- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
The Insertion of Fluorescent Proteins in a Variable Region of Respiratory Syncytial Virus L Polymerase Results in Fluorescent and Functional Enzymes But with Reduced Activities
Jenna Fix, Marie Galloux, Marie-Lise Blondot, Jean-François Eléouët*
Abstract
The respiratory syncytial virus (RSV) Large protein L is the catalytic subunit of the RNA-dependent RNA polymerase complex. Currently, no structural information is available for RSV L. Sequence alignments of L protein from human and bovine strains of RSV revealed the existence of two variable regions, VR1 and VR2. Following comparison with morbillivirus and rhabdovirus L genes, VR2, which is located between domains V and VI, was chosen as an insertion site for sequences encoding the epitope tag HA or the fluorescent proteins eGFP and mCherry. Recombinant tagged-L proteins co-localized with RSV N and P proteins in transfected cells. These recombinant polymerases were shown to be functional using a viral minigenome system assay, their activities being reduced by ~70% compared to the unmodified L polymerase. We have also shown by site-directed mutagenesis that the GDNQ motif (residues 810-813 for the Long strain of HRSV) is essential for L activity.
Article Information
Identifiers and Pagination:
Year: 2011Volume: 5
First Page: 103
Last Page: 108
Publisher Id: TOVJ-5-103
DOI: 10.2174/1874357901105010103
Article History:
Received Date: 16/5/2011Revision Received Date: 29/6/2011
Acceptance Date: 13/7/2011
Electronic publication date: 6/9/2011
Collection year: 2011
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http: //creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the INRA, Unité de Virologie Immunologie Moléculaires UR892, F-78350 Jouy-en-Josas, France; Tel: (33) 1 34 65 26 04; Fax: (33) 1 34 65 26 21; E-mail: jean-francois.eleouet@jouy.inra.fr
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 16-5-2011 |
Original Manuscript | The Insertion of Fluorescent Proteins in a Variable Region of Respiratory Syncytial Virus L Polymerase Results in Fluorescent and Functional Enzymes But with Reduced Activities | |
Human and bovine respiratory syncytial viruses (HRSV and BRSV) are two closely related, highly infectious, worldwide prevalent viruses that are the leading cause of acute lower respiratory tract disease in children and calves, respectively [1Collins PL, Crowe JE. Respiratory Syncytial Virus and Metapneumovirus In: Knipe DM, Howley PM, Eds. In Fields Virology. 5th. Philadelphia: Lippincott Williams & Wilkins 2007; pp. 1601-46.]. RSV is a negative strand RNA virus that belongs to the Pneumovirus genus within the Paramyxoviridae family. The 15 kb negative-strand genomic RNA is encapsidated by the nucleoprotein (N) forming a ribonucleoprotein complex (RNP) which is the template for the viral RNA-dependent RNA polymerase (RdRp) complex [2Cowton VM, McGivern DR, Fearns R. Unravelling the complexities of respiratory syncytial virus RNA synthesis J Gen Virol 2006; 87(Pt 7): 1805-21.]. The RdRp minimum complex is composed of the large catalytic subunit L and its cofactor the phosphoprotein P. The replicase activity also requires N, while efficient transcriptase activity needs the M2-1 anti-termination factor [3Collins PL, Hill MG, Camargo E, Grosfeld H, Chanock RM, Murphy BR. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5' proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development Proc Natl Acad Sci USA 1995; 92(25): 11563-7.]. Cell infection by RSV induces the formation of cytoplasmic inclusion bodies [4Norrby E, Marusyk H, Orvell C. Morphogenesis of respiratory syncytial virus in a green monkey kidney cell line (Vero) J Virol 1970; 6(2): 237-42.] containing N, P, M2-1, L, the matrix protein (M), the non-structural protein 2 (NS2), low amounts of the small hydrophobic SH protein, viral RNA, and the cellular proteins Hsp70 and actin [5Garcia J, Garcia-Barreno B, Vivo A, Melero JA. Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22K protein Virology 1993; 195(1): 243-7.