- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Detection of HIV-1 in Saliva: Implications for Case-Identification, Clinical Monitoring and Surveillance for Drug Resistance§
Maya Balamane*, 1, Mark A Winters1, Sudeb C Dalai1, Alexandra H Freeman2, Mark W Traves3, Dennis M Israelski3, David A Katzenstein1, Jeffrey D Klausner2
Abstract
Background:
Saliva tests that detect antibodies are used to diagnose HIV infection. The goal of this study was to determine whether saliva could be used for nucleic acid-based tests to measure HIV-1 virus load (VL) and detect drug resistance.
Methods:
69 HIV infected individuals provided 5-10 ml of saliva and blood samples. Viral RNA was isolated from saliva and dried blood spots using the Nuclisens extraction. Saliva VL was measured using a modified Amplicor assay, and genotyping was performed using an in-house RT-PCR/sequencing protocol. Plasma VLs were obtained from concurrently drawn clinical tests.
Results:
Thirty-six of 47 (77%) plasma viremic patients had measurable saliva HIV-1 RNA. Paired plasma and saliva HIV RNA levels were significantly correlated (Spearman’s correlation = .6532, p<.0001), but saliva VL was typically lower. Three of 22 patients with undetectable plasma VL (<50 copies/ml) had detectable saliva HIV RNA. Eleven of 30 patients with undetectable saliva RNA had detectable plasma HIV-1 RNA. Comparison of the protease and reverse transcriptase gene sequences from paired saliva and plasma of 20 patients showed less than 1% difference overall, and few resistance-related amino acid differences
Conclusions:
Most patients with plasma virus >50 copies/mL had detectable saliva HIV RNA, and the genotypic data was highly concordant between saliva and plasma. In patients with high levels of plasma HIV RNA, saliva might be useful in identifying viremia and evaluating drug resistance.
Article Information
Identifiers and Pagination:
Year: 2010Volume: 4
First Page: 88
Last Page: 93
Publisher Id: TOVJ-4-88
DOI: 10.2174/1874357901004010088
Article History:
Received Date: 1/4/2010Revision Received Date: 19/4/2010
Acceptance Date: 22/4/2010
Electronic publication date: 28/5/2010
Collection year: 2010
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http: //creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Division of Infectious Diseases, Stanford University, 300 Pasteur Drive, Room S-146 Grant Bldg., Stanford, CA 94305, USA; Tel: 650-723-8291; Fax: 650-725-2395; Email: balamane@stanford.edu
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 1-4-2010 |
Original Manuscript | Detection of HIV-1 in Saliva: Implications for Case-Identification, Clinical Monitoring and Surveillance for Drug Resistance§ | |
INTRODUCTION
The current standard of care for HIV-infected individuals includes routine monitoring of plasma virus load (VL) and genotypic testing for antiretroviral (ARV) drug resistance. In resource-rich as well as resource-limited public health settings, there is increasing demand for lower-cost and more feasible community-level virologic testing strategies for clinical monitoring and management of combination ARV therapy. The use of analyte alternatives to blood, particularly those that are easier to collect, have the potential to meet these demands.
Both HIV and anti-HIV antibodies can be detected in saliva [1Freel SA, Williams JM, Nelson JA, et al. Characterization of human immunodeficiency virus type 1 in saliva and blood plasma by V3-specific heteroduplex tracking assay and genotype analyses J Virol 2001; 75: 4936-0.], providing an alternative to blood to detect HIV antibodies to diagnose HIV infection [2Hodinka RL, Nagashunmugam T, Malamud D. Detection of human immunodeficiency virus antibodies in oral fluids Clin Diagn Lab Immunol 1998; 5: 419-26.]. Saliva HIV antibody tests with specialized devices for proper saliva specimen collection are currently licensed by the U.S. Food and Drug Administration (FDA) [3Granade TC, Phillips SK, Parekh B, et al. Detection of antibodies to human immunodeficiency virus type 1 in oral fluids: a large-scale evaluation of immunoassay performance Clin Diagn Lab Immunol 1998; 5: 171-5.]. These tests are used in resource-limited settings as well as domestic public health clinics where rapid testing and ease of use are preferred [4Pascoe SJ, Langhaug LF, Mudzori J, Burke E, Hayes R, Cowan FM. Field evaluation of diagnostic accuracy of an oral fluid rapid test for HIV, tested at point-of-service sites in rural Zimbabwe AIDS Patient Care STDS 2009; 23: 571-6.]. The Centers for Disease Control (CDC) has included rapid HIV testing as part of its policy recommendations for HIV prevention programs [5Advancing HIV prevention: new strategies for a changing epidemic--United States, 2003 MMWR Morb Mortal Wkly Rep 2003; 52: 329-.]. The CDC has also recently promoted provider-initiated testing with reduced consent and counseling barriers, in order to increase accessibility to HIV testing and decrease the continued transmission of HIV [5Advancing HIV prevention: new strategies for a changing epidemic--United States, 2003 MMWR Morb Mortal Wkly Rep 2003; 52: 329-., 6Roberts KJ, Grusky O, Swanson AN. Outcomes of blood and oral fluid rapid HIV testing: a literature review, 2000-2006 AIDS Patient Care STDS 2007; 21: 621-37.].
