- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Epidemiologic Status of Picobirnavirus in India, A Less Explored Viral Disease
Yashpal Singh Malik1, *, Shubhankar Sircar1, Sharad Saurabh1, Jobin Jose Kattoor1, Rashmi Singh2, Balasubramanian Ganesh3, Souvik Ghosh4, Kuldeep Dhama1, Raj Kumar Singh1
Abstract
Since the unexpected discovery of picobirnaviruses (PBV) in 1988, they have been reported in many animals including mammals and birds, which comprises both terrestrial and marine species. Due to their divergent characteristics to other viral taxa they are classified into a new family Picobirnaviridae. Although their pathogenicity and role in causing diarrhea still remains a question since they have been discovered in symptomatic and asymptomatic cases both. Recent studies employing state-of-art molecular tools have described their presence in various clinical samples, like stool samples from different mammals and birds, respiratory tracts of pigs and humans, sewage water, different foods, etc. Furthermore, their epidemiological status from different parts of the world in different hosts has also increased. Due to their diverse host and irregular host pattern their role in causing diarrhea remains alien. The heterogeneity nature can be ascribed to segmented genome of PBV, which renders them prone to continuous reassortment. Studies have been hampered on PBVs due to their non-adaptability to cell culture system. Here, we describe the molecular epidemiological data on PBVs in India and discusses the overall status of surveillance studies carried out till date in India.
Article Information
Identifiers and Pagination:
Year: 2018Volume: 12
Issue: Suppl-2, M6
First Page: 99
Last Page: 109
Publisher Id: TOVJ-12-99
DOI: 10.2174/1874357901812010099
Article History:
Received Date: 7/7/2017Revision Received Date: 14/3/2018
Acceptance Date: 15/5/2018
Electronic publication date: 31/08/2018
Collection year: 2018
open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution 4.0 International Public License (CC-BY 4.0), a copy of which is available at: https://creativecommons.org/licenses/by/4.0/legalcode. This license permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
* Address correspondence to this author at the ICAR-Indian Veterinary Research Institute, Izatnagar, Bareilly 243122, Uttar Pradesh, India; Tel: 7500777999; E-mail: malikyps@gmail.com
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 7-7-2017 |
Original Manuscript | Epidemiologic Status of Picobirnavirus in India, A Less Explored Viral Disease | |
1. Introduction
Picobirnavirus (PBV) is connected with acute gastroenteritis cases in humans as well as animals. Although they were identified as birna-like viruses in 1988, its pathobiology is still uncertain. Further, viruses of this group remained in unclaimed virus family until 2009 when PBVs were recognized distinctly as members of the family Picobirnaviridae possessing double stranded bi-segmented RNA genome with human picobirnavirus as prototype species. Its genome is approximately 4.2 kbp, where the large segment, also known as genomic segment 1, is of 2.2-2.7 kbp and encodes the capsid protein, while another segment, also known as genomic segment 2, is of 1.2-1.9 kbp size and encodes the RNA dependent RNA polymerase (RdRp). The virion is non-enveloped, small, spherical, 33-41 nm in diameter and consists of a simple core capsid with a distinctive icosahedral arrangement [1Malik YS, Kumar N, Sharma K, et al. Epidemiology, phylogeny, and evolution of emerging enteric Picobirnaviruses of animal origin and their relationship to human strains 2014.].
Information gathered as of now on the epidemiology of PBVs rely primarily on the detection of viral dsRNA by Polyacrylamide Gel Electrophoresis (PAGE), electron microscopy or reverse transcription PCR (RT-PCR) detection assay based on two pairs of primers targeting the genomic segment 2 of PBV strains viz.; “4-GA-91” and “1-CHN-97” isolated in the USA and China, respectively [2Rosen BI, Fang Z-Y, Glass RI, Monroe SS. Cloning of human picobirnavirus genomic segments and development of an RT-PCR detection assay. Virology 2000; 277(2): 316-29.
[http://dx.doi.org/10.1006/viro.2000.0594] [PMID: 11080479] ]. PBVs are classified into genogroup I (prototype strain: GI/PBV/human/China/1-CHN-97/1997) and II (prototype strain: GII/PBV/human/USA/4-GA-91/1991). Based on the migration pattern of the bi-segmented dsRNA, PBV appears either with large genome profile or small genome profile. Likewise, based on differences in the genomic sequences of segment 2, PBV is further classified into genogroups I, II, non-I and non-II. Hitherto reports indicate the presence of divergent PBVs pointing towards the emergence of putative new genogroups in humans as well as animals [3Smits SL, Schapendonk CM, van Beek J, et al. New viruses in idiopathic human diarrhea cases, the Netherlands. Emerg Infect Dis 2014; 20(7): 1218-22.
[http://dx.doi.org/10.3201/eid2007.140190] [PMID: 24964003] , 4Li L, Giannitti F, Low J, et al. Exploring the virome of diseased horses. J Gen Virol 2015; 96(9): 2721-33.
[http://dx.doi.org/10.1099/vir.0.000199] [PMID: 26044792] ]. Culture of PBVs in vitro has not yielded any positive signal yet and there exit no animal model to study the pathobiology of virus infection, as well. Of the note, PBV has not been established yet as an etiological agent of diarrhea. Rather these are more often isolated as co-infecting agents with a number of diarrheal causes indicating their role in synergistic effect in association with the primary enteric cause. PBVs have been shown to cause gastroenteritis in immune-compromised individuals suggesting that immune status of the host plays an important role in establishing PBV infection as an opportunistic pathogen [5Giordano MO, Martinez LC, Rinaldi D, et al. Diarrhea and enteric emerging viruses in HIV-infected patients. AIDS Res Hum Retroviruses 1999; 15(16): 1427-32.
[http://dx.doi.org/10.1089/088922299309937] [PMID: 10555105] -8Giordano MO, Martinez LC, Rinaldi D, et al. Detection of picobirnavirus in HIV-infected patients with diarrhea in Argentina. J Acquir Immune Defic Syndr Hum Retrovirol 1998; 18(4): 380-3.
[http://dx.doi.org/10.1097/00042560-199808010-00010] [PMID: 9704944] ].
1.1. Epidemiology of Picobirnavirus
Picobirnaviruses were initially discovered by the finding of two nucleic acid segments through PAGE during investigations of acute gastroenteritis in children and from feces of black-footed pigmy rice rats [9Pereira HG, Fialho AM, Flewett TH, Teixeira JM, Andrade ZP. Novel viruses in human faeces. Lancet 1988; 2(8602): 103-4.
[http://dx.doi.org/10.1016/S0140-6736(88)90032-3] [PMID: 2898672] , 10Pereira HG, Flewett TH, Candeias JA, Barth OM. A virus with a bisegmented double-stranded RNA genome in rat (Oryzomys nigripes) intestines. J Gen Virol 1988; 69(Pt 11): 2749-54.
[http://dx.doi.org/10.1099/0022-1317-69-11-2749] [PMID: 3053986] ]. The virus has subsequently been detected in faecal samples from domestic and wild animal species, including rats, giant anteaters, guinea pigs, rabbits, chickens, pigs, calves, foals, cat and dogs [4Li L, Giannitti F, Low J, et al. Exploring the virome of diseased horses. J Gen Virol 2015; 96(9): 2721-33.
[http://dx.doi.org/10.1099/vir.0.000199] [PMID: 26044792] , 9Pereira HG, Fialho AM, Flewett TH, Teixeira JM, Andrade ZP. Novel viruses in human faeces. Lancet 1988; 2(8602): 103-4.
[http://dx.doi.org/10.1016/S0140-6736(88)90032-3] [PMID: 2898672] -34Volotäo EM, Soares CC, Albuquerque MC, et al. First evidence of a trisegmented double-stranded RNA virus in canine faeces. Vet J 2001; 161(2): 205-7.
[http://dx.doi.org/10.1053/tvjl.2000.0532] [PMID: 11243690] ]. Epidemiological studies on PBVs in India are still in its early stages, with limited reports only. Here, we converse with the epidemiological studies carried out as on date in India on PBVs and present the current knowledge of PBVs in different hosts emphasizing the Indian perspective.
1.2. Human Picobirnaviruses
The actual journey of PBV discovery started in 1988, when Pereira and colleagues described a novel virus in human faeces serendipitously [9Pereira HG, Fialho AM, Flewett TH, Teixeira JM, Andrade ZP. Novel viruses in human faeces. Lancet 1988; 2(8602): 103-4.
[http://dx.doi.org/10.1016/S0140-6736(88)90032-3] [PMID: 2898672] ]. Subsequently, in 1995 PBVs were detected as co-infecting agents with Cryptosporidium in human faeces [15Gallimore CI, Green J, Casemore DP, Brown DW. Detection of a picobirnavirus associated with Cryptosporidium positive stools from humans. Arch Virol 1995; 140(7): 1275-8.
[http://dx.doi.org/10.1007/BF01322752] [PMID: 7646357] ] and the virus was characterized by two extraction methods i.e. phenol/chloroform and guanidinium thiocyanate (GTC)/silica method followed by electron microscopy to know its physical properties [35Gallimore CI, Appleton H, Lewis D, Green J, Brown DW. Detection and characterisation of bisegmented double-stranded RNA viruses (picobirnaviruses) in human faecal specimens. J Med Virol 1995; 45(2): 135-40.
