cells for 3 days, cytopathic effects were observed on PAMs but not on Marc-145 cells for two inoculates and on both cell types for four inoculates. These six PRRSV inoculates were sequenced. Two isolates, NVDC-ZJ and NVDC-JS, exhibited higher homology with NADC30 than other PRRSV Type 2 isolates and were included in subsequent analyses. Excluding the poly (A) tails, complete genomes of NVDC-ZJ and NVDC-JS were 15,016 nucleotides in length. These two isolates shared 99.9% homology with each other, 95.4% homology with NADC30, 85.3% homology with VR2332 (representative Type 2 PRRSV), and 83.8% homology with JXA1 (representative highly pathogenic PRRSV).
Next, we analyzed the genomic characteristics of NVDC-ZJ in detail. Compared to other NADC30-like representative isolates, including HENAN-XINX, JL580, HEN-HB, CHsx1401 MN184A, and NADC30, the 5′-UTR and 3′-UTR of NVDC-ZJ had nucleotide homologies of 95.3–100% and 95.4–100%, respectively. Predicted nonstructural proteins encoded by ORF1a and ORF1b of NVDC-ZJ shared nucleotide homologies of 83.7–99.9% and 89.9–99.9%, respectively. Among all nonstructural proteins, the most variable protein within ORF1a, Nsp2, of NVDC-ZJ exhibited an amino acid identify of 67.0–99.9% with other isolates. Detailed identities for each coding region of NCDC-ZJ with other NADC30-like strains and representative North American isolates are summarized in (Table 1).
Sequencing Analysis of 5’-UTR and 3’-UTR of Two NADC30-like Isolates
The 5′-UTR and 3′-UTR are documented to be important regulatory elements in the PRRSV genome [7Snijder EJ, Meulenberg JJ. The molecular biology of arteriviruses. J Gen Virol 1998; 79: 961-79.
[http://dx.doi.org/10.1099/0022-1317-79-5-961] ]. In this study, we compared the 5′-UTR and 3′-UTR nucleotide sequences of these isolates and other PRRSV isolates. The 5′-UTRs of the two isolates were 190 nucleotides in length, with no new deletions or insertions in this region. However, the 3′-UTRs were 148 nucleotides in length and contained a 3-nucleotide deletion at position 118–120 Fig. (1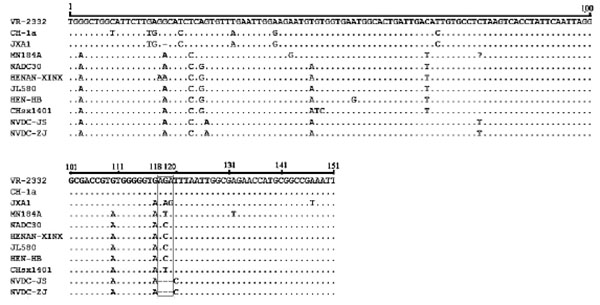 ).
).
 |
Fig. (1) Alignment of 3’-UTR sequences of two NADC30-like PRRSVs. Deleted region is indicated by the black box. |
Amino Acid Variation in Nsp2 of the Two NADC30-like Isolates
The most important and variable nonstructural protein of PRRSV, Nsp2 exhibited the lowest amino acid identity among the viral proteins. Compared to the classical PRRSV and HP-PRRSV, these two isolates exhibited discontinuous deletion of 131 amino acids within the Nsp2 protein, including an 111-aa deletion at position 322–432, 1-aa deletion at position 483, and 19-aa deletion at position 504–522 Fig. (2 ), which are similar to other NADC30-like strains. These discontinuous deletions were also observed in NADC30 and MN184 isolated in the United States [14Zhou L, Wang ZC, Ding YP, et al. NADC30-like strain of porcine reproductive and respiratory syndrome virus, china. Emerg Infect Dis 2015; 21(12): 2256-7.
), which are similar to other NADC30-like strains. These discontinuous deletions were also observed in NADC30 and MN184 isolated in the United States [14Zhou L, Wang ZC, Ding YP, et al. NADC30-like strain of porcine reproductive and respiratory syndrome virus, china. Emerg Infect Dis 2015; 21(12): 2256-7.
[http://dx.doi.org/10.3201/eid2112.150360] ].
