- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Glycoprotein C Gene of Caprine Herpesvirus Type 1 Contains Short Sequence Repeats (SSR)
Elvira Tarsitano, Michele Camero, Anna Lucia Bellacicco, Nicola Decaro, Vito Martella, Canio Buonavoglia, Maria Tempesta*
Abstract
Caprine herpesvirus 1 (CpHV-1) is responsible for vaginal and respiratory disease in goats. Infection by vaginal route is usually restricted to the genital tract whereas by nasal route the virus can spread throughout the body. In order to evaluate genomic diversity, nucleotide sequences of glycoprotein C (gC) of 13 (n.8 vaginal, n.5 nasal) CpHV-1 strains were analyzed. Amino acid (aa) sequences showed a variable number of short sequence repeats (SSR). Nucleotide and amino acid sequences of amplified products showed to contain a variable number of short sequence repeats among the examined strains. These results indicated that CpHV-1 isolates had genetic diversity in the gC gene regarding the number of SSR: 4 SSR of 60 bp in one strain, 2 SSR of 30 bp in seven strains and 1 SSR of 15 bp in three strains. Two strains had no SSR.
Article Information
Identifiers and Pagination:
Year: 2010Volume: 4
First Page: 85
Last Page: 87
Publisher Id: TOVJ-4-85
DOI: 10.2174/1874357901004010085
Article History:
Received Date: 17/3/2010Revision Received Date: 15/4/2010
Acceptance Date: 19/4/2010
Electronic publication date: 25/5/2010
Collection year: 2010
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Department of Veterinary Public Health, Faculty of Veterinary Medicine, University of Bari, Str. Prov. Casamassima Km 3, 70010, Valenzano, Bari, Italy; Tel: +39-080-4679838; Fax: +39-080-4679843; E-mail: m.tempesta@veterinaria.uniba.it
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 17-3-2010 |
Original Manuscript | Glycoprotein C Gene of Caprine Herpesvirus Type 1 Contains Short Sequence Repeats (SSR) | |
INTRODUCTION
Caprine herpesvirus type 1 (CpHV-1) is an α-herpesvirus of goats sharing several biological behaviours with human herpesvirus type 2 (HHV-2), i.e., genital lesions, latency in sacral ganglia [1Tempesta M, Pratelli A, Greco G, Martella V, Buonavoglia C. Detection of caprine herpesvirus 1 in sacral ganglia of latently infected goats by PCR J Clin Microbiol 1999; 37: 1598-9.]. CpHV-1 is responsible for systemic lethal infections in kids [2Roperto F, Pratelli A, Guarino G, et al. Natural caprine herpesvirus 1 (CpHV-1) infection in kids J Comp Pathol 2000; 122: 298-302.] and respiratory or genital infections in adult goats [3Horner GW, Hunter R, Day AM. An outbreak of vulvovaginitis in goats caused by a Caprine herpesvirus N Z Vet J 1982; 30: 150-2.]. CpHV-1 can be isolated from the nose and/or vagina of infected animals. Natural or experimental infection may occur both via nasal and/or genital route [4Tempesta M, Pratelli A, Corrente M, Buonavoglia C. A preliminary study on the pathogenicity of a strain of caprine herpesvirus-1 Comp Immun Microbiol Infect Dis 1999; 22: 137-47., 5Tempesta M, Pratelli A, Normanno G, et al. Experimental intravaginal infection of goats with caprine herpesvirus 1 J Vet Med B Infect Dis Vet Public Health 2000; 47: 197-201.]. When the virus infects goats by the nose, the infection can spread throughout the body and reach the genital mucosa. The infection by the vaginal route remains restricted to the genital tract [4Tempesta M, Pratelli A, Corrente M, Buonavoglia C. A preliminary study on the pathogenicity of a strain of caprine herpesvirus-1 Comp Immun Microbiol Infect Dis 1999; 22: 137-47., 6Tempesta M, Buonavoglia D, Greco G, et al. A study on the infection and reactivation of caprine herpesvirus 1 in goats In: Proceedings of the 7th International Conference on Goat; May 15-18; Tours, France. 2000.]. No differences have been observed among nasal or vaginal isolates using virus neutralization test. Italian, Swiss, American and Spanish CpHV-1 strains, have been analysed by Restriction Fragment Length Polymorphism (RFLP), showing differences in the restriction patterns: i.e. altered sizes of single fragments, lacking or additional fragments [7Keuser V, Espejo-Serrano J, Schynts F, Georgin JP, Thiry E. Isolation of Caprine herpesvirus 1 in Spain Vet Rec 2004; 154: 395-9.-9Rocchigiani AM, Puggioni G, Murtino AP, et al. Isolamento e caratterizzazione di un ceppo di Caprine Herpesvirus 1 in Sardegna In: Proceedings III Workshop Virologia Veterinaria; June 11-12; Bari, Italy. 2009.].