[http://dx.doi.org/10.1002/jmv.1890450204] [PMID: 7775930] ]. In 2000, two prototype strains for GG-I (1-CHN-97) and GG-II (4-GA-91) were cloned and sequenced, obtained from human HIV infected patient and non-HIV infected patient from China and USA respectively [2Rosen BI, Fang Z-Y, Glass RI, Monroe SS. Cloning of human picobirnavirus genomic segments and development of an RT-PCR detection assay. Virology 2000; 277(2): 316-29.
[http://dx.doi.org/10.1006/viro.2000.0594] [PMID: 11080479] ]. Their presence has been reported in human faeces from Argentina [5Giordano MO, Martinez LC, Rinaldi D, et al. Diarrhea and enteric emerging viruses in HIV-infected patients. AIDS Res Hum Retroviruses 1999; 15(16): 1427-32.
[http://dx.doi.org/10.1089/088922299309937] [PMID: 10555105] , 8Giordano MO, Martinez LC, Rinaldi D, et al. Detection of picobirnavirus in HIV-infected patients with diarrhea in Argentina. J Acquir Immune Defic Syndr Hum Retrovirol 1998; 18(4): 380-3.
[http://dx.doi.org/10.1097/00042560-199808010-00010] [PMID: 9704944] , 36Martínez LC, Giordano MO, Isa MB, et al. Molecular diversity of partial-length genomic segment 2 of human picobirnavirus. Intervirology 2003; 46(4): 207-13.
[http://dx.doi.org/10.1159/000072429] [PMID: 12931028] , 37Giordano MO, Masachessi G, Martinez LC, et al. Two instances of large genome profile picobirnavirus occurrence in Argentinian infants with diarrhea over a 26-year period (1977-2002). J Infect 2008; 56(5): 371-5.
[http://dx.doi.org/10.1016/j.jinf.2008.02.017] [PMID: 18403022] ], Brazil [38Barreto ML, Milroy CA, Strina A, et al. Community-based monitoring of diarrhea in urban Brazilian children: incidence and associated pathogens. Trans R Soc Trop Med Hyg 2006; 100(3): 234-42.
[http://dx.doi.org/10.1016/j.trstmh.2005.03.010] [PMID: 16303156] ], USA [7Grohmann GS, Glass RI, Pereira HG, et al. Enteric viruses and diarrhea in HIV-infected patients. N Engl J Med 1993; 329(1): 14-20.
[http://dx.doi.org/10.1056/NEJM199307013290103] [PMID: 8099429] , 39Cunliffe NA, Glass RI. Gastrointestinal manifestations of HIV infection. Lancet 1996; 348(9033): 1037.
[http://dx.doi.org/10.1016/S0140-6736(05)64970-7] [PMID: 8855890] , 40Zhang T, Breitbart M, Lee WH, et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol 2006; 4(1): e3.
[http://dx.doi.org/10.1371/journal.pbio.0040003] [PMID: 16336043] ], Venezuela [41Ludert JE, Liprandi F. Identification of viruses with bi- and trisegmented double-stranded RNA genome in faeces of children with gastroenteritis. Res Virol 1993; 144(3): 219-24.
[http://dx.doi.org/10.1016/S0923-2516(06)80032-4] [PMID: 8356343] , 42González GG, Pujol FH, Liprandi F, Deibis L, Ludert JE. Prevalence of enteric viruses in human immunodeficiency virus seropositive patients in Venezuela. J Med Virol 1998; 55(4): 288-92.
[http://dx.doi.org/10.1002/(SICI)1096-9071(199808)55:4<288::AID-JMV6>3.0.CO;2-X] [PMID: 9661837] ], Hungary [43Bányai K, Jakab F, Reuter G, et al. Sequence heterogeneity among human picobirnaviruses detected in a gastroenteritis outbreak. Arch Virol 2003; 148(12): 2281-91.
[http://dx.doi.org/10.1007/s00705-003-0200-z] [PMID: 14648286] ], Italy [44Cascio A, Bosco M, Vizzi E, Giammanco A, Ferraro D, Arista S. Identification of picobirnavirus from faeces of Italian children suffering from acute diarrhea. Eur J Epidemiol 1996; 12(5): 545-7.
[http://dx.doi.org/10.1007/BF00144011] [PMID: 8905320] ], Netherlands [45van Leeuwen M, Williams MM, Koraka P, Simon JH, Smits SL, Osterhaus AD. Human picobirnaviruses identified by molecular screening of diarrhea samples. J Clin Microbiol 2010; 48(5): 1787-94.
[http://dx.doi.org/10.1128/JCM.02452-09] [PMID: 20335418] ], Russia [46Novikova NA, Epifanova NV, Fedorova OF, Golitsyna LN, Kupriianova NV. [Detection of picobirnaviruses by electrophoresis of RNA in polyacrylamide gel]. Vopr Virusol 2003; 48(6): 41-3.
[PMID: 14708231] ], Thailand [47Wakuda M, Pongsuwanna Y, Taniguchi K. Complete nucleotide sequences of two RNA segments of human picobirnavirus. J Virol Methods 2005; 126(1-2): 165-9.
[http://dx.doi.org/10.1016/j.jviromet.2005.02.010] [PMID: 15847933] ] and Australia [48Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog 2008; 4(2): e1000011.
[http://dx.doi.org/10.1371/journal.ppat.1000011] [PMID: 18398449] ] that advocates circulation of the virus in several developed and developing countries of the world.
The available reports from India on PBVs in humans specify one geographical location i.e. Kolkata [49Bhattacharya R, Sahoo GC, Nayak MK, et al. Molecular epidemiology of human picobirnaviruses among children of a slum community in Kolkata, India. Infect Genet Evol 2006; 6(6): 453-8.
[http://dx.doi.org/10.1016/j.meegid.2006.02.005] [PMID: 16616879] ], the eastern part of the country. It was first reported in faecal specimens of children from a slum community [49Bhattacharya R, Sahoo GC, Nayak MK, et al. Molecular epidemiology of human picobirnaviruses among children of a slum community in Kolkata, India. Infect Genet Evol 2006; 6(6): 453-8.
[http://dx.doi.org/10.1016/j.meegid.2006.02.005] [PMID: 16616879] ], where out of the 56 diarrheic and 607 non-diarrheic samples screened, around 3.57% and 1.64% samples were found positive for PBV, respectively. Among the positive faecal specimens, both large (n=11) and small (n=1) genome pattern were observed in the PAGE analysis. Subsequently, from the same region, sporadic occurrence of this virus in humans was reported from 1999 to 2003 with detection of 21 (2.47%) positive out of 850 tested [50Bhattacharya R, Sahoo GC, Nayak MK, et al. Detection of Genogroup I and II human picobirnaviruses showing small genomic RNA profile causing acute watery diarrhoea among children in Kolkata, India. Infect Genet Evol 2007; 7(2): 229-38.
[http://dx.doi.org/10.1016/j.meegid.2006.09.005] [PMID: 17049316] ].All the positive samples were mostly taken from children which were showing acute watery diarrhea. These two reports showed the presence of both short genome profile as well as large genome profile PBV strains in India. Additionally, both reports showed diversity in human PBVs. In 2007, a report confirmed PBV infection in 2.06% of samples collected from children aged below 5 years [51Ganesh B, Nataraju SM, Rajendran K, et al. Detection of closely related Picobirnaviruses among diarrhoeic children in Kolkata: evidence of zoonoses? Infect Genet Evol 2010; 10(4): 511-6.
[http://dx.doi.org/10.1016/j.meegid.2010.02.008] [PMID: 20178864] ]. Majority of the PBVs however, showed large genomic profile (n=22) as compared to the virus with small profile (n=1). These diverse sequence heterogeneity among the human PBVs belongto both GG-I and GG-II.
On molecular characterization of PBV positive samples from India in different studies, it was evident that PBV genogroup I is predominant in humans. Initial report of PBV in India, confirmed that genogroup I was more in asymptomatic (n=4) and symptomatic (n=1) cases and genogroup II was only diarrheic case [49Bhattacharya R, Sahoo GC, Nayak MK, et al. Molecular epidemiology of human picobirnaviruses among children of a slum community in Kolkata, India. Infect Genet Evol 2006; 6(6): 453-8.
[http://dx.doi.org/10.1016/j.meegid.2006.02.005] [PMID: 16616879] ]. Whereas, the subsequent study showed almost equal distribution of genogroup I (n=6) and genogroup II (n=5) but the study performed in 2007 showed higher prevalence of genogroup I (13/23) than genogroup II [50Bhattacharya R, Sahoo GC, Nayak MK, et al. Detection of Genogroup I and II human picobirnaviruses showing small genomic RNA profile causing acute watery diarrhoea among children in Kolkata, India. Infect Genet Evol 2007; 7(2): 229-38.
[http://dx.doi.org/10.1016/j.meegid.2006.09.005] [PMID: 17049316] ]. Although the presence of genogroup I as mixed infection was also reported from a diarrheic sample PBV/Human/INDIA/GPBV6/2007, in which 4 strains were from genogroup I and one strain from genogroup II was observed, suggesting intricate genetic reassortment and recombination episodes within the human PBV population [52Ganesh B, Nagashima S, Ghosh S, et al. Detection and molecular characterization of multiple strains of Picobirnavirus causing mixed infection in a diarrhoeic child: Emergence of prototype Genogroup II-like strain in Kolkata, India. Int J Mol Epidemiol Genet 2011; 2(1): 61-72.