Amino Acid Alteration in GP3 of the Two NADC30-like Isolates
GP3 is a minor structural protein that is important for assembly of infectious PRRSV. GP3 is highly antigenic and has two fewer conserved epitopes at amino acids 67–74 (67YEPGRSLW74) and 74–85 (74WCRIGHDRCGED85) [21Li B, Fang LR, Liu SY, et al. The genomic diversity of Chinese porcine reproductive and respiratory syndrome virus isolates from 1996 to 2009. Vet Microbiol 2010; 146: 226-37.
[http://dx.doi.org/10.1016/j.vetmic.2010.05.011] ]. To understand the variations of the two epitopes, we studied alignment of the amino acid sequences of the two isolates in this study. We identified a 1-aa substitution at location 67 (Y67→F67) in the minimal epitope 67–74 and a 3-aa substitution at positions 83 and 85 (Y83→H83, G83→E83, D85→S85) in epitope 74–85 Fig. (3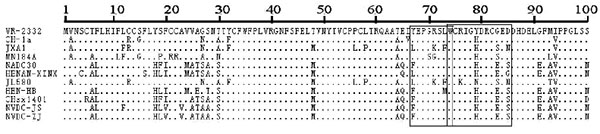 ).
).
Amino Acid Variation in GP5 of the Two NADC30-like Isolates
We further explored the genetic divergence in the two NADC30-like isolates of PRRSV. GP5, the most variable gene of PRRSV, was analyzed with other representative PRRSV Type 2 isolates. The GP5 amino acid sequence for each strain consisted of 200 residues with no deletions or insertions. Strains shared amino acid identities of 100% with each other, 92% with NADC30, 88% with MN184A, 95.4–96% with other NADC30-like strains, 85.5% with the representative HP-PRRSV JXA1 (isolated after 2006 in China), and 85.5% with VR-2332.
GP5 contains a signal peptide, three transmembrane proteins, an extravirion region, and an intravirion region [22Han J, Wang Y, Faaberg KS. Complete genome analysis of RFLP184 isolates of porcine reproductive and respiratory syndrome virus. Virus Res 2006; 122: 175-82.
[http://dx.doi.org/10.1016/j.virusres.2006.06.003] ]. The hypervariable region was observed in the signal peptide, extravirion region, and intravirion region. Compared to VR-2332, the signal peptide had four concurrent mutations at positions 3, 8, 14, and 16 (E3→G8, A8→V8, L14 →S14, and S16→F16) Fig. (4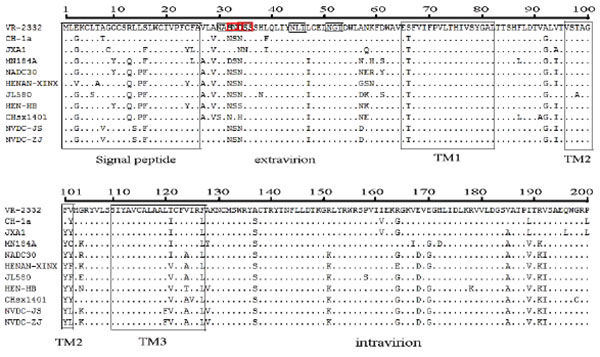 ). We analyzed some known functional domains in the extravirion region of GP5, such as the primary neutralizing epitope B, decoy epitope A, and potential glycosylation sites. Decoy epitope A, comprised of A27/V27LVN near the primary neutralizing epitope, may delude most antibodies against GP5 and delay production of viral neutralizing antibodies [23Ostrowski M, Galeota JA, Jar AM, et al. Identification of neutralizing and nonneutralizing epitopes in the porcine reproductive and respiratory syndrome virus GP5 ectodomain. J Virol 2002; 76: 4241-50.
). We analyzed some known functional domains in the extravirion region of GP5, such as the primary neutralizing epitope B, decoy epitope A, and potential glycosylation sites. Decoy epitope A, comprised of A27/V27LVN near the primary neutralizing epitope, may delude most antibodies against GP5 and delay production of viral neutralizing antibodies [23Ostrowski M, Galeota JA, Jar AM, et al. Identification of neutralizing and nonneutralizing epitopes in the porcine reproductive and respiratory syndrome virus GP5 ectodomain. J Virol 2002; 76: 4241-50.