In voluntary counseling and testing centers, there is a growing interest in detecting acute infection among high-risk individuals. During acute infection, there is a brief period where high levels of HIV replication occur before the development of anti-HIV antibodies. Because VL levels are often greater than 100,000 copies/ml, methods to identify acutely-infected individuals include pooled screening of antibody negative plasma samples, which has substantially mitigated testing costs [7Pilcher CD, Fiscus SA, Nguyen TQ, et al. Detection of acute infections during HIV testing in North Carolina N Engl J Med 2005; 352: 1873-83.]. However, minimally invasive and lower cost sample collection methods have additional potential for increasing community-level identification of acute infection.
VL and drug resistance testing using saliva can help meet the increasing demands to identify acute seroconverters, identify transmitted drug resistance in public health screening for acute infection, and scale up community-based treatment monitoring. Although saliva has been successfully used for antibody testing and for field diagnosis of HIV infection, the feasibility of nucleic acid testing to both detect acute infection and to monitor individuals receiving ARV treatment has not been thoroughly examined. We investigated the feasibility of using saliva to detect and quantify HIV-1 RNA and to genotype HIV-1.
METHODS AND MATERIALS
Study Population
Sixty-nine HIV infected individuals were enrolled in this study under informed consent: 40 recruited from the San Francisco City Clinic (approved by the UCSF Committee for Human Research) and 29 from the Peninsula AIDS Research Consortium clinic at the San Mateo County AIDS Program (approved by the Mills-Peninsula/San Mateo Clinical Research Committee IRB). These were patients attending clinic for HIV care or sexually transmitted disease services. Among 40 patients at the San Francisco City Clinic, a questionnaire was administered for inclusion and exclusion purposes. Questions included presence of oral ulcers or sores, current bleeding in mouth, and whether there was eating, drinking, brushing teeth, flossing or smoking in the past hour prior to saliva collections. A yes answer to any of the questions excluded the individual from participating in the study.
Sample Collection
Patients provided 5-10 ml of saliva by spitting into a sterile cup. Dried blood spots (DBS) were prepared from finger sticks onto Whatman 903 filter paper (approximately 50ul per DBS) and dried for several hours at room temperature prior to packaging and transporting to the processing lab. Saliva was frozen in the clinic and transported to the testing lab on dry ice. Both DBS and saliva were stored at -70c for up to two weeks prior to testing.
Sample Processing
RNA Extraction from Saliva and DBS
HIV-1 RNA plasma VLs were obtained from concurrently drawn clinical tests (Roche AMPLICOR HIV-1 Monitor assay, version 1.5), when available, or performed in house from DBS. Sixty-four virus loads were obtained from plasma while 5 were obtained from DBS. Saliva was homogenized (500uL) and centrifuged at 800g for 10 min to pellet debris, and the resulting supernatant was ultra centrifuged at 23,885 x g for 30 min to pellet virus to match the identical ultracentrifugation performed to pellet virus in corresponding patient plasma samples. The pellet was resuspended in 9mL of Nuclisens lysis buffer. For DBS, spots were placed directly into 9mL of Nuclisens lysis buffer and agitated for 2 hours at room temperature. The Roche Amplicor internal quantification standard (QS) was added to each saliva or DBS sample dissolved in lysis buffer, and RNA extraction completed according to the Nuclisens protocol (Biomerieux, Durham NC, USA). From this point, HIV VL was quantified using the ultrasensitive Roche AMPLICOR HIV-1 Monitor assay, version 1.5 (Roche Molecular Diagnostics, Branchberg NJ, USA), according to the manufacturer’s instructions. DBS VL values were adjusted to account for sample volume input (50ul DBS). All VL values were normalized to 1 ml of fluid and expressed as RNA copies per ml. The lower limit of quantification was defined as 50 RNA copies/ml.