[PMID: 21537403] ]. Further, based on the sequence analysis and phylogenetic studies of several human PBV of genogroup I origin, it was found that they are closely related to the genogroup I strains of porcine origin which were earlier reported from Hungary, Venezuela and Argentina. This report also pointed towards the possibility of viral zoonoses from different animal species to human [52Ganesh B, Nagashima S, Ghosh S, et al. Detection and molecular characterization of multiple strains of Picobirnavirus causing mixed infection in a diarrhoeic child: Emergence of prototype Genogroup II-like strain in Kolkata, India. Int J Mol Epidemiol Genet 2011; 2(1): 61-72.
[PMID: 21537403] ]. The presence of PBV as co-infection was also reported in the Indian studies discussed above, where PBV was associated with rotavirus (n = 3) or astrovirus (n = 3) or both (n=1) [49Bhattacharya R, Sahoo GC, Nayak MK, et al. Molecular epidemiology of human picobirnaviruses among children of a slum community in Kolkata, India. Infect Genet Evol 2006; 6(6): 453-8.
[http://dx.doi.org/10.1016/j.meegid.2006.02.005] [PMID: 16616879] , 50Bhattacharya R, Sahoo GC, Nayak MK, et al. Detection of Genogroup I and II human picobirnaviruses showing small genomic RNA profile causing acute watery diarrhoea among children in Kolkata, India. Infect Genet Evol 2007; 7(2): 229-38.
[http://dx.doi.org/10.1016/j.meegid.2006.09.005] [PMID: 17049316] ] and Salmonella spp. (n = 1) [50Bhattacharya R, Sahoo GC, Nayak MK, et al. Detection of Genogroup I and II human picobirnaviruses showing small genomic RNA profile causing acute watery diarrhoea among children in Kolkata, India. Infect Genet Evol 2007; 7(2): 229-38.
[http://dx.doi.org/10.1016/j.meegid.2006.09.005] [PMID: 17049316] ]. However, no co-infection with norovirus, sapovirus or adenovirus or with parasites (Cryptosporidium spp., Giardia spp., Entamoeba spp., helminths) or bacteria (Vibrio spp., Shigella spp., Escherichia coli) was seen.
1.3. Bovine Picobirnaviruses
Though first report of PBV infection in bovines emerged from Russia in the year 1989 [22Vanopdenbosch E, Wellemans G. Birna-type virus in diarrhoeic calf faeces. Vet Rec 1989; 125(24): 610.
[PMID: 2609497] ], reports on their prevalence remained obscured for a long time. PBVs were found in neonatal calves as sub-clinical infections along with Cryptosporidium parvum [53Villacorta I, Peeters JE, Vanopdenbosch E, Ares-Mazás E, Theys H. Efficacy of halofuginone lactate against Cryptosporidium parvum in calves. Antimicrob Agents Chemother 1991; 35(2): 283-7.
[http://dx.doi.org/10.1128/AAC.35.2.283] [PMID: 2024962] ]. In the year 2003, Buzinaro and colleagues while determining the incidence of group A rotavirus (RVA) in dairy herds of Sao Paulo Brazil came to know the existence of this bi-segmented virus in four samples [23Buzinaro MG, Freitas PP, Kisiellius JJ, Ueda M, Jerez JA. Identification of a bisegmented double-stranded RNA virus (picobirnavirus) in calf faeces. Vet J 2003; 166(2): 185-7.
[http://dx.doi.org/10.1016/S1090-0233(03)00031-5] [PMID: 12902184] ]. In 2003, Novikova and co-workers discovered this virus from faeces of a calf along with 3 cases of diarrhea in children in Russia [46Novikova NA, Epifanova NV, Fedorova OF, Golitsyna LN, Kupriianova NV. [Detection of picobirnaviruses by electrophoresis of RNA in polyacrylamide gel]. Vopr Virusol 2003; 48(6): 41-3.
[PMID: 14708231] ]. A recent study in the year 2016 in Brazil reported small genome profile and the first sequence report from the American continent in bovine host [29Takiuchi E, Macedo R, Kunz AF, et al. Electrophoretic RNA genomic Profiles of Brazilian Picobirnavirus (PBV) strains and molecular characterization of a PBV isolated from diarrheic calf. Virus Res 2016; 211: 58-63.
[http://dx.doi.org/10.1016/j.virusres.2015.09.022] [PMID: 26435337] ].
The first detection of bovine PBV from India is attributed to the unanticipated discovery of this bi-segmented dsRNA virus strain, RUBV-P, in a one month old diarrheic calf. Molecular characterization of the strain RUBV-P for its full length gene segment 2 revealed distant genetic relatedness to other genogroup I PBVs of other animal species. Due to lack of sequence availability it was not possible to assess the genetic heterogeneity among bovine PBVs [24Ghosh S, Kobayashi N, Nagashima S, Naik TN. Molecular characterization of full-length genomic segment 2 of a bovine picobirnavirus (PBV) strain: evidence for high genetic diversity with genogroup I PBVs. J Gen Virol 2009; 90(Pt 10): 2519-24.
[http://dx.doi.org/10.1099/vir.0.013987-0] [PMID: 19587136] ]. Subsequently, in one of our study carried out during 2007-2010 from subtropical (central India) and western Himalayan area of Uttarakhand, 136 faecal samples from buffalo (n=122) and cow calves (n=14) were screened for PBV. Among the five PBV samples identified, three were from buffalo calves and one from cow calf exhibiting acute diarrhea, while one sample was from a non-diarrheic buffalo calf. This report of PBV infection in diarrheic calves was based on RNA-PAGE analysis therefore sequence based assessment cannot be established with these samples [25Malik YS, Chandrashekar KM, Sharma K, et al. Picobirnavirus detection in bovine and buffalo calves from foothills of Himalaya and Central India. Trop Anim Health Prod 2011; 43(8): 1475-8.
[http://dx.doi.org/10.1007/s11250-011-9834-0] [PMID: 21479844] ]. Later on, we detected a buffalo PBV isolate from Maharashtra, India (PBV-B18) of genogroup I exhibiting very less homology of 44.5% and 45.1% at nucleotide and amino acid levels, respectively, with the prototype human PBV strain of GG I (1-CHN-97/AF246939). To note, this buffalo PBV isolate (B18) showed less homology with the previously reported bovine PBV strain from India (RUBV-P/GG-I/2005/GQ221268) suggesting this unique PBV isolate may represent an emerging heterogeneous group of virus with a new lineage [26Malik YS, Kumar N, Sharma K, et al. Molecular characterization of a genetically diverse bubaline picobirnavirus strain, India. Wetchasan Sattawaphaet 2013; 43: 609-13.]. In 2013, Mondal and co-workers also reported the presence of PBV infection in diarrheic buffalo and cattle calves along with co-infection of rotavirus though RNA-PAGE analysis [27Mondal A, Chakravarti S, Majee SB, Bannalikar AS. Detection of picobirnavirus and rotavirus in diarrhoeic faecal samples of cattle and buffalo calves in Mumbai metropolis, Western India. Vet Ital 2013; 49(4): 357-60.
[PMID: 24362776] ]. The same author described the presence of GG-I type PBV in diarrheic calves from West Bengal, India [28Mondal A, Joardar SN. Detection of Genogroup I Picobirnavirus in Diarrhoeic Faecal Samples of Calves from West Bengal, India. J Immunol Immunopathol 2014; 16: 40-3.
[http://dx.doi.org/10.5958/0973-9149.2014.01076.4] ]. These reports of PBV infection in diarrheic buffalo calves expanded our knowledge regarding the host range and their geographical distribution. In March 2012, a surveillance study on enteric viruses reported a unique bovine PBV isolate (HP-7) from diarrheic calves which carried dual infection of GG-I and GG-II genogroup [54Malik YS, Sharma AK, Kumar N, Sharma K, Ganesh B, Kobayashi N. Identification and characterisation of a novel genogroup II picobirnavirus in a calf in India. Vet Rec 2014; 174(11): 278.
[http://dx.doi.org/10.1136/vr.102065] [PMID: 24570405] ]. Upon sequence analysis, it was found that HP-7-I was closer to Hungarian human PBV (Accession number AJ504796) and Brazilian canine PBV (Accession number FJ164032) isolates respectively. Whereas, HP-7-II was alike Indian human PBV (Accession number AB517738). This report supported the fact of finding frequent mixed infection of PBVs comprising both genotypes in humans and porcine population [52Ganesh B, Nagashima S, Ghosh S, et al. Detection and molecular characterization of multiple strains of Picobirnavirus causing mixed infection in a diarrhoeic child: Emergence of prototype Genogroup II-like strain in Kolkata, India. Int J Mol Epidemiol Genet 2011; 2(1): 61-72.
[PMID: 21537403] , 55Smits SL, Poon LL, van Leeuwen M, et al. Genogroup I and II picobirnaviruses in respiratory tracts of pigs. Emerg Infect Dis 2011; 17(12): 2328-30.
[http://dx.doi.org/10.3201/eid1712.110934] [PMID: 22172405] ].