-12Lindquist ME, Lifland AW, Utley TJ, Santangelo PJ, Crowe JE Jr. Respiratory syncytial virus induces host RNA stress granules to facilitate viral replication J Virol 2010; 84(23): 12274-84.]. In rhabdovirus-infected cells, inclusion bodies were shown to be sites of viral RNA synthesis [13Lahaye X, Vidy A, Pomier C, et al. Functional characterization of Negri bodies (NBs) in rabies virus-infected cells: Evidence that NBs are sites of viral transcription and replication J Virol 2009; 83(16): 7948-58., 14Heinrich BS, Cureton DK, Rahmeh AA, Whelan SP. Protein expression redirects vesicular stomatitis virus RNA synthesis to cytoplasmic inclusions PLoS Pathog 2010; 6(6): e1000958.]. For this reason, it is thought that these structures are likely to be sites of replication and/or transcription for RSV [15Santangelo P, Nitin N, LaConte L, Woolums A, Bao G. Live-cell characterization and analysis of a clinical isolate of bovine respiratory syncytial virus, using molecular beacons J Virol 2006; 80(2): 682-8.]. Until recently, detection of RSV L in cells has been hampered by the lack of suitable antibodies [9Carromeu C, Simabuco FM, Tamura RE, Farinha Arcieri LE, Ventura AM. Intracellular localization of human respiratory syncytial virus L protein Arch Virol 2007; 152(12): 2259-63.].
As for all of the polymerases of non-segmented negative-stranded viruses (NNSV), the L of RSV is believed to perform all of the catalytic functions, e.g. RNA synthesis, capping, methylation and polyadenylation [16Poch O, Blumberg BM, Bougueleret L, Tordo N. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains J Gen Virol 1990; 71(Pt 5): 1153-62., 17Stec DS, Hill MG 3rd, Collins PL. Sequence analysis of the polymerase L gene of human respiratory syncytial virus and predicted phylogeny of nonsegmented negative-strand viruses Virology 1991; 183(1): 273-87.]. Analysis of the primary sequences of L revealed the presence of six blocks of conserved regions (CRs I-VI) shared among all Mononegavirales L proteins [16Poch O, Blumberg BM, Bougueleret L, Tordo N. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains J Gen Virol 1990; 71(Pt 5): 1153-62.]. Recent structural studies of the L encoded by the rhabdovirus vesicular stomatitis virus (VSV) have shown that these CRs correspond to physical distinct functional domains, domains I-IV forming a ring domain, and domains V-VI forming an appendage of three globular domains [18Rahmeh AA, Schenk AD, Danek EI, et al. Molecular architecture of the vesicular stomatitis virus RNA polymerase Proc Natl Acad Sci USA 2010; 107(46): 20075-80.]. A polymerase signature motif GDNQ, which is thought to be the active site for phosphodiester bond formation, lies in the CR III (residues 810-813) of the HRSV Long strain [17Stec DS, Hill MG 3rd, Collins PL. Sequence analysis of the polymerase L gene of human respiratory syncytial virus and predicted phylogeny of nonsegmented negative-strand viruses Virology 1991; 183(1): 273-87.].
Sequence alignments of Morbillivirus (also members of the Paramyxoviridae family) L proteins revealed three conserved domains (D1, D2, D3) separated by two highly variable regions termed « hinges » H1 and H2 [19Duprex WP, Collins FM, Rima BK. Modulating the function of the measles virus RNA-dependent RNA polymerase by insertion of green fluorescent protein into the open reading frame J Virol 2002; 76(14): 7322-8.]. It was shown that insertion of eGFP in the second hinge (H2) had an attenuation effect on the virus replication but did not abolish the polymerase activity [19Duprex WP, Collins FM, Rima BK. Modulating the function of the measles virus RNA-dependent RNA polymerase by insertion of green fluorescent protein into the open reading frame J Virol 2002; 76(14): 7322-8., 20Brown DD, Rima BK, Allen IV, et al. Rational attenuation of a morbillivirus by modulating the activity of the RNA-dependent RNA polymerase J Virol 2005; 79(22): 14330-8.]. However, the sequence of L RSV doesn’t share enough sequence homology with those of the Morbilliviruses L proteins to assign equivalent domains.