[http://dx.doi.org/10.1128/JVI.76.9.4241-4250.2002] ]. We did not find any mutation of amino acids of epitope A. Primary neutralizing epitope B comprised amino acids at residues 37 to 44 (SHLQLIYN). This epitope is conserved and recognized by neutralizing monoclonal antibodies. No amino acid mutations of epitope B were found in either isolate Fig. (4 ).
).
There were at least three N-glycosylation sites (N-X-S/T) located in the extravirion region of GP5. The first potential N-glycosylation site was located in the hypervariable region (N32 or N33 or N34) of GP5 extravirion. The second and third potential N-glycosylation sites were located in residues N44 and N51. There were two concurrent mutations at positions 32 and 34 (S32→N32, D34→N34) within the two isolates Fig. (4 ). In the intravirion domain, there were nine concurrent amino acid mutations at positions 137(A→S), 151(R→K), 164(R→G), 168(E→D), 170(E→G), 185(V→A), 189(I→V), 191(R→K), and 192(V→I) Fig. (4
). In the intravirion domain, there were nine concurrent amino acid mutations at positions 137(A→S), 151(R→K), 164(R→G), 168(E→D), 170(E→G), 185(V→A), 189(I→V), 191(R→K), and 192(V→I) Fig. (4 ).
).
Phylogenetic Analyses of The Two NADC30-like Isolates with Other PRRSVs
Phylogenetic analysis was performed on the full-length genomic sequences of 17 PRRSV strains, including 14 Type 2 and one Type 1 strain from the GenBank database Table 1 and the two NADC30-like isolates in this study. In the phylogenetic tree, the two NADC30-like isolates formed a new branch within PRRSV Type 2 Subtype 2. They were closely related to CHsx1401 and NADC30 Fig. (5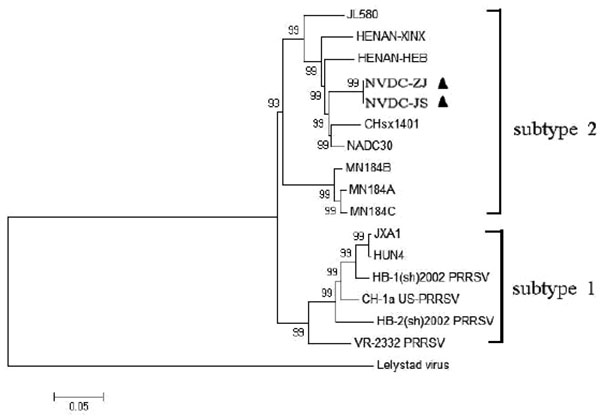 ).
).
DISCUSSION
PPRS is one of the most important infectious diseases in swine, causing reproductive failure in sows and boars and respiratory disease in pigs of all ages. North American PRRSV strains were first reported in China in 1996, followed by isolation of some epidemic strains in Chinese swine herds [12Guo B, Chen Z, Liu W, et al. Isolation and identification of porcine reproductive and respiratory syndrome (PRRS) virus from fetuses suspicious of PRRS. Chin J Anim Poult Infect Dis 1996; 17: 1-4.]. In 2006, unparalleled outbreaks of HP-PRRSV led to huge economic losses of pig husbandry and an extensive pandemic in China [24Tian KG, Yu XL, Zhao TZ, et al. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2007; (6): 1-10.]. NADC30-like isolates were first reported in Henan province in 2012 and then isolated from other provinces in China [14Zhou L, Wang ZC, Ding YP, et al. NADC30-like strain of porcine reproductive and respiratory syndrome virus, china. Emerg Infect Dis 2015; 21(12): 2256-7.
[http://dx.doi.org/10.3201/eid2112.150360] , 25Sun Z, Wang J, Bai XF, et al. Pathogenicity comparison between highly pathogenic and NADC30-like porcine reproductive and respiratory syndrome virus. Arch Virol 2016; 161(8): 2257-61.
[http://dx.doi.org/10.1007/s00705-016-2883-y] , 26Zhao K, Ye C, Chang XB, et al. Importation and recombination are responsible for the latest emergence of highly pathogenic porcine reproductive and respiratory syndrome virus in china. J Virol 2015; 89(20): 10712-6.