1.4. Porcine Picobirnaviruses
The earliest report of PBV infection in pigs dates back in 1989 when this dsRNA virus was discovered by polyacrylamide gel electrophoresis in Brazil [17Gatti MS, de Castro AF, Ferraz MM, Fialho AM, Pereira HG. Viruses with bisegmented double-stranded RNA in pig faeces. Res Vet Sci 1989; 47(3): 397-8.
[PMID: 2687991] ]. Bi-segmented banding pattern confirmed the presence of PBV infection which was also observed during the investigation of intestinal content from human, pigmy rice rats and guinea pig. Consecutively, in successive years detection of this bi-segmented pathogen was also observed in different diagnostic labs of UK [18Chasey D. Porcine picobirnavirus in UK? Vet Rec 1990; 126(18): 465.
[PMID: 2356603] ] followed by various Latin American studies on porcine PBV in Venezuela [19Ludert JE, Hidalgo M, Gil F, Liprandi F. Identification in porcine faeces of a novel virus with a bisegmented double stranded RNA genome. Arch Virol 1991; 117(1-2): 97-107.
[http://dx.doi.org/10.1007/BF01310495] [PMID: 2006903] , 56Carruyo GM, Mateu G, Martínez LC, et al. Molecular characterization of porcine picobirnaviruses and development of a specific reverse transcription-PCR assay. J Clin Microbiol 2008; 46(7): 2402-5.
[http://dx.doi.org/10.1128/JCM.00655-08] [PMID: 18508933] , 57Núñez GMC, Aristiguieta ACA, Fraire FL, Leon JEL. Porcine picobirnavirus infection in Venezuelan farms 2014; 24], Brazil [58Alfieri AA, Alfieri AF, de Freitas JC, et al. Ocurrence of Escherichia Coli, rotavirus, picobirnavirus and Cryptosporidium Parvum in a postweaning diarrhoea focus in swine. Semin Cienc Agrar 1994; 15: 5-7.], Argentina [59Martínez LC, Masachessi G, Carruyo G, et al. Picobirnavirus causes persistent infection in pigs. Infect Genet Evol 2010; 10(7): 984-8.
[http://dx.doi.org/10.1016/j.meegid.2010.06.004] [PMID: 20601172] , 60Giordano MO, Martinez LC, Masachessi G, et al. Evidence of closely related picobirnavirus strains circulating in humans and pigs in Argentina. J Infect 2011; 62(1): 45-51.
[http://dx.doi.org/10.1016/j.jinf.2010.09.031] [PMID: 20888858] ] which has also reported this virus in due course of detecting various other enteric pathogens or working specifically for PBV diagnosis. A study in Hungary also elaborated the possibility of interspecies transmission among the porcine and human PBV strains [20Bányai K, Martella V, Bogdán A, et al. Genogroup I picobirnaviruses in pigs: Evidence for genetic diversity and relatedness to human strains. J Gen Virol 2008; 89(Pt 2): 534-9.
[http://dx.doi.org/10.1099/vir.0.83134-0] [PMID: 18198385] ]. The PBV isolates detected in Venezuela, Argentina and Hungary were closely related to human GG-I PBVs [20Bányai K, Martella V, Bogdán A, et al. Genogroup I picobirnaviruses in pigs: Evidence for genetic diversity and relatedness to human strains. J Gen Virol 2008; 89(Pt 2): 534-9.
[http://dx.doi.org/10.1099/vir.0.83134-0] [PMID: 18198385] , 56Carruyo GM, Mateu G, Martínez LC, et al. Molecular characterization of porcine picobirnaviruses and development of a specific reverse transcription-PCR assay. J Clin Microbiol 2008; 46(7): 2402-5.
[http://dx.doi.org/10.1128/JCM.00655-08] [PMID: 18508933] , 60Giordano MO, Martinez LC, Masachessi G, et al. Evidence of closely related picobirnavirus strains circulating in humans and pigs in Argentina. J Infect 2011; 62(1): 45-51.
[http://dx.doi.org/10.1016/j.jinf.2010.09.031] [PMID: 20888858] ]. In 2011, Smits and colleagues reported the PBV infection in respiratory tracts of pigs, signifying their role as respiratory pathogens [55Smits SL, Poon LL, van Leeuwen M, et al. Genogroup I and II picobirnaviruses in respiratory tracts of pigs. Emerg Infect Dis 2011; 17(12): 2328-30.
[http://dx.doi.org/10.3201/eid1712.110934] [PMID: 22172405] ]. While working on a Porcine PBV strain 221/04-16, Banyai and co-workers also demonstrated that due to high degree of insertions/deletions in the PBV genome are responsible for short and long type of genome segments resulting in sequence length heterogeneity [61Bányai K, Potgieter C, Gellért Á, et al. Genome sequencing identifies genetic and antigenic divergence of porcine picobirnaviruses. J Gen Virol 2014; 95(Pt 10): 2233-9.
[http://dx.doi.org/10.1099/vir.0.057984-0] [PMID: 24584476] ]. These reports regarding the presence of multiple PBV strains present in pigs also emerged from China in the year 2014 [62Chen M, Sun H, Lan D, et al. Molecular detection of genogroup I and II picobirnaviruses in pigs in China. Virus Genes 2014; 48(3): 553-6.
[http://dx.doi.org/10.1007/s11262-014-1058-8] [PMID: 24682937] ]. Reports emerged in Thailand where diarrheic and non-diarrheic piglets were found to possess the PBV infection along with co-infection of group A rotavirus (RVA) and bocavirus (BoV), all PBV strains were belonged to GG-I genogroup and phylogenetically they were close to Chinese PBV strains [63Wilburn L, Yodmeeklin A, Kochjan P, et al. Molecular detection and characterization of picobirnaviruses in piglets with diarrhea in Thailand. Arch Virol 2017; 162(4): 1061-6.
[http://dx.doi.org/10.1007/s00705-016-3190-3] [PMID: 28032197] ].
The first detection and molecular characterization of porcine PBV (BG-Por-2/2010 and BG-Por-7/ 2010) was reported from a piggery located in an urban slum at Kolkata, India [64Ganesh B, Bányai K, Kanungo S, Sur D, Malik YS, Kobayashi N. Detection and molecular characterization of porcine picobirnavirus in feces of domestic pigs from kolkata, India. Indian J Virol 2012; 23(3): 387-91.
[http://dx.doi.org/10.1007/s13337-012-0106-z] [PMID: 24293831] ]. The phylogenetic analysis revealed genetic similarity to different porcine and human GG-I PBVs from different geographical regions. In 2016, Porcine PBV infection was reported from North Eastern Region of India with strains having varied diversity and were not found to be closely associated with any other Indian isolates of PBVs so far [21Kylla H, Dutta TK, Roychoudhury P, Malik YS, Mandakini R, Subudhi PK. Prevalence and molecular characterization of porcine Picobirnavirus in piglets of North East Region of India. Trop Anim Health Prod 2017; 49(2): 417-22.
[http://dx.doi.org/10.1007/s11250-016-1210-7] [PMID: 27987110] ]. The study also claims that PBV infections are higher in acute summer and winter seasons compared to spring and autumn seasons with high prevalence recorded in cross breed pigs in contrast to local breeds.
1.5. Picobirnaviruses in Other Species
With the availability of high end instruments and state-of-art techniques, PBVs have been detected in several un-common animal species throughout the world. In 2015, researchers explored the virome of 458 rhesus macaques collected from different geographical areas of neighboring country Bangladesh [65Anthony SJ, Islam A, Johnson C, et al. Non-random patterns in viral diversity. Nat Commun 2015; 6: 8147.
[http://dx.doi.org/10.1038/ncomms9147] [PMID: 26391192] ]. They applied the combination of consensus PCR (cPCR) along with High Throughput Sequencing (HTS) to characterize 184 viruses from 14 virus families. Out of them all, 120 were PBV of the 184 viruses found in these animals which showed the presence of this bipartite virus in the population of nearby primate population. Moreover these free ranging macaques are usually found near the human settlements and therefore the risk of zoonoses cannot be ruled out. Earlier, high incidence of PBV infection in asymptomatic primates was also observed in Argentina where individually caged orangutans were diagnosed with persistent PBV infections over the span of three years [66Masachessi G, Ganesh B, Martinez LC, et al. Maintenance of picobirnavirus (PBV) infection in an adult orangutan (Pongo pygmaeus) and genetic diversity of excreted viral strains during a three-year period. Infect Genet Evol 2015; 29: 196-202.
[http://dx.doi.org/10.1016/j.meegid.2014.11.019] [PMID: 25435283] ]. Application of metagenomics sometime reveals unknown pathogens in unusual hosts and gives a comprehensive picture regarding genetic map of those pathogens, similar study was performed in Portugal in 2015 where PBV infection was found in wolves [67Conceição-Neto N, Mesquita JR, Zeller M, et al. Reassortment among picobirnaviruses found in wolves. Arch Virol 2016; 161(10): 2859-62.
[http://dx.doi.org/10.1007/s00705-016-2987-4] [PMID: 27438074] ]. This study also described the reassortment events going among PBVs in which identical capsid segments together with diverse RdRp segments were found.
Alike, a PBV strain, PBV/Horse/India/BG-Eq-3/2010, was identified in the faeces of a 10 month old weaned female foal with diarrhoea in January 2010 from Kolkata, India [31Ganesh B, Banyai K, Masachessi G, et al. Genogroup I picobirnavirus in diarrhoeic foals: can the horse serve as a natural reservoir for human infection? Vet Res (Faisalabad) 2011; 42: 52.