To obtain preliminary data on the organization of the RSV L protein, we aligned L sequences from human RSV (HRSV) and bovine RSV (BRSV) strains and found that these L proteins are highly conserved except for two highly variable regions, that we designated VR1 and VR2 (Fig. 1A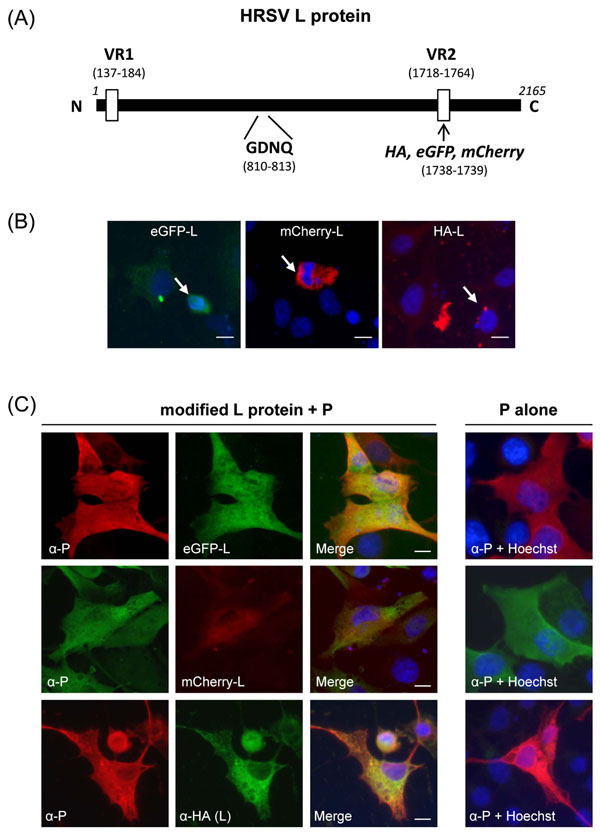 ). The VR2 region includes residues 1717 to 1764 in human RSV Long strain and is located between CRs V and VI, which contain putative capping and cap methylation activities, respectively. We investigated whether the RSV L VR2 region could tolerate sequence insertions, as has been previously shown for morbilliviruses. In this study, we used plasmids pN, pP, pM2-1 and pL coding for HRSV (strain Long) N, P, M2-1 and L proteins, respectively, under the control of the T7 promoter [21Tran TL, Castagne N, Dubosclard V, et al. The respiratory syncytial virus M2-1 protein forms tetramers and interacts with RNA and P in a competitive manner J Virol 2009; 83(13): 6363-74.]. An encephalomyocarditis virus internal ribosome entry site (IRES) sequence was placed between the T7 promoter and the inserted ORF to enhance protein expression in BSR/T7-5 cells, a BHK21 clone stably expressing T7 RNA polymerase [22Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter J Virol 1999; 73(1): 251-9.], as previously described [23Jin H, Clarke D, Zhou HZ, et al. Recombinant human respiratory syncytial virus (RSV) from cDNA and construction of subgroup A and B chimeric RSV Virology 1998; 251(1): 206-14.]. We introduced a unique restriction enzyme site at nucleotide 5212 of the L gene in the T7 polymerase-driven expression pL vector, by site directed mutagenesis (Quickchange, Stratagene). Codons 1738 and 1739 (GTT GAC) were substituted with GTC GAC, creating a SalI site without any change in the L amino acid sequence. We then generated three L protein expression plasmids, pHA-L, pmCherry-L and peGFP-L, encoding the L protein tagged with an HA epitope (42 bp), mCherry (Clontech, 708 bp), and eGFP (Clontech, 718 bp), respectively. To do so, complementary oligonucleotides encoding a HA tag epitope (sequences available on request) were annealed to generate SalI-compatible ends. The resulting fragment was inserted into the SalI restriction site present in VR2 in frame with the L sequence. EGFP and mCherry genes were amplified by standard techniques and cloned in-frame into the SalI site of VR2.