Genotyping and Sequence Analysis
Genotyping was performed on samples with a VL greater than 200 copies/ml as previously described [8Winters MA, Coolley KL, Girard YA, et al. A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside inhibitors J Clin Invest 1998; 102: 1769-75.] when sufficient sample was available. Drug resistance mutations were identified using the Stanford HIV Drug Resistance Database (http://HIVDB.stanford.edu). Sequences were aligned with ClustalW [9Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice Nucleic Acids Res 1994; 22: 4673-80.] and a best-fit nucleotide substitution model was estimated using the program Modeltest [10Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution Bioinformatics 1998; 14: 817-.] and manual examination in PAUP [11Swofford DL. PAUP: Phylogenetic Analysis Using Parsimony (*and Other Methods) 0d64. 1998; v4]. A maximum likelihood (ML) phylogenetic tree (Fig.1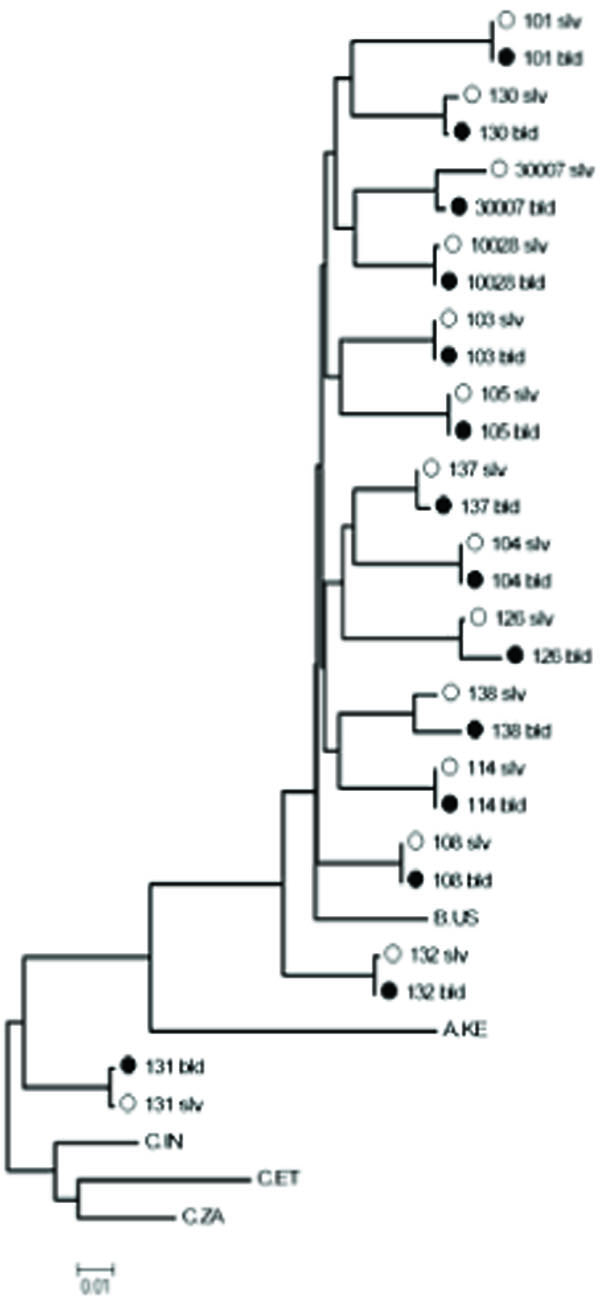 ) was constructed under this model with the program PhyML. Genetic distances between paired saliva and plasma sequences were estimated from this tree.