[http://dx.doi.org/10.1186/1297-9716-42-52] [PMID: 21414192] ]. Surprisingly, sequence comparison and phylogenetic analysis revealed close genetic relatedness to a human genogroup I PBV strain (Hu/GPBV1) detected earlier from the same part of India.
Using RT-PCR assay, PBV has been detected in 2.4% (3/125) and 3% (3/100) diarrheic kids and lambs fecal samples, respectively, from July, 2013 – February 2014 in Northern parts of India. Three PBV isolates, each from caprine and ovine species were typed as genogroup I (GGI) while two ovine isolates were typed as genogroup II (GGII). Even in one of the ovine samples both genogroups were found as a mixed infection (Unpublished data).
PBVs have also been defined to be present as persistent infection in captive animal species which expanded our knowledge regarding the host range of this virus [68Masachessi G, Martínez LC, Giordano MO, et al. Picobirnavirus (PBV) natural hosts in captivity and virus excretion pattern in infected animals. Arch Virol 2007; 152(5): 989-98.
[http://dx.doi.org/10.1007/s00705-006-0900-2] [PMID: 17245535] ]. Although the diagnosis method applied in that study was RNA-PAGE which has lower sensitivity then RT-PCR therefore the number of PBV positive species could have been more if RT-PCR assay would have been applied. Similarly, PBV infection in captive feline species was also described in 2012 in Uruguay where four different feline species were found to possess the PBV infection [69Gillman L, Sánchez AM, Arbiza J. Picobirnavirus in captive animals from Uruguay: identification of new hosts. Intervirology 2013; 56(1): 46-9.
[http://dx.doi.org/10.1159/000338275] [PMID: 22759924] ]. In 2009, a report emerged from Brazil in which further investigation on captive species like Wistor rats, snakes and domestic dogs was carried out to characterize PBV from these species and they applied the RT-PCR diagnosis method to diagnose the infection [70Fregolente MCD, de Castro-Dias E, Martins SS, Spilki FR, Allegretti SM, Gatti MSV. Molecular characterization of picobirnaviruses from new hosts. Virus Res 2009; 143(1): 134-6.
[http://dx.doi.org/10.1016/j.virusres.2009.03.006] [PMID: 19463731] ]. This study shed more light on co-circulation of PBV in different hosts and identified snakes as a new host for PBVs. Following reports of PBV infection in different hosts Woo and colleagues discovered a novel PBV in feces of Calfornia Sea Lions and prosed a new species as otarine PBV for which they published the whole genome [71Woo PC, Lau SK, Bai R, et al. Complete genome sequence of a novel picobirnavirus, otarine picobirnavirus, discovered in California sea lions. J Virol 2012; 86(11): 6377-8.
[http://dx.doi.org/10.1128/JVI.00686-12] [PMID: 22570247] ]. These reports contributed in the understanding that PBV infections are persistent in symptomatic as well as asymptomatic animals which includes both mammals and birds [72Masachessi G, Martinez LC, Ganesh B, et al. Establishment and maintenance of persistent infection by picobirnavirus in greater rhea (Rhea Americana). Arch Virol 2012; 157(11): 2075-82.
[http://dx.doi.org/10.1007/s00705-012-1400-1] [PMID: 22782138] ]. Studies on domesticated birds like turkey and broilers have also reported the diagnosis of PBV infection in USA and Brazil [73Verma H, Mor SK, Erber J, Goyal SM. Prevalence and complete genome characterization of turkey picobirnaviruses. Infect Genet Evol 2015; 30: 134-9.
[http://dx.doi.org/10.1016/j.meegid.2014.12.014] [PMID: 25530436] , 74Ribeiro Silva R, Bezerra DAM, Kaiano JHL, et al. Genogroup I avian picobirnavirus detected in Brazilian broiler chickens: a molecular epidemiology study. J Gen Virol 2014; 95(Pt 1): 117-22.
[http://dx.doi.org/10.1099/vir.0.054783-0] [PMID: 24108140] ]. In 2014, a thorough NGS study on camel feces revealed huge abundance of PBV infection co-infecting the camel population along with other viral pathogens [75Woo PC, Lau SK, Teng JL, et al. Metagenomic analysis of viromes of dromedary camel fecal samples reveals large number and high diversity of circoviruses and picobirnaviruses. Virology 2014; 471-473: 117-25.
[http://dx.doi.org/10.1016/j.virol.2014.09.020] [PMID: 25461537] ]. Keeping in mind the findings of Woo and colleagues, in 2016 we diagnosed PBV infection in captive camel population of Gujarat state where 93.10% of samples were found positive with no diarrheic symptoms (Unpublished data). Likewise, a comprehensive molecular epidemiological study was conducted in several mammalian species of terrestrial and marine habitat in Hong Kong revealed high diversity among different mammals [76Woo PC, Teng JL, Bai R, et al. High diversity of genogroup I picobirnaviruses in mammals. Front Microbiol 2016; 7: 1886.
[http://dx.doi.org/10.3389/fmicb.2016.01886] [PMID: 27933049] ]. These studies expanded our knowledge regarding the increase in host range of PBV’s and their greater susceptibility to domesticated, terrestrial as well as marine mammalian species. The risk of species jumping among different PBV strains is an alarming condition which can be addressed by continuous surveillance of this virus in different mammalian and avian species.
1.6. Evolutionary Analysis of Picobirnaviruses
The total number of nucleotide submission of PBV from different species are very few from India (n=87) (Table 1). Out of the 87 sequences only one sequence is having complete information of RdRp gene (Segment 2).
Sequences from different species of Indian PBV population were phylogenetically analyzed using the MEGA 6.0 software applying the General Time Reversible model along with Gamma distribution which was used to model evolutionary rate differences among sites [77Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013; 30(12): 2725-9.
[http://dx.doi.org/10.1093/molbev/mst197] [PMID: 24132122] ]. The analysis included 53 sequences comprising the reference sequences of PBV for GG-I and GG-II (Fig. 1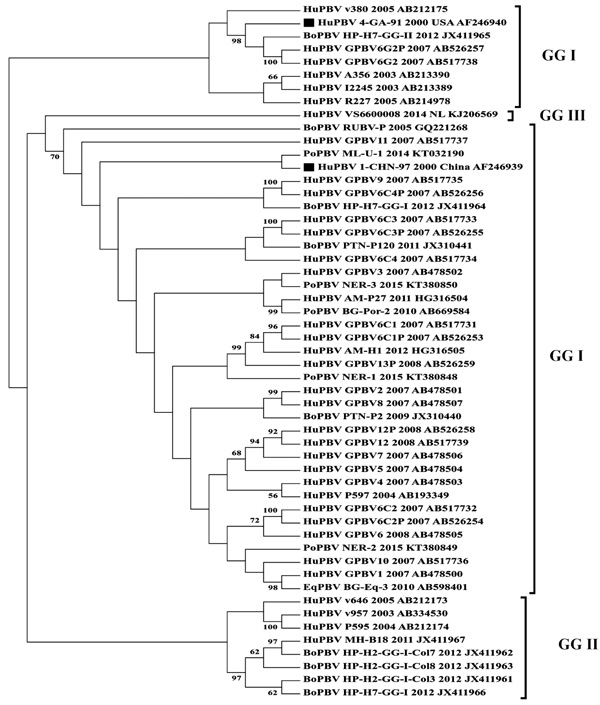 ). Huge diversity was observed within the Indian PBV sequences when they were phylogenetically analyzed suggesting species jumping with no specific host among the Indian PBV population. Although the available GG-I and GG-II sequences had formed a separate clade (Fig. 1
). Huge diversity was observed within the Indian PBV sequences when they were phylogenetically analyzed suggesting species jumping with no specific host among the Indian PBV population. Although the available GG-I and GG-II sequences had formed a separate clade (Fig. 1 ). The bipartite nature of PBVs renders them as possible candidates for further recombination and reassortment among the different isolates [20Bányai K, Martella V, Bogdán A, et al. Genogroup I picobirnaviruses in pigs: Evidence for genetic diversity and relatedness to human strains. J Gen Virol 2008; 89(Pt 2): 534-9.
). The bipartite nature of PBVs renders them as possible candidates for further recombination and reassortment among the different isolates [20Bányai K, Martella V, Bogdán A, et al. Genogroup I picobirnaviruses in pigs: Evidence for genetic diversity and relatedness to human strains. J Gen Virol 2008; 89(Pt 2): 534-9.
[http://dx.doi.org/10.1099/vir.0.83134-0] [PMID: 18198385] , 43Bányai K, Jakab F, Reuter G, et al. Sequence heterogeneity among human picobirnaviruses detected in a gastroenteritis outbreak. Arch Virol 2003; 148(12): 2281-91.
[http://dx.doi.org/10.1007/s00705-003-0200-z] [PMID: 14648286] ]. A recent report has also confirmed and speculated that animal and human PBV strains can re-assort independently [67Conceição-Neto N, Mesquita JR, Zeller M, et al. Reassortment among picobirnaviruses found in wolves. Arch Virol 2016; 161(10): 2859-62.