). The VR2 region includes residues 1717 to 1764 in human RSV Long strain and is located between CRs V and VI, which contain putative capping and cap methylation activities, respectively. We investigated whether the RSV L VR2 region could tolerate sequence insertions, as has been previously shown for morbilliviruses. In this study, we used plasmids pN, pP, pM2-1 and pL coding for HRSV (strain Long) N, P, M2-1 and L proteins, respectively, under the control of the T7 promoter [21Tran TL, Castagne N, Dubosclard V, et al. The respiratory syncytial virus M2-1 protein forms tetramers and interacts with RNA and P in a competitive manner J Virol 2009; 83(13): 6363-74.]. An encephalomyocarditis virus internal ribosome entry site (IRES) sequence was placed between the T7 promoter and the inserted ORF to enhance protein expression in BSR/T7-5 cells, a BHK21 clone stably expressing T7 RNA polymerase [22Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter J Virol 1999; 73(1): 251-9.], as previously described [23Jin H, Clarke D, Zhou HZ, et al. Recombinant human respiratory syncytial virus (RSV) from cDNA and construction of subgroup A and B chimeric RSV Virology 1998; 251(1): 206-14.]. We introduced a unique restriction enzyme site at nucleotide 5212 of the L gene in the T7 polymerase-driven expression pL vector, by site directed mutagenesis (Quickchange, Stratagene). Codons 1738 and 1739 (GTT GAC) were substituted with GTC GAC, creating a SalI site without any change in the L amino acid sequence. We then generated three L protein expression plasmids, pHA-L, pmCherry-L and peGFP-L, encoding the L protein tagged with an HA epitope (42 bp), mCherry (Clontech, 708 bp), and eGFP (Clontech, 718 bp), respectively. To do so, complementary oligonucleotides encoding a HA tag epitope (sequences available on request) were annealed to generate SalI-compatible ends. The resulting fragment was inserted into the SalI restriction site present in VR2 in frame with the L sequence. EGFP and mCherry genes were amplified by standard techniques and cloned in-frame into the SalI site of VR2.
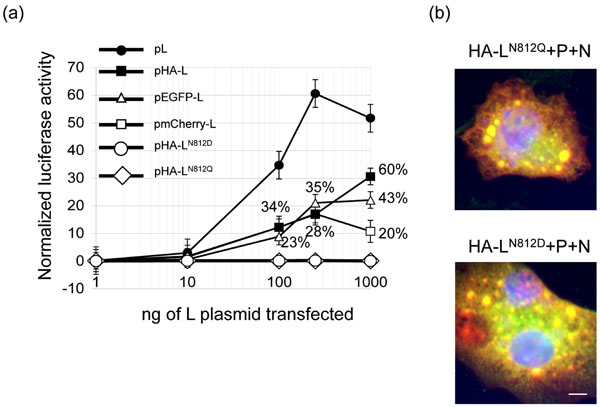 |
Fig. (3) Activity of tagged L polymerases. (A) Relative activities of wild type, HA-, mCherry-, eGFP-tagged and N812Q and N812D mutant HA-tagged L polymerases in a series of minigenome rescue experiments. Various amounts of pL (filled circles), pHA-L (crosses), pmCherry-L (open squares), peGFP-L (open triangles), and pHA-LN812D (open circles) or pHA-LN812Q (filled diamonds) were used in minigenome transfection assays. Each luciferase minigenome activity value was normalized 24 h after transfection based on β-galactosidase expression and is the average of three independent experiments assayed in triplicate. The indicated percentages represent the relative activity of the structurally modified RdRp compared to the wild type control. Error bars denote standard deviations. (B) HA-LN812Q and HA-LN812D co-localize with inclusion bodies formed by P and N. BSRT7/5 cells were co-transfected with pP, pN and either pHA-LN812D or pHA-LN812Q, fixed and labeled with anti-P (red) and anti-HA (green) antibodies as described in the Fig. (1 |