[http://dx.doi.org/10.1128/JVI.01446-15] ].
To explore the genetic divergence of NADC30-like strains in Chinese swine herds, we investigated the complete nucleotide sequences of two isolates and analyzed their phylogenetic relationships with other PRRSV strains. Nucleotide sequences of the two isolates were highly homologous with sequences of other NADC30-like strains, and highly variable regions existed in NSP1β, NSP2, GP3, and GP5. Phylogenetic analysis divided the 16 PRRSV Type 2 strains into two subtypes: subtype 1 contained classical PRRSV and HP-PRRSV, and subtype 2 contained MN184 PRRSV and other NADC30-like isolates (including the two studied here). Moreover, the strains were closely related to and may have evolved from NADC30 by gradual variation and accumulation of genomic changes.
The Nsp2 gene is highly variable and includes naturally occurring mutations, insertions, and deletions, which might be the most important marker for monitoring genetic variation and evolution of PRRSV [27Fang Y, Schneider P, Zhang WP, et al. Diversity and evolution of a newly emerged North American Type 1 porcine arterivirus: analysis of isolates collected between 1999 and 2004. Arch Virol 2007; 152: 1009-17.
[http://dx.doi.org/10.1007/s00705-007-0936-y] ]. In 2006, an atypical PRRS outbreak in China was caused by a highly pathogenic PRRSV strain that contained a unique discontinuous deletion of 30 amino acids in the Nsp2 hypervariable region [24Tian KG, Yu XL, Zhao TZ, et al. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2007; (6): 1-10.]. Our two studied isolates had a discontinuous deletion of 131 amino acids in the Nsp2 region, but the deletion differed from those of the HP-PRRSV strains and were more similar to those of the NADC30 strains. Some NADC30-like strains have HP-PRRSV virulence [14Zhou L, Wang ZC, Ding YP, et al. NADC30-like strain of porcine reproductive and respiratory syndrome virus, china. Emerg Infect Dis 2015; 21(12): 2256-7.
[http://dx.doi.org/10.3201/eid2112.150360] , 26Zhao K, Ye C, Chang XB, et al. Importation and recombination are responsible for the latest emergence of highly pathogenic porcine reproductive and respiratory syndrome virus in china. J Virol 2015; 89(20): 10712-6.
[http://dx.doi.org/10.1128/JVI.01446-15] ]. Pathogenic analysis indicated the two isolates in this study were milder in virulence than the HP-PRRSV strains (data not show). Therefore, we conclude that the 131-aa deletion was not related to the virulence of PRRSV in China. Other research has associated PRRSV virulence with multiple factors. This issue is a complex question that will require further study.
GP3 and GP5 are important targets for analyzing the virulence and genetic variation of PRRSV, which is involved in viral immune evasion and minimization of the virus-neutralizing antibody response [28Ansar IH, Kwon BJ, Osorio FA, et al. Influence of N-linked glycosylation of porcine reproductive and respiratory syndrome virus GP5 on virus infectivity, antigenicity, and ability to induce neutralizing antibodies. J Virol 2006; 80: 3994-4004.
[http://dx.doi.org/10.1128/JVI.80.8.3994-4004.2006] -30Mardassi H, Mounir S, Dea S. Molecular analysis of the ORFs 3 to 7 of porcine reproductive and respiratory syndrome virus, Quebec reference strain. Arch Virol 1995; 140: 1405-18.
[http://dx.doi.org/10.1007/BF01322667] ]. Compared to VR-2332, two epitopes of GP3 had four substitutions. Substitutions were observed in the signal peptide, extravirion, intravirion, and transmembrane regions. We also observed two potential N-glycosylation sites on GP5 in the two isolates. However, it is unknown whether these mutations or glycosylation sites are related to the virulence or immunogenicity changes of PRRSV. Nevertheless, our data indicate that the genetic diversity of PRRSV should be considered as a serious issue for PRRSV control and prevention.
CONCLUSION
We have detailed the genomic sequences of two NADC30-like strains of PRRSV and explored their phylogenetic relationships with other PRRSV strains. The two NADC30-like isolates of PRRSV were introduced to mainland China from North America, thereafter gradually accumulating genomic changes.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies that are base of this research.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors confirm that they have no conflicts of interest related to any content in this article.