[http://dx.doi.org/10.1007/s00705-016-2987-4] [PMID: 27438074] ].
 |
Fig. (1) Phylogram display genetic diversity between different Picobirnavirus populations in India. |
Further, to confirm the species diversity among Indian PBV isolates, the dataset was analyzed using the SplitsTree4 software package [78Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 2006; 23(2): 254-67.
[http://dx.doi.org/10.1093/molbev/msj030] [PMID: 16221896] ]. The Minimum Spanning Network applied to the dataset bifurcated the isolates into two major Indian PBV populations which shows a bigger network comprising of human, bovine, porcine and equine isolates reported till date whereas, the smaller population comprises majorly of human PBV isolates with only one bovine PBV strain RUBV-P also clustering along with these human strains (Fig. 2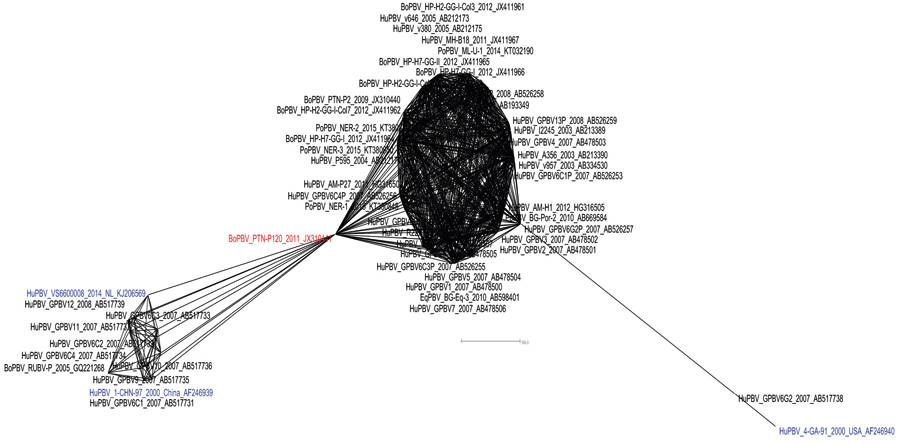 ). The recently proposed GG-III genogroup isolate from Netherlands (VS6600008) also clustered within this bunch of human origin PBV isolates.
). The recently proposed GG-III genogroup isolate from Netherlands (VS6600008) also clustered within this bunch of human origin PBV isolates.
 |
Fig. (2) Minimum spanning network exhibiting diversity between Indian Picobirnavirus isolates. |
Interestingly, in this analysis a bovine PBV (PTN-P120) isolate emerged as a pivotal strain from where two major clusters were seemed to have emerged in the Indian PBV population. The reference strain for GG-II PBV (4-GA-91) was again distantly placed beside the Indian human PBV strain (GPBV6G2) from Kolkata which also belong to GG-II genogroup. Rest all other strains pertaining to GG-I and GG-II were clustered inside the major constellation of Indian PBV isolates. The analysis suggested that these all isolates are resultant of species jumping, where GG-I and GG-II sequences were seemed to have co-evolved with each other having no clear ancestor among them.
Nevertheless, due to limited epidemiological data available on PBVs in Indian human and animal population we can’t approach to a concrete conclusion regarding the evolution, host-virus relationship and the infection pattern of PBV in Indian perspective.
CONCLUSION AND FUTURE PROSPECTS
Since their emergence in 1988, picobirnaviruses have established themselves as cause of great concern worldwide. However, PBVs have formally not yet been associated with diarrhea. An adequate cell culture system or an animal model has not been established till date, which implicates several drawbacks in understanding the biology and pathogenicity of PBVs. They have shown association with gastroenteritis and respiratory infections. The grave notch is that they can inflict wide range of susceptible hosts. The genetic relatedness between human and animal PBV warrant further studies on zoonotic potential. Therefore, continued surveillance is needed to assess the emergence of new PBV in different host, its origin, diversity and public health implications. Moreover, the tropism of this virus is still in question and its relevance as a major causative agent for neonatal or adult diarrhea is still a matter of speculations. The persistence of PBV and its diagnosis in a host with or without the signs of diarrhea always contradicts the different studies which claim it as notable cause of diarrhea. Also, the detection of PBVs from sewage and surface water, indicates that the virus continues to exist in various diverse environmental conditions and also being shed from asymptomatic carrier host animals. A clear picture of the virus-host-environment is to be studied under the “One Health” concept. With only few records of sequence data available on the GenBank database and negligible number of complete gene information from India the need of active surveillance based research becomes important in future. Improved management practices should be adopted on animal farms for prevention and control of picobirnavirus infection.
CONFLICT OF INTEREST
All authors declare that there exist no commercial or financial relationships that could in any way lead to a potential conflict of interest.
ACKNOWLEDGEMENTS
All the authors acknowledge and thank their respective Institutes and Universities.
REFERENCES
| [1] | Malik YS, Kumar N, Sharma K, et al. Epidemiology, phylogeny, and evolution of emerging enteric Picobirnaviruses of animal origin and their relationship to human strains 2014. |
| [2] | Rosen BI, Fang Z-Y, Glass RI, Monroe SS. Cloning of human picobirnavirus genomic segments and development of an RT-PCR detection assay. Virology 2000; 277(2): 316-29. [http://dx.doi.org/10.1006/viro.2000.0594] [PMID: 11080479] |
| [3] | Smits SL, Schapendonk CM, van Beek J, et al. New viruses in idiopathic human diarrhea cases, the Netherlands. Emerg Infect Dis 2014; 20(7): 1218-22. [http://dx.doi.org/10.3201/eid2007.140190] [PMID: 24964003] |
| [4] | Li L, Giannitti F, Low J, et al. Exploring the virome of diseased horses. J Gen Virol 2015; 96(9): 2721-33. [http://dx.doi.org/10.1099/vir.0.000199] [PMID: 26044792] |
| [5] | Giordano MO, Martinez LC, Rinaldi D, et al. Diarrhea and enteric emerging viruses in HIV-infected patients. AIDS Res Hum Retroviruses 1999; 15(16): 1427-32. [http://dx.doi.org/10.1089/088922299309937] [PMID: 10555105] |
| [6] | Martínez LC, Giordano MO, Isa MB, et al. Molecular diversity of partial-length genomic segment 2 of human picobirnavirus. Intervirology 2003; 46(4): 207-13. [http://dx.doi.org/10.1159/000072429] [PMID: 12931028] |
| [7] | Grohmann GS, Glass RI, Pereira HG, et al. Enteric viruses and diarrhea in HIV-infected patients. N Engl J Med 1993; 329(1): 14-20. [http://dx.doi.org/10.1056/NEJM199307013290103] [PMID: 8099429] |
| [8] | Giordano MO, Martinez LC, Rinaldi D, et al. Detection of picobirnavirus in HIV-infected patients with diarrhea in Argentina. J Acquir Immune Defic Syndr Hum Retrovirol 1998; 18(4): 380-3. [http://dx.doi.org/10.1097/00042560-199808010-00010] [PMID: 9704944] |
| [9] | Pereira HG, Fialho AM, Flewett TH, Teixeira JM, Andrade ZP. Novel viruses in human faeces. Lancet 1988; 2(8602): 103-4. [http://dx.doi.org/10.1016/S0140-6736(88)90032-3] [PMID: 2898672] |
| [10] | Pereira HG, Flewett TH, Candeias JA, Barth OM. A virus with a bisegmented double-stranded RNA genome in rat (Oryzomys nigripes) intestines. J Gen Virol 1988; 69(Pt 11): 2749-54. [http://dx.doi.org/10.1099/0022-1317-69-11-2749] [PMID: 3053986] |
| [11] | Haga IR, Martins SS, Hosomi ST, Vicentini F, Tanaka H, Gatti MS. Identification of a bisegmented double-stranded RNA virus (Picobirnavirus) in faeces of giant anteaters (Myrmecophaga tridactyla). Vet J 1999; 158(3): 234-6. [http://dx.doi.org/10.1053/tvjl.1999.0369] [PMID: 10558845] |
| [12] | Pereira H, De Araujo H, Fialho A. Castro Ld, Monteiro S. A virus with bi-segmented double-stranded RNA gerome in guinea pig intestines. Mem Inst Oswaldo Cruz 1989; 84: 137-40. [http://dx.doi.org/10.1590/S0074-02761989000100025] [PMID: 2157131] |
| [13] | Ludert JE, Abdul-Latiff L, Liprandi A, Liprandi F. Identification of picobirnavirus, viruses with bisegmented double stranded RNA, in rabbit faeces. Res Vet Sci 1995; 59(3): 222-5. [http://dx.doi.org/10.1016/0034-5288(95)90006-3] [PMID: 8588095] |
| [14] | Gallimore C, Lewis D, Brown D. Detection and characterization of a novel bisegmented double-stranded RNA virus (picobirnavirus) from rabbit faeces. Arch Virol 1993; 133(1-2): 63-73. [http://dx.doi.org/10.1007/BF01309744] [PMID: 8240018] |