The expression of HA-L, mCherry-L and eGFP-L and their ability to co-localize with RSV P and N proteins in cells was analyzed by fluorescence microscopy. BSR/T7-5 cells grown on coverslips in 24 well plates were independently transfected either with 0.4 µg of pHA-L, pmCherry-L, or peGFP-L vector alone, or together with 0.3 µg of pP, or 0.1 µg of pN, or with both pP and pN, using Lipofectamine 2000 (Invitrogen). Twenty four hours post-transfection cells were fixed with 4% paraformaldehyde in PBS for 30 min, permeabilized for 5 min with 0.1% Triton X-100 and 0.5% BSA in PBS and incubated for 1 h at room temperature with the primary antibodies diluted 1:100 in PBS. For HA-L, we used a mouse anti-HA monoclonal antibody (Sigma, clone HA-7), a rabbit anti-P antiserum previously described [24Castagne N, Barbier A, Bernard J, et al. Biochemical characterization of the respiratory syncytial virus P-P and P-N protein complexes and localization of the P protein oligomerization domain J Gen Virol 2004; 85(Pt 6): 1643-53.] and a mouse anti-N (Serotec) for P and N labeling, respectively. Cells were then washed with PBS and incubated for an additional hour with either FITC-conjugated sheep anti-mouse (P.A.R.I.S), Alexa Fluor-488 goat anti-rabbit, Alexa Fluor-594 rabbit anti-mouse, or Alexa594 goat anti-rabbit (Invitrogen) IgG, depending of the presence of eGFP- or mCherry- recombinant L constructs. Nuclei were stained with Hoechst 33342 (Invitrogen). Cells were observed with a Nikon TE200 microscope and images were processed using MetaVue software (Molecular Devices). When HA-L, eGFP-L, and mCherry-L were expressed alone, no or very low levels of fluorescence were observed except in a few cells that were showing an apoptotic phenotype, i.e. rounded cells with condensed nuclei (Fig. 1B ). Similar results were obtained when the various recombinant L constructs were co-expressed with N in the absence of P (data not shown). These results suggest that when expressed alone or with N, the L protein is toxic to the cells. Interestingly, when P was co-transfected with each L construct, a low but specific fluorescence level was observed in the cytoplasm of HA-L, eGFP-L and mCherry-L transfected cells (Fig. 1C
). Similar results were obtained when the various recombinant L constructs were co-expressed with N in the absence of P (data not shown). These results suggest that when expressed alone or with N, the L protein is toxic to the cells. Interestingly, when P was co-transfected with each L construct, a low but specific fluorescence level was observed in the cytoplasm of HA-L, eGFP-L and mCherry-L transfected cells (Fig. 1C ). Although co-localization of P and L within the cytoplasm was not clear, these results indicate that co-expression of P together with L in the absence of other RSV proteins may stabilize L or render it less toxic to the cells. Indeed, it was previously shown that the presence of P induces VSV L rearrangements, the formation of VSV L dimers [18Rahmeh AA, Schenk AD, Danek EI, et al. Molecular architecture of the vesicular stomatitis virus RNA polymerase Proc Natl Acad Sci USA 2010; 107(46): 20075-80.] and seems to stabilize VSV L [25Canter DM, Jackson RL, Perrault J. Faithful and efficient in vitro reconstitution of vesicular stomatitis virus transcription using plasmid-encoded L and P proteins Virology 1993; 194(2): 518-29., 26Canter DM, Perrault J. Stabilization of vesicular stomatitis virus L polymerase protein by P protein binding: a small deletion in the C-terminal domain of L abrogates binding Virology 1996; 219(2): 376-86.].