In order to evaluate genomic diversity of the isolates, nucleotide and amino acid sequences of glycoprotein C (gC), were analyzed in thirteen CpHV-1 strains.
MATERIAL AND METHODS
Strains
Thirteen CpHV-1 strains were analyzed in this study: eight vaginal and five nasal isolates. As reported in Table 1, the viruses were isolated in natural or experimental reactivations of CpHV-1 in geographically and timely different caprine flocks.
Extraction of Viral DNA
DNA was extracted from the original nasal and vaginal samples using the Qiamp Tissue Kit (Qiagen GmbH, Germany) according to the instructions of the manufacturer and stored at +4°C.
Amplification of gC Gene
The glycoprotein C gene was amplified by polymerase chain reaction (PCR). Primers (Nik1 and Nik2) specific for CpHV-1 [10Hecht P, Engels M, Loepfe E, Ackermann M. Comparison of the glycoprotein gC genes of bovine and caprine herpesviruses In: Proceedings of the 3rd Congress European Society Veterinary Virology; September 4-7; Interlaken, Switzerland. 1995.] and internal primers (CB3F and CB3R) were used for the amplification of 1800 bp and 400 bp fragment respectively of the gC gene. The internal primers were used to point out the short sequence repeats (SSR), as reported in Table 2.
The total volume of the PCR reaction was 50µl, which contained 10-50ng extracted DNA, 1X buffer (100mM Tris-HCl, pH 8.3 and 500mM KCl), 25mM MgCl2, 1.25mM dNTP, 50pmol of each primer and 1.25 units of TaKaRa LA TaqTM (TAKARA BIO INC.) in distilled water. The PCR programme was 94°C for 1 min (TaKaRa LA TaqTM activation temperature), followed by 35 cycles of 1 min and 30 sec at 95°C, 1,00 min at 60°C and 2 min and 30 sec at 72°C, and it was terminated with an extension at 72°C for 10 min (Thermocycler Applied Biosystems Gene Amp PCR system 9700). The PCR products were electrophoresed in 1.4% agarose gels (Ambion) in TAE buffer and visualised under UV light after ethidium bromide staining (10mg/ml). GeneRulerTM 50 bp DNA Ladder (Fermentas) was used as molecular weight standard.
Sequence Analysis
The PCR products were purified in Ultrafree-DA columns (Amicon, Millipore) and sequenced using the Taq DyeTerminator Cycle Sequencing Kit (Applied Biosystems) with an ABIPRISM 377. Sequences were aligned for the same strain. The nucleotide and amino acid sequences were read using the analytical tools by the National Center for Biotechnology Information (NCBI) and align by the program CLUSTALW and FASTA [11Thompson JD, Higgins DG, Gibson TJ, Clustal W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice Nucleic Acids Res 1994; 22: 4673-80.].
RESULTS
PCR products had a size ranging from 400 bp to 464 bp using primers CB3F and CB3R. The amplified fragments show that the nucleotide sequences had a different molecular weight as described in Fig. (1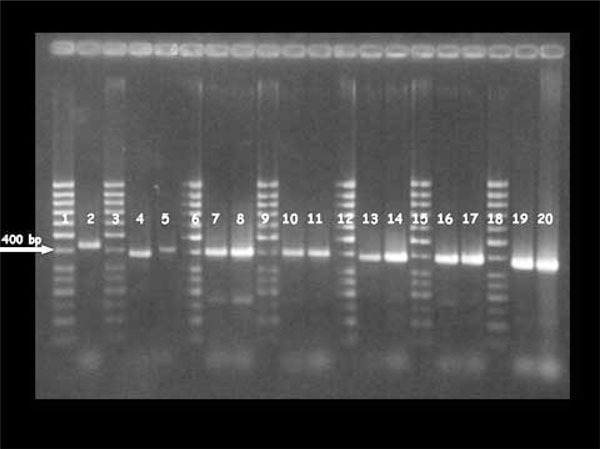 ).
).
The nucleotide sequences of the amplified products showed a variable number of short sequence repeats (SSR), each containing 15 nucleotides, among the examined strains: 4 SSR in one strain (nasal), 2 SSR in seven strains (n.5 vaginal, n.2 nasal) and 1 SSR in three strains (n.2 vaginal, n.1 nasal). Two strains had no SSR (n.1 vaginal, n.1 nasal), (Table 3). At the protein level, the SSR corresponded to the amino acid stretch FEDSA, ENDGA, KEDGA or KEDSA on the basis of the analyzed strain (Fig. 2 ).