) was constructed under this model with the program PhyML. Genetic distances between paired saliva and plasma sequences were estimated from this tree.
 |
Fig. (1) Maximum likelihood phylogenetic tree of HIV-1 protease and reverse transcriptase genes comparing blood and saliva samples. (942 nucleotides total; PR codons 22-99 and RT codons 1-238). |
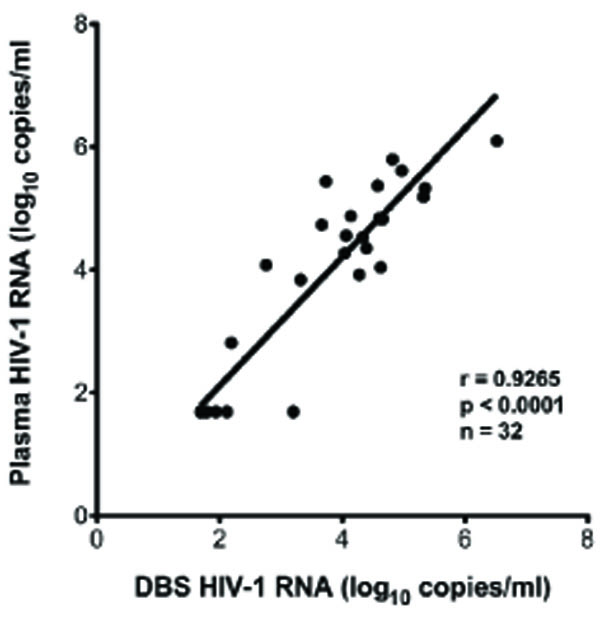 |
Fig. (2a) Correlation of plasma HIV-1 RNA with DBS HIV-1 RNA. |
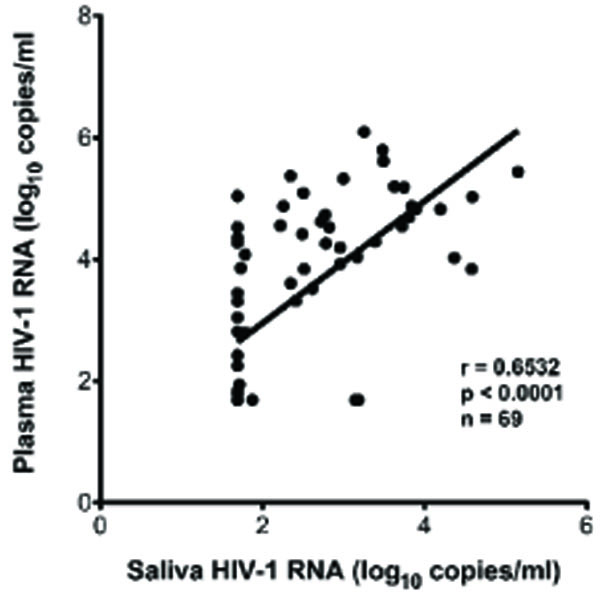 |
Fig. (2b) Correlation of plasma HIV-1 RNA with saliva HIV-1 RNA. |
RESULTS
Validation of VL Determination from DBS and Saliva
Virus spiking experiments were performed to determine the best method for isolating RNA from saliva. High VL HIV-1 plasma of known copy was diluted to 50,000 copies/mL, 10,000 copies/mL and 2,000 copies/mL in normal human plasma and saliva. HIV-1 RNA was successfully recovered from saliva and plasma using Ultrasensitive Roche AMPLICOR HIV-1 Monitor assay Extraction or the Ultrasensitive Nuclisens Extraction Kit. However, the Roche QS was not efficiently recovered from saliva using the Amplicor extraction method [12Shepard RN, Schock J, Robertson K, et al. Quantitation of human immunodeficiency virus type 1 RNA in different biological compartments J Clin Microbiol 2000; 38: 1414-8.] (data not shown), resulting in inaccurate VL measurements. The Ultrasensitive Nuclisens Kit was able to recover both HIV-1 and QS from spiked plasma and saliva as well as negative control samples with equal efficiency, and this method was used to extract RNA for both VL and genotype determinations.
In five cases, the timing and location of specimen acquisition did not include an appropriate plasma sample for VL determination. In these cases, DBS were assayed to determine plasma VL. To validate the use of DBS to measure VL, matched pairs of plasma and DBS were collected from 32 subjects, and VL (median DBS VL=5419, range=49-3,301,925copies/mL) determined on each specimen as described in Materials and Methods. Results (Fig. 2a ) indicate a significant correlation between plasma-derived and DBS derived VL values (p<0.0001). There was a median 0.18 log10 RNA copies/ml difference between plasma and DBS VL values. Based on the strong correlation and small differences between plasma and DBS VL, the five subjects with DBS-derived VL measurements were included in the study.