| [15] | Gallimore CI, Green J, Casemore DP, Brown DW. Detection of a picobirnavirus associated with Cryptosporidium positive stools from humans. Arch Virol 1995; 140(7): 1275-8. [http://dx.doi.org/10.1007/BF01322752] [PMID: 7646357] |
| [16] | Leite JPG, Monteiro SP, Fialho AM, Pereira HG. A novel avian virus with trisegmented double-stranded RNA and further observations on previously described similar viruses with bisegmented genome. Virus Res 1990; 16(2): 119-26. [http://dx.doi.org/10.1016/0168-1702(90)90016-5] [PMID: 2385956] |
| [17] | Gatti MS, de Castro AF, Ferraz MM, Fialho AM, Pereira HG. Viruses with bisegmented double-stranded RNA in pig faeces. Res Vet Sci 1989; 47(3): 397-8. [PMID: 2687991] |
| [18] | Chasey D. Porcine picobirnavirus in UK? Vet Rec 1990; 126(18): 465. [PMID: 2356603] |
| [19] | Ludert JE, Hidalgo M, Gil F, Liprandi F. Identification in porcine faeces of a novel virus with a bisegmented double stranded RNA genome. Arch Virol 1991; 117(1-2): 97-107. [http://dx.doi.org/10.1007/BF01310495] [PMID: 2006903] |
| [20] | Bányai K, Martella V, Bogdán A, et al. Genogroup I picobirnaviruses in pigs: Evidence for genetic diversity and relatedness to human strains. J Gen Virol 2008; 89(Pt 2): 534-9. [http://dx.doi.org/10.1099/vir.0.83134-0] [PMID: 18198385] |
| [21] | Kylla H, Dutta TK, Roychoudhury P, Malik YS, Mandakini R, Subudhi PK. Prevalence and molecular characterization of porcine Picobirnavirus in piglets of North East Region of India. Trop Anim Health Prod 2017; 49(2): 417-22. [http://dx.doi.org/10.1007/s11250-016-1210-7] [PMID: 27987110] |
| [22] | Vanopdenbosch E, Wellemans G. Birna-type virus in diarrhoeic calf faeces. Vet Rec 1989; 125(24): 610. [PMID: 2609497] |
| [23] | Buzinaro MG, Freitas PP, Kisiellius JJ, Ueda M, Jerez JA. Identification of a bisegmented double-stranded RNA virus (picobirnavirus) in calf faeces. Vet J 2003; 166(2): 185-7. [http://dx.doi.org/10.1016/S1090-0233(03)00031-5] [PMID: 12902184] |
| [24] | Ghosh S, Kobayashi N, Nagashima S, Naik TN. Molecular characterization of full-length genomic segment 2 of a bovine picobirnavirus (PBV) strain: evidence for high genetic diversity with genogroup I PBVs. J Gen Virol 2009; 90(Pt 10): 2519-24. [http://dx.doi.org/10.1099/vir.0.013987-0] [PMID: 19587136] |
| [25] | Malik YS, Chandrashekar KM, Sharma K, et al. Picobirnavirus detection in bovine and buffalo calves from foothills of Himalaya and Central India. Trop Anim Health Prod 2011; 43(8): 1475-8. [http://dx.doi.org/10.1007/s11250-011-9834-0] [PMID: 21479844] |
| [26] | Malik YS, Kumar N, Sharma K, et al. Molecular characterization of a genetically diverse bubaline picobirnavirus strain, India. Wetchasan Sattawaphaet 2013; 43: 609-13. |
| [27] | Mondal A, Chakravarti S, Majee SB, Bannalikar AS. Detection of picobirnavirus and rotavirus in diarrhoeic faecal samples of cattle and buffalo calves in Mumbai metropolis, Western India. Vet Ital 2013; 49(4): 357-60. [PMID: 24362776] |
| [28] | Mondal A, Joardar SN. Detection of Genogroup I Picobirnavirus in Diarrhoeic Faecal Samples of Calves from West Bengal, India. J Immunol Immunopathol 2014; 16: 40-3. [http://dx.doi.org/10.5958/0973-9149.2014.01076.4] |
| [29] | Takiuchi E, Macedo R, Kunz AF, et al. Electrophoretic RNA genomic Profiles of Brazilian Picobirnavirus (PBV) strains and molecular characterization of a PBV isolated from diarrheic calf. Virus Res 2016; 211: 58-63. [http://dx.doi.org/10.1016/j.virusres.2015.09.022] [PMID: 26435337] |
| [30] | Browning GF, Chalmers RM, Snodgrass DR, et al. The prevalence of enteric pathogens in diarrhoeic thoroughbred foals in Britain and Ireland. Equine Vet J 1991; 23(6): 405-9. [http://dx.doi.org/10.1111/j.2042-3306.1991.tb03751.x] [PMID: 1663866] |
| [31] | Ganesh B, Banyai K, Masachessi G, et al. Genogroup I picobirnavirus in diarrhoeic foals: can the horse serve as a natural reservoir for human infection? Vet Res (Faisalabad) 2011; 42: 52. [http://dx.doi.org/10.1186/1297-9716-42-52] [PMID: 21414192] |
| [32] | Navarro R, Yibin C, Nair R, et al. Molecular characterization of complete genomic segment-2 of picobirnavirus strains detected in a cat and a dog. Infect Genet Evol 2017; 54: 200-4. [http://dx.doi.org/10.1016/j.meegid.2017.07.006] [PMID: 28688978] |
| [33] | Costa A, Cubel Garcia R, Labarthe N, Leite J. Detection of double-stranded RNA viruses in fecal samples of dogs with gastroenteritis in Rio de Janeiro, Brazil. Arq Bras Med Vet Zootec 2004; 56: 554-7. [http://dx.doi.org/10.1590/S0102-09352004000400020] |
| [34] | Volotäo EM, Soares CC, Albuquerque MC, et al. First evidence of a trisegmented double-stranded RNA virus in canine faeces. Vet J 2001; 161(2): 205-7. [http://dx.doi.org/10.1053/tvjl.2000.0532] [PMID: 11243690] |
| [35] | Gallimore CI, Appleton H, Lewis D, Green J, Brown DW. Detection and characterisation of bisegmented double-stranded RNA viruses (picobirnaviruses) in human faecal specimens. J Med Virol 1995; 45(2): 135-40. [http://dx.doi.org/10.1002/jmv.1890450204] [PMID: 7775930] |
| [36] | Martínez LC, Giordano MO, Isa MB, et al. Molecular diversity of partial-length genomic segment 2 of human picobirnavirus. Intervirology 2003; 46(4): 207-13. [http://dx.doi.org/10.1159/000072429] [PMID: 12931028] |
| [37] | Giordano MO, Masachessi G, Martinez LC, et al. Two instances of large genome profile picobirnavirus occurrence in Argentinian infants with diarrhea over a 26-year period (1977-2002). J Infect 2008; 56(5): 371-5. [http://dx.doi.org/10.1016/j.jinf.2008.02.017] [PMID: 18403022] |
| [38] | Barreto ML, Milroy CA, Strina A, et al. Community-based monitoring of diarrhea in urban Brazilian children: incidence and associated pathogens. Trans R Soc Trop Med Hyg 2006; 100(3): 234-42. [http://dx.doi.org/10.1016/j.trstmh.2005.03.010] [PMID: 16303156] |
| [39] | Cunliffe NA, Glass RI. Gastrointestinal manifestations of HIV infection. Lancet 1996; 348(9033): 1037. [http://dx.doi.org/10.1016/S0140-6736(05)64970-7] [PMID: 8855890] |
| [40] | Zhang T, Breitbart M, Lee WH, et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol 2006; 4(1): e3. [http://dx.doi.org/10.1371/journal.pbio.0040003] [PMID: 16336043] |
| [41] | Ludert JE, Liprandi F. Identification of viruses with bi- and trisegmented double-stranded RNA genome in faeces of children with gastroenteritis. Res Virol 1993; 144(3): 219-24. [http://dx.doi.org/10.1016/S0923-2516(06)80032-4] [PMID: 8356343] |
| [42] | González GG, Pujol FH, Liprandi F, Deibis L, Ludert JE. Prevalence of enteric viruses in human immunodeficiency virus seropositive patients in Venezuela. J Med Virol 1998; 55(4): 288-92. [http://dx.doi.org/10.1002/(SICI)1096-9071(199808)55:4<288::AID-JMV6>3.0.CO;2-X] [PMID: 9661837] |
| [43] | Bányai K, Jakab F, Reuter G, et al. Sequence heterogeneity among human picobirnaviruses detected in a gastroenteritis outbreak. Arch Virol 2003; 148(12): 2281-91. [http://dx.doi.org/10.1007/s00705-003-0200-z] [PMID: 14648286] |
| [44] | Cascio A, Bosco M, Vizzi E, Giammanco A, Ferraro D, Arista S. Identification of picobirnavirus from faeces of Italian children suffering from acute diarrhea. Eur J Epidemiol 1996; 12(5): 545-7. [http://dx.doi.org/10.1007/BF00144011] [PMID: 8905320] |
| [45] | van Leeuwen M, Williams MM, Koraka P, Simon JH, Smits SL, Osterhaus AD. Human picobirnaviruses identified by molecular screening of diarrhea samples. J Clin Microbiol 2010; 48(5): 1787-94. [http://dx.doi.org/10.1128/JCM.02452-09] [PMID: 20335418] |
| [46] | Novikova NA, Epifanova NV, Fedorova OF, Golitsyna LN, Kupriianova NV. [Detection of picobirnaviruses by electrophoresis of RNA in polyacrylamide gel]. Vopr Virusol 2003; 48(6): 41-3. [PMID: 14708231] |