ACKNOWLEDGEMENTS
This work was supported by grants from "The National Key Research and Development Program of China (2016YFD0500705)" Thank you.
REFERENCES
| [1] | Wensvoort G, Terpstra C. et al. Mystery swine disease in The Netherlands, the isolation of Lelystad virusVet Q 1991;13(3):121-130. |
| [2] | Meulenberg JJ, Hulst MM, de Meijer EJ, et al. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology 1993; 192: 62-72. [http://dx.doi.org/10.1006/viro.1993.1008] |
| [3] | Allende R, Lewis TL, Lu Z, et al. North American and European porcine reproductive and respiratory syndrome viruses differ in non-structuralprotein coding regions. J Gen Virol 1999; 80: 307-15. [http://dx.doi.org/10.1099/0022-1317-80-2-307] |
| [4] | Firth AE, Zevenhoven-Dobbe JC, Wills NM, et al. Discovery of a small arterivirus gene that overlaps the GP5 coding sequence and is important for virus production. J Gen Virol 2011; 92(Pt5): 1097-106. [http://dx.doi.org/10.1099/vir.0.029264-0] |
| [5] | Johnson CR, Griggs TF, Gnanandarajah J, et al. Novel structural protein in porcine reproductive and respiratory syndrome virus encoded by an alternative ORF5present in all arteriviruses. J Gen Virol 2011; 92: 1107-16. [http://dx.doi.org/10.1099/vir.0.030213-0] |
| [6] | Meulenberg JJ, Petersen-den Besten A, De Kluyver EP, et al. Characterization of proteins encoded by ORFs 2 to 7 of Lelystad virus. Virology 1995; 206: 155-63. [http://dx.doi.org/10.1016/S0042-6822(95)80030-1] |
| [7] | Snijder EJ, Meulenberg JJ. The molecular biology of arteriviruses. J Gen Virol 1998; 79: 961-79. [http://dx.doi.org/10.1099/0022-1317-79-5-961] |
| [8] | Oleksiewicz MB, Bøtner A, Madsen KG, et al. Sensitive detection and typing of porcine reproductive and respiratory syndrome virus by RT-PCR amplification of whole viral genes. Vet Microbiol 1998; 64(1): 7-22. [http://dx.doi.org/10.1016/S0378-1135(98)00254-5] |
| [9] | Madsen KG, Hansen CM, Madsen ES, et al. Sequence analysis of porcine reproductive and respiratory syndrome virus of the American type collected from Danish swine herds. Arch Virol 1998; 143(9): 1683-700. [http://dx.doi.org/10.1007/s007050050409] |
| [10] | Collins JE, Benfield DA, Christianson WT, et al. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J Vet Diagn Invest 1992; 4: 117-26. [http://dx.doi.org/10.1177/104063879200400201] |
| [11] | Balka G. Genetic diversity of porcine reproductive and respiratory syndrome virus strains circulating in Hungarian swine herds. Vet Microbiol 2008; 127: 128-35. [http://dx.doi.org/10.1016/j.vetmic.2007.08.001] |
| [12] | Guo B, Chen Z, Liu W, et al. Isolation and identification of porcine reproductive and respiratory syndrome (PRRS) virus from fetuses suspicious of PRRS. Chin J Anim Poult Infect Dis 1996; 17: 1-4. |
| [13] | Chen NH, Cao Z, Yu XL, et al. Emergence of novel European genotype porcine reproductive and respiratory syndrome virus in mainland China. J Gen Virol 2011; 92: 880-92. [http://dx.doi.org/10.1099/vir.0.027995-0] |
| [14] | Zhou L, Wang ZC, Ding YP, et al. NADC30-like strain of porcine reproductive and respiratory syndrome virus, china. Emerg Infect Dis 2015; 21(12): 2256-7. [http://dx.doi.org/10.3201/eid2112.150360] |
| [15] | Kleiboeker SB, Schommer SK, Lee SM, et al. Simultaneous detection of North American and European porcine reproductive and respiratory syndrome virus using real-time quantitative reverse transcriptase-PCR. Vet Diagn Invest 2005; 17: 165-70. [http://dx.doi.org/10.1177/104063870501700211] |