) indicate a significant correlation between plasma-derived and DBS derived VL values (p<0.0001). There was a median 0.18 log10 RNA copies/ml difference between plasma and DBS VL values. Based on the strong correlation and small differences between plasma and DBS VL, the five subjects with DBS-derived VL measurements were included in the study.
Saliva Virus Load
Overall, HIV-1 RNA was detected in 47 of 69 (68%) plasma and 39 of 69 (56%) saliva specimens. Of the 47 subjects with plasma viremia (median 22,514 copies/ml, range 68 to 1,244,229 copies/ml); 36 patients (77%) had measurable HIV RNA in saliva (median 75 copies/ml, range 52 to 141,621 copies/ml). There was a significant correlation between plasma and saliva VL in these subjects (Fig. 2b ), p<.0001). Patients with detectable saliva VL had a median plasma VL of 33,530 copies/ml, range 0 to 1,244,229 copies/ml. Patients without detectable saliva had a median plasma VL of 0 copies/ml, and range 0 to 109,000 copies/ml Plasma VLs were higher than saliva VL levels by an average 0.9 log10 RNA copies/ml. Two subjects exhibited a higher VL in saliva than in plasma, while three subjects had detectable HIV-1 RNA in saliva but had undetectable plasma VL.
), p<.0001). Patients with detectable saliva VL had a median plasma VL of 33,530 copies/ml, range 0 to 1,244,229 copies/ml. Patients without detectable saliva had a median plasma VL of 0 copies/ml, and range 0 to 109,000 copies/ml Plasma VLs were higher than saliva VL levels by an average 0.9 log10 RNA copies/ml. Two subjects exhibited a higher VL in saliva than in plasma, while three subjects had detectable HIV-1 RNA in saliva but had undetectable plasma VL.
Sequence Conservation and Drug Resistance Between Plasma and Saliva
To our knowledge, there is no previous literature reporting the sequencing of the gag-pol gene of HIV-1 from saliva specimens. A gag-pol fragment of nearly 1,000 base-pairs was sequenced from both plasma and saliva in 20 subjects. A phylogenetic tree showed that saliva and plasma sequences within individuals were 1) closely related (average genetic distance 0.0043 nucleotide substitutions/site; range 0.0011-0.0196); 2) highly concordant (average genotypic concordance >99% at both the nucleotide and AA level); and 3) segregated by subject. Thirteen subjects had genotypic resistance mutations in either plasma, saliva, or both (Table 1). In the protease (PR) gene, nine patients had minor resistance mutations (L10I, A71AV and A71V) with no major resistance mutations. Discordance in the presence of PR mutations between plasma and saliva was found in five subjects; in each case a minor PR mutation was found in the plasma that was not present in saliva. In the reverse transcriptase (RT) region, six patients were found to harbor resistance mutations (D67N, K103N, V108I, V118I, E138A, G190A and the revertant mutation T215S). Differences in RT resistance mutations were seen between plasma and saliva genotypes in four of six subjects; in two subjects mutations were identified in saliva but not plasma, and in two subjects mutations were found in plasma but not in saliva. In the PR gene 7/19 sequences demonstrated at least one difference between saliva and plasma in polymorphic residues associated with protease inhibitor resistance (secondary or minor mutations), while 4/19 RT sequences differed at major drug resistance codons.
DISCUSSION
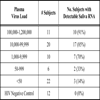
Previous studies have shown that non-plasma samples, including oral transudate, urine, and DBS, are appropriate analytes for the detection of antibodies to HIV to diagnose HIV infection [2Hodinka RL, Nagashunmugam T, Malamud D. Detection of human immunodeficiency virus antibodies in oral fluids Clin Diagn Lab Immunol 1998; 5: 419-26.]. As a candidate diagnostic analyte, saliva can be easily obtained at lower cost and without the discomfort, invasiveness, and subsequent risks associated with needle use and disposal in conventional phlebotomy. Similarly, DBS provide an alternative means of blood collection for VL testing [13Reigadas S, Schrive MH, Aurillac-Lavignolle V, Fleury HJ. Quantitation of HIV-1 RNA in dried blood and plasma spots J Virol Methods 2009; 161: 177-80.]. We have found that among public health clinic patients, HIV-1 RNA could be successfully isolated, extracted, reverse transcribed and amplified from a relatively small volume of saliva (500ul). Comparison of viral RNA detection and quantification from saliva and plasma demonstrated that most subjects (83%) with more than 1,000 RNA copies/ml in plasma had detectable viral RNA in their oral secretions (Table 2a).