). Although co-localization of P and L within the cytoplasm was not clear, these results indicate that co-expression of P together with L in the absence of other RSV proteins may stabilize L or render it less toxic to the cells. Indeed, it was previously shown that the presence of P induces VSV L rearrangements, the formation of VSV L dimers [18Rahmeh AA, Schenk AD, Danek EI, et al. Molecular architecture of the vesicular stomatitis virus RNA polymerase Proc Natl Acad Sci USA 2010; 107(46): 20075-80.] and seems to stabilize VSV L [25Canter DM, Jackson RL, Perrault J. Faithful and efficient in vitro reconstitution of vesicular stomatitis virus transcription using plasmid-encoded L and P proteins Virology 1993; 194(2): 518-29., 26Canter DM, Perrault J. Stabilization of vesicular stomatitis virus L polymerase protein by P protein binding: a small deletion in the C-terminal domain of L abrogates binding Virology 1996; 219(2): 376-86.].
Fig. (2B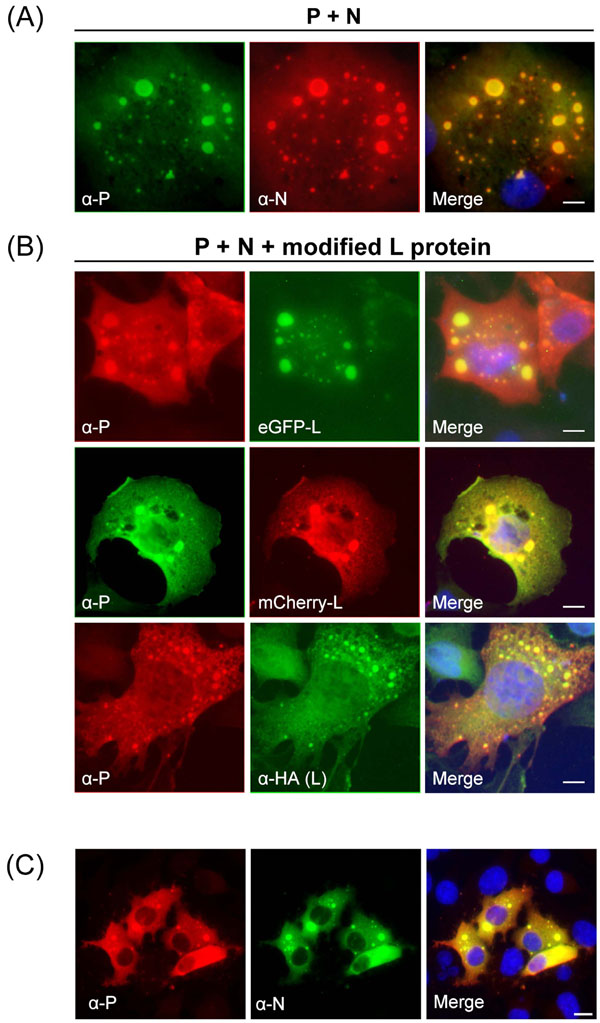 ) shows co-expression of HA-L, eGFP-L or L-mCherry together with P and N, which resulted in the formation of cytoplasmic inclusions bodies containing L, N and P, that were similar to those observed in RSV-infected cells (Fig. 2C
) shows co-expression of HA-L, eGFP-L or L-mCherry together with P and N, which resulted in the formation of cytoplasmic inclusions bodies containing L, N and P, that were similar to those observed in RSV-infected cells (Fig. 2C ) [9Carromeu C, Simabuco FM, Tamura RE, Farinha Arcieri LE, Ventura AM. Intracellular localization of human respiratory syncytial virus L protein Arch Virol 2007; 152(12): 2259-63.]. The computer-generated merged images showed yellow patches, indicative of co-localization of L and P. Co-transfection of BSRT7/5 cells with pP and pN in the absence of L also induced cytoplasmic inclusions bodies containing both P and N (Fig. 2A
) [9Carromeu C, Simabuco FM, Tamura RE, Farinha Arcieri LE, Ventura AM. Intracellular localization of human respiratory syncytial virus L protein Arch Virol 2007; 152(12): 2259-63.]. The computer-generated merged images showed yellow patches, indicative of co-localization of L and P. Co-transfection of BSRT7/5 cells with pP and pN in the absence of L also induced cytoplasmic inclusions bodies containing both P and N (Fig. 2A ). This is consistent with previous studies [5Garcia J, Garcia-Barreno B, Vivo A, Melero JA. Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22K protein Virology 1993; 195(1): 243-7.]. To compare structures seen inside transfected cells with those seen during virus infection, HEp-2 cells were infected with hRSV, fixed 24 h after infection and stained with anti-N and anti-P antibodies. As shown in Fig. (2C