| [47] | Wakuda M, Pongsuwanna Y, Taniguchi K. Complete nucleotide sequences of two RNA segments of human picobirnavirus. J Virol Methods 2005; 126(1-2): 165-9. [http://dx.doi.org/10.1016/j.jviromet.2005.02.010] [PMID: 15847933] |
| [48] | Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog 2008; 4(2): e1000011. [http://dx.doi.org/10.1371/journal.ppat.1000011] [PMID: 18398449] |
| [49] | Bhattacharya R, Sahoo GC, Nayak MK, et al. Molecular epidemiology of human picobirnaviruses among children of a slum community in Kolkata, India. Infect Genet Evol 2006; 6(6): 453-8. [http://dx.doi.org/10.1016/j.meegid.2006.02.005] [PMID: 16616879] |
| [50] | Bhattacharya R, Sahoo GC, Nayak MK, et al. Detection of Genogroup I and II human picobirnaviruses showing small genomic RNA profile causing acute watery diarrhoea among children in Kolkata, India. Infect Genet Evol 2007; 7(2): 229-38. [http://dx.doi.org/10.1016/j.meegid.2006.09.005] [PMID: 17049316] |
| [51] | Ganesh B, Nataraju SM, Rajendran K, et al. Detection of closely related Picobirnaviruses among diarrhoeic children in Kolkata: evidence of zoonoses? Infect Genet Evol 2010; 10(4): 511-6. [http://dx.doi.org/10.1016/j.meegid.2010.02.008] [PMID: 20178864] |
| [52] | Ganesh B, Nagashima S, Ghosh S, et al. Detection and molecular characterization of multiple strains of Picobirnavirus causing mixed infection in a diarrhoeic child: Emergence of prototype Genogroup II-like strain in Kolkata, India. Int J Mol Epidemiol Genet 2011; 2(1): 61-72. [PMID: 21537403] |
| [53] | Villacorta I, Peeters JE, Vanopdenbosch E, Ares-Mazás E, Theys H. Efficacy of halofuginone lactate against Cryptosporidium parvum in calves. Antimicrob Agents Chemother 1991; 35(2): 283-7. [http://dx.doi.org/10.1128/AAC.35.2.283] [PMID: 2024962] |
| [54] | Malik YS, Sharma AK, Kumar N, Sharma K, Ganesh B, Kobayashi N. Identification and characterisation of a novel genogroup II picobirnavirus in a calf in India. Vet Rec 2014; 174(11): 278. [http://dx.doi.org/10.1136/vr.102065] [PMID: 24570405] |
| [55] | Smits SL, Poon LL, van Leeuwen M, et al. Genogroup I and II picobirnaviruses in respiratory tracts of pigs. Emerg Infect Dis 2011; 17(12): 2328-30. [http://dx.doi.org/10.3201/eid1712.110934] [PMID: 22172405] |
| [56] | Carruyo GM, Mateu G, Martínez LC, et al. Molecular characterization of porcine picobirnaviruses and development of a specific reverse transcription-PCR assay. J Clin Microbiol 2008; 46(7): 2402-5. [http://dx.doi.org/10.1128/JCM.00655-08] [PMID: 18508933] |
| [57] | Núñez GMC, Aristiguieta ACA, Fraire FL, Leon JEL. Porcine picobirnavirus infection in Venezuelan farms 2014; 24 |
| [58] | Alfieri AA, Alfieri AF, de Freitas JC, et al. Ocurrence of Escherichia Coli, rotavirus, picobirnavirus and Cryptosporidium Parvum in a postweaning diarrhoea focus in swine. Semin Cienc Agrar 1994; 15: 5-7. |
| [59] | Martínez LC, Masachessi G, Carruyo G, et al. Picobirnavirus causes persistent infection in pigs. Infect Genet Evol 2010; 10(7): 984-8. [http://dx.doi.org/10.1016/j.meegid.2010.06.004] [PMID: 20601172] |
| [60] | Giordano MO, Martinez LC, Masachessi G, et al. Evidence of closely related picobirnavirus strains circulating in humans and pigs in Argentina. J Infect 2011; 62(1): 45-51. [http://dx.doi.org/10.1016/j.jinf.2010.09.031] [PMID: 20888858] |
| [61] | Bányai K, Potgieter C, Gellért Á, et al. Genome sequencing identifies genetic and antigenic divergence of porcine picobirnaviruses. J Gen Virol 2014; 95(Pt 10): 2233-9. [http://dx.doi.org/10.1099/vir.0.057984-0] [PMID: 24584476] |
| [62] | Chen M, Sun H, Lan D, et al. Molecular detection of genogroup I and II picobirnaviruses in pigs in China. Virus Genes 2014; 48(3): 553-6. [http://dx.doi.org/10.1007/s11262-014-1058-8] [PMID: 24682937] |
| [63] | Wilburn L, Yodmeeklin A, Kochjan P, et al. Molecular detection and characterization of picobirnaviruses in piglets with diarrhea in Thailand. Arch Virol 2017; 162(4): 1061-6. [http://dx.doi.org/10.1007/s00705-016-3190-3] [PMID: 28032197] |
| [64] | Ganesh B, Bányai K, Kanungo S, Sur D, Malik YS, Kobayashi N. Detection and molecular characterization of porcine picobirnavirus in feces of domestic pigs from kolkata, India. Indian J Virol 2012; 23(3): 387-91. [http://dx.doi.org/10.1007/s13337-012-0106-z] [PMID: 24293831] |
| [65] | Anthony SJ, Islam A, Johnson C, et al. Non-random patterns in viral diversity. Nat Commun 2015; 6: 8147. [http://dx.doi.org/10.1038/ncomms9147] [PMID: 26391192] |
| [66] | Masachessi G, Ganesh B, Martinez LC, et al. Maintenance of picobirnavirus (PBV) infection in an adult orangutan (Pongo pygmaeus) and genetic diversity of excreted viral strains during a three-year period. Infect Genet Evol 2015; 29: 196-202. [http://dx.doi.org/10.1016/j.meegid.2014.11.019] [PMID: 25435283] |
| [67] | Conceição-Neto N, Mesquita JR, Zeller M, et al. Reassortment among picobirnaviruses found in wolves. Arch Virol 2016; 161(10): 2859-62. [http://dx.doi.org/10.1007/s00705-016-2987-4] [PMID: 27438074] |
| [68] | Masachessi G, Martínez LC, Giordano MO, et al. Picobirnavirus (PBV) natural hosts in captivity and virus excretion pattern in infected animals. Arch Virol 2007; 152(5): 989-98. [http://dx.doi.org/10.1007/s00705-006-0900-2] [PMID: 17245535] |
| [69] | Gillman L, Sánchez AM, Arbiza J. Picobirnavirus in captive animals from Uruguay: identification of new hosts. Intervirology 2013; 56(1): 46-9. [http://dx.doi.org/10.1159/000338275] [PMID: 22759924] |
| [70] | Fregolente MCD, de Castro-Dias E, Martins SS, Spilki FR, Allegretti SM, Gatti MSV. Molecular characterization of picobirnaviruses from new hosts. Virus Res 2009; 143(1): 134-6. [http://dx.doi.org/10.1016/j.virusres.2009.03.006] [PMID: 19463731] |
| [71] | Woo PC, Lau SK, Bai R, et al. Complete genome sequence of a novel picobirnavirus, otarine picobirnavirus, discovered in California sea lions. J Virol 2012; 86(11): 6377-8. [http://dx.doi.org/10.1128/JVI.00686-12] [PMID: 22570247] |
| [72] | Masachessi G, Martinez LC, Ganesh B, et al. Establishment and maintenance of persistent infection by picobirnavirus in greater rhea (Rhea Americana). Arch Virol 2012; 157(11): 2075-82. [http://dx.doi.org/10.1007/s00705-012-1400-1] [PMID: 22782138] |
| [73] | Verma H, Mor SK, Erber J, Goyal SM. Prevalence and complete genome characterization of turkey picobirnaviruses. Infect Genet Evol 2015; 30: 134-9. [http://dx.doi.org/10.1016/j.meegid.2014.12.014] [PMID: 25530436] |
| [74] | Ribeiro Silva R, Bezerra DAM, Kaiano JHL, et al. Genogroup I avian picobirnavirus detected in Brazilian broiler chickens: a molecular epidemiology study. J Gen Virol 2014; 95(Pt 1): 117-22. [http://dx.doi.org/10.1099/vir.0.054783-0] [PMID: 24108140] |
| [75] | Woo PC, Lau SK, Teng JL, et al. Metagenomic analysis of viromes of dromedary camel fecal samples reveals large number and high diversity of circoviruses and picobirnaviruses. Virology 2014; 471-473: 117-25. [http://dx.doi.org/10.1016/j.virol.2014.09.020] [PMID: 25461537] |
| [76] | Woo PC, Teng JL, Bai R, et al. High diversity of genogroup I picobirnaviruses in mammals. Front Microbiol 2016; 7: 1886. [http://dx.doi.org/10.3389/fmicb.2016.01886] [PMID: 27933049] |
| [77] | Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013; 30(12): 2725-9. [http://dx.doi.org/10.1093/molbev/mst197] [PMID: 24132122] |
| [78] | Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 2006; 23(2): 254-67. [http://dx.doi.org/10.1093/molbev/msj030] [PMID: 16221896] |