).
DISCUSSION
The SSR in the family of Herpesviridae genomes play diverse roles, including modulating gene expression as contingency loci, facilitating genome rearrangements via recombination and affecting protein structure and possibly protein-protein interactions [12Riley DE, Krieger JN. Diverse eukaryotic transcripts suggest short tandem repeats have cellular functions Biochem Biophys Res Commun 2002; 298: 581-6.-16Tan BT, Warnke RA, Arber DA. The frequency of B- and T-cell gene rearrangements and epstein-barr virus in T-cell lymphomas: a comparison between angioimmunoblastic T-cell lymphoma and peripheral T-cell lymphoma, unspecified with and without associated B-cell proliferations J Mol Diagn 2006; 8: 466-75.]. Genetic variations and classification into different genogroups have been described for other herpesviruses, such as bovine herpesvirus type 1 (BHV-1), [17Wang J, Horner GW, O'keefe JS. Genetic characterisation of Bovine Herpesvirus 1 in New Zealand NZ Vet J 2006; 54: 61-.], varicella-zoster virus [18Muir WB, Nichols R, Breuer J. Phylogenetic analysis of varicella-zoster virus: evidence of intercontinental spread of genotypes and recombination J Virol 2002; 76: 1971-9.], simian varicella virus [19Mahalingam R, Gray WL. The simian varicella virus genome contains an invertible 665 base pair terminal element that is absent in the varicella zoster virus genome Virology 2007; 366: 387-93.] Epstein-Barr virus [20Sample J, Young L, Martin B, Chatman T, Kieff E, Rickinson A. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes J Virol 1990; 64: 4084-92., 21Kim IJ Burkum CE, Cookenham T, Schwartzberg Pl, Woodland Dl, Blackman MA. Perturbation of B cell activation in SLAM-associated protein-deficient mice is associated with changes in gammaherpesvirus latency reservoirs J Immunol 2007; 178: 1692-701.], cytomegalovirus [22Chou S. Comparative analysis of sequence variation in gp116 and gp55 components of glycoprotein B of human cytomegalovirus Virology 1992; 188: 388-90., 23Chou SW, Dennison KM. Analysis of interstrain variation in cytomegalovirus glycoprotein B sequences encoding neutralization-related epitopes J Infect Dis 1991; 163: 1229-34.], and human herpesviruses 6 and 7 [24Clark D A. Human herpesvirus 6 Rev Med Virol 2000; 10: 155-73., 25Franti M, Aubin JT, Poirel L, et al. Definition and distribution analysis of glycoprotein B gene alleles of human herpesvirus 7 J Virol 1998; 72: 8725-30.]. As reported by Norberg et al. [26Norberg P, Bergström T, Rekabdar E, Lindh M, Liljeqvist JA. Phylogenetic analysis of clinical herpes simplex virus type 1 isolates identified three genetic groups and recombinant viruses J Virol 2004; 78: 10755-64.], DNA sequencing of large regions from the clinical HHV-1 isolates may be helpful in revealing genetic alterations associated with HHV-1 pathogenesis and may be used for molecular epidemiology studies of different viral populations as well as for studies of single patient isolates and to investigate the role of homologous recombination in the evolution of the HHV-1 genome.
Nucleotide and amino acid sequence analysis indicate that CpHV-1 isolates have genetic diversity in the gC gene according to the number of SSR detected (4 SSR of 60 bp in one strain, 2 SSR of 30 bp in seven strains and 1 SSR of 15 bp in three strains). Variable regions containing SSR have been also reported in HHV-1 [27Umene K. Mechanism and application of genetic recombination in herpesviruses Rev Med Virol 1990; 9: 171-82., 28Umene K. Herpesvirus: genetic variability and recombination Touka Shobou, Fukuoka 1998.]. On the basis of few information available, it may be supposed that CpHV-1 isolates are characterized by high G+C content and presence of nucleotides repeats in the sequences of glycoprotein C gene. Genetic rearrangements (SSR) in the gC gene, may be useful for differentiation of CpHV-1 strains as reported for feline herpesvirus type 1 (FeHV-1) [29Hamano M, Maeda K, Mizukoshi F, et al. Genetic rearrangements in the gC gene of the feline herpesvirus type 1 Virus Genes 2004; 28: 55-60.] and further studies will also elucidate whether the SSR number is related to any biological feature of CpHV-1 i.e. the nasal or the vaginal tropism.
ACKNOWLEDGEMENT
This work was supported by grants from University of Bari, Italy project PRIN2007.