| [16] | Kim HS, Kwang J, Yoon IJ, et al. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch Virol 1993; 133: 477-83. [http://dx.doi.org/10.1007/BF01313785] |
| [17] | Zeman D, Neiger R, Yaeger M, et al. Laboratory investigation of PRRS virus infection in three swine herds. J Vet Diagn Invest 1993; 5(4): 522-8. [http://dx.doi.org/10.1177/104063879300500404] |
| [18] | Han W, Wu JJ, Deng XY, et al. Molecular mutations associated with the in vitro passage of virulent porcine reproductive and respiratory syndrome virus. Virus Genes 2009; 38: 276-84. [http://dx.doi.org/10.1007/s11262-008-0322-1] |
| [19] | Lu G, Moriyama EN. Vector NTI, a balanced all-in-one sequence analysis suite. Brief Bioinform 2004; 5(4): 378-88. [http://dx.doi.org/10.1093/bib/5.4.378] |
| [20] | Tamura K, Dudley J, Nei M, et al. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 2007; 24(8): 1596-9. [http://dx.doi.org/10.1093/molbev/msm092] |
| [21] | Li B, Fang LR, Liu SY, et al. The genomic diversity of Chinese porcine reproductive and respiratory syndrome virus isolates from 1996 to 2009. Vet Microbiol 2010; 146: 226-37. [http://dx.doi.org/10.1016/j.vetmic.2010.05.011] |
| [22] | Han J, Wang Y, Faaberg KS. Complete genome analysis of RFLP184 isolates of porcine reproductive and respiratory syndrome virus. Virus Res 2006; 122: 175-82. [http://dx.doi.org/10.1016/j.virusres.2006.06.003] |
| [23] | Ostrowski M, Galeota JA, Jar AM, et al. Identification of neutralizing and nonneutralizing epitopes in the porcine reproductive and respiratory syndrome virus GP5 ectodomain. J Virol 2002; 76: 4241-50. [http://dx.doi.org/10.1128/JVI.76.9.4241-4250.2002] |
| [24] | Tian KG, Yu XL, Zhao TZ, et al. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2007; (6): 1-10. |
| [25] | Sun Z, Wang J, Bai XF, et al. Pathogenicity comparison between highly pathogenic and NADC30-like porcine reproductive and respiratory syndrome virus. Arch Virol 2016; 161(8): 2257-61. [http://dx.doi.org/10.1007/s00705-016-2883-y] |
| [26] | Zhao K, Ye C, Chang XB, et al. Importation and recombination are responsible for the latest emergence of highly pathogenic porcine reproductive and respiratory syndrome virus in china. J Virol 2015; 89(20): 10712-6. [http://dx.doi.org/10.1128/JVI.01446-15] |
| [27] | Fang Y, Schneider P, Zhang WP, et al. Diversity and evolution of a newly emerged North American Type 1 porcine arterivirus: analysis of isolates collected between 1999 and 2004. Arch Virol 2007; 152: 1009-17. [http://dx.doi.org/10.1007/s00705-007-0936-y] |
| [28] | Ansar IH, Kwon BJ, Osorio FA, et al. Influence of N-linked glycosylation of porcine reproductive and respiratory syndrome virus GP5 on virus infectivity, antigenicity, and ability to induce neutralizing antibodies. J Virol 2006; 80: 3994-4004. [http://dx.doi.org/10.1128/JVI.80.8.3994-4004.2006] |
| [29] | Cancel-Tirado SM, Evans RB, Yoon KJ. Monoclonal antibody analysis of porcine reproductive and respiratory syndrome virus epitopes associated with antibody-dependent enhancement and neutralization of virus infection. Veterinary Immunology and Immunopathology 2004; 102(3): 249-62. [http://dx.doi.org/10.1016/j.vetimm.2004.09.017] |
| [30] | Mardassi H, Mounir S, Dea S. Molecular analysis of the ORFs 3 to 7 of porcine reproductive and respiratory syndrome virus, Quebec reference strain. Arch Virol 1995; 140: 1405-18. [http://dx.doi.org/10.1007/BF01322667] |