Comparison of DBS and plasma for the detection and quantification of HIV-1 RNA showed good correlation (Fig.2a ). This is consistent with a growing literature in which dried blood spots have been evaluated in comparison to plasma [14Hamers RL, Smit PW, Stevens W, Schuurman R, Rinke de Wit TF. Dried fluid spots for HIV type-1 viral load and resistance genotyping: a systematic review Antivir Ther 2009; 14: 619-29.]. As expected from previous studies [15Liuzzi G, Chirianni A, Clementi M, et al. Analysis of HIV-1 load in blood, semen and saliva: evidence for different viral compartments in a cross-sectional and longitudinal study AIDS 1996; 10: F51-6., 16Shugars DC, Slade GD, Patton LL, Fiscus SA. Oral and systemic factors associated with increased levels of human immunodeficiency virus type 1 RNA in saliva Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2000; 89: 432-0.], VL in saliva contained less HIV-1 RNA compared to plasma, however most viremic patients in our study had detectable saliva RNA. Overall, the sensitivity of saliva VL was found to be 77% compared to plasma (Table 2b). Sensitivity could be low for technical or biological reasons. There might not be any HIV present in the saliva compartment for some patients due to no localized replication. While this sensitivity may be insufficient to monitor patients on suppressive ARV, the high proportion of saliva RNA-positivity when plasma VL is higher (95% at plasma VL > 33,500 copies/ml) may make saliva a suitable analyte for evaluating untreated or acutely-infected individuals.
). This is consistent with a growing literature in which dried blood spots have been evaluated in comparison to plasma [14Hamers RL, Smit PW, Stevens W, Schuurman R, Rinke de Wit TF. Dried fluid spots for HIV type-1 viral load and resistance genotyping: a systematic review Antivir Ther 2009; 14: 619-29.]. As expected from previous studies [15Liuzzi G, Chirianni A, Clementi M, et al. Analysis of HIV-1 load in blood, semen and saliva: evidence for different viral compartments in a cross-sectional and longitudinal study AIDS 1996; 10: F51-6., 16Shugars DC, Slade GD, Patton LL, Fiscus SA. Oral and systemic factors associated with increased levels of human immunodeficiency virus type 1 RNA in saliva Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2000; 89: 432-0.], VL in saliva contained less HIV-1 RNA compared to plasma, however most viremic patients in our study had detectable saliva RNA. Overall, the sensitivity of saliva VL was found to be 77% compared to plasma (Table 2b). Sensitivity could be low for technical or biological reasons. There might not be any HIV present in the saliva compartment for some patients due to no localized replication. While this sensitivity may be insufficient to monitor patients on suppressive ARV, the high proportion of saliva RNA-positivity when plasma VL is higher (95% at plasma VL > 33,500 copies/ml) may make saliva a suitable analyte for evaluating untreated or acutely-infected individuals.
Although HIV-1 is transmitted through blood, semen, breast milk, and cervical secretions, the evidence for transmissible virus in saliva is less convincing [17Robinson EK, Evans BG. Oral sex and HIV transmission AIDS 1999; 13: 737-8.]. In the past, researchers have found that VL from non blood sources can be 1 to 2 log values lower than VL from corresponding plasma samples. The relatively low level of virus in saliva (median saliva VL= 75copies/mL) may be one explanation for the apparent infrequency of transmission of virus from saliva [18Campo J, Perea MA, del Romero J, Cano J, Hernando V, Bascones A. Oral transmission of HIV, reality or fiction? An update Oral Dis 2006; 12: 219-8., 19Shugars DC, Wahl SM. The role of the oral environment in HIV-1 transmission J Am Dent Assoc 1998; 129: 851-.]. However, transmission rates are higher in other non blood analytes than in saliva regardless of lower VLs in those compartments. There is proposed evidence to suggest existence of anti viral activity in the oral environment preventing HIV-1 transmission [19Shugars DC, Wahl SM. The role of the oral environment in HIV-1 transmission J Am Dent Assoc 1998; 129: 851-.]. We were surprised to identify three individuals who had detectable saliva VL while having a plasma VL of < 50 copies/ml. In addition we found two individuals with saliva HIV-1 RNA levels (> 20,000 copies/ml) that were 2 to 5-fold greater than their respective plasma levels. It is possible that undetected inflammatory responses in the oral cavity may have increased the VL in the patients’ respective saliva. Although multiple studies of HIV-1 RNA in virus particles from non-blood fluids have shown evidence for compartmentalized viral replication [12Shepard RN, Schock J, Robertson K, et al. Quantitation of human immunodeficiency virus type 1 RNA in different biological compartments J Clin Microbiol 2000; 38: 1414-8.], in our study, it is unclear if independent replication is occurring in different biological compartments. In the present analysis, differences in the detection and quantification of viral RNA in a small number of subjects suggest that the oral cavity may support compartmental replication and shedding in HIV-infected individuals and these rates of replication may be different for different patients. However, further studies need to be performed to further evaluate this as inflammatory conditions of the mouth were not closely examined.
Several viral characteristics may be inferred from sequence analysis of HIV RNA including subtype, diversity [20Delwart EL, Mullins JI, Gupta P, et al. Human immunodeficiency virus type 1 populations in blood and semen J Virol 1998; 72: 617-23.], drug resistance [21Kassaye S, Lee E, Kantor R, et al. Drug resistance in plasma and breast milk after single-dose nevirapine in subtype C HIV type 1: population and clonal sequence analysis AIDS Res Hum Retroviruses 2007; 23: 1055-61.] and envelope tropism [22Freel SA, Fiscus SA, Pilcher CD, et al. Envelope diversity, coreceptor usage and syncytium-inducing phenotype of HIV-1 variants in saliva and blood during primary infection AIDS 2003; 17: 2025-33.]. Our phylogenetic comparisons of gag-pol gene sequences obtained from DBS, plasma and saliva showed considerable sequence homology and provided assurance that sample-mix-up or contamination did not occur. These findings are in agreement with previous studies where sequencing of the envelope gene in saliva and plasma was concordant [22Freel SA, Fiscus SA, Pilcher CD, et al. Envelope diversity, coreceptor usage and syncytium-inducing phenotype of HIV-1 variants in saliva and blood during primary infection AIDS 2003; 17: 2025-33.]. However, while comparison of plasma and saliva HIV-1 pol-genes demonstrated only modest overall differences (<1%) in nucleic and amino acid sequences from each compartment, examination of specific codons suggest potentially important differences between saliva and plasma virus. In 8/20 patients, drug-associated resistance mutations in the PR and/or RT gene were found in only one compartment. These differences were primarily found at minor mutation sites in our population of primarily untreated patients, except for one treated patient where a K103N mutation was found in plasma-associated virus but not in saliva. While it is possible that sampling bias may have affected our ability to detect mutations in different compartments, all of the samples genotyped met similar viral load criteria (>200 copies/ml) to be included in the analysis. Our findings of different genotype profiles in plasma vs saliva suggest variations in selective pressure between these two compartments. Further analysis of resistance mutations between saliva and plasma in highly ARV experienced patients is needed to further evaluate this hypothesis.
In summary, saliva may be a useful analyte for detection of HIV-1 when HIV-1 RNA levels are high, such as in acute HIV infection. In monitoring responses to ARV therapy, saliva VL measures have reduced sensitivity and may not accurately reflect VL in plasma, but nevertheless could identify high-level viremia during treatment failure. The use of HIV-1 sequences to monitor drug resistance may be possible, although additional analysis of compartmental differences is needed. Further studies on compartmental differences in HIV viremia and drug resistance can help optimize saliva testing as an alternative analyte for monitoring patients in clinical settings.
NOTES
§ Poster presented at International AIDS Conference, Mexico City, Mexico, August 2008.
ACKNOWLEDGEMENT
California HIV Research Program (CHRP) grant: CH05-SMCH-612.