). This is consistent with previous studies [5Garcia J, Garcia-Barreno B, Vivo A, Melero JA. Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22K protein Virology 1993; 195(1): 243-7.]. To compare structures seen inside transfected cells with those seen during virus infection, HEp-2 cells were infected with hRSV, fixed 24 h after infection and stained with anti-N and anti-P antibodies. As shown in Fig. (2C ), P and N co-localized in cytoplasmic inclusion bodies similar to those observed in BSRT7 co-transfected with pN and pP. Taken together our data show that BSRT7/5 cells transfected with pN and pP form cytoplasmic inclusion bodies that are morphologically indistinguishable from those seen in virus infected cells. Moreover transfection of plasmids coding for L proteins harboring short (HA tag) or longer (mCherry and eGFP) in-frame insertions in their VR2 region resulted in the expression of tagged L proteins that can interact with the RSV N and P proteins, leading to their incorporation in those cytoplasmic inclusion bodies.
), P and N co-localized in cytoplasmic inclusion bodies similar to those observed in BSRT7 co-transfected with pN and pP. Taken together our data show that BSRT7/5 cells transfected with pN and pP form cytoplasmic inclusion bodies that are morphologically indistinguishable from those seen in virus infected cells. Moreover transfection of plasmids coding for L proteins harboring short (HA tag) or longer (mCherry and eGFP) in-frame insertions in their VR2 region resulted in the expression of tagged L proteins that can interact with the RSV N and P proteins, leading to their incorporation in those cytoplasmic inclusion bodies.
Finally, the activity of HA-L, mCherry-L and eGFP-L modified polymerases was assessed using an RSV minigenome encoding a luciferase (LUC) reporter gene. As residues D and N in the GDNQ motif within the L gene of Rinderpest virus were previously demonstrated to be critically important [27Chattopadhyay A, Raha T, Shaila MS. Effect of single amino acid mutations in the conserved GDNQ motif of L protein of Rinderpest virus on RNA synthesis in vitro and in vivo Virus Res 2004; 99(2): 139-45.], we changed the putative catalytic site GDNQ motif (residues 810-813) of the RSV L protein to GDDQ and GDQQ in the pHA-L vector, using site-directed mutagenesis (Quickchange, Stratagene) in order to generate an inactive L protein. BSR/T7-5 cells in 24 well plates were co-transfected with 0.5 µg of pM/Luc encoding the RSV minigenome containing the firefly LUC reporter gene [21Tran TL, Castagne N, Dubosclard V, et al. The respiratory syncytial virus M2-1 protein forms tetramers and interacts with RNA and P in a competitive manner J Virol 2009; 83(13): 6363-74.], 0.25µg of pRSV-β-Gal (Promega) to normalize transfection efficiencies, 0.5 µg of pN, 0.5 µg of pP, 0.5 µg of pM2-1 and increasing amounts of one of the L expression plasmids, peGFP-L, pmCherry-L, pHA-L, pHA-LN812D, pHA-LN812Q, or pL encoding the wild-type polymerase (wt L). Cells were harvested 24 h post-transfection, lyzed in 100 µl of luciferase lysis buffer (30 mM Tris pH 7.9, 10 mM MgCl2, 1 mM DTT, 1% (v/v) Triton X-100, and 15% (v/v) glycerol), and luciferase (LUC) activity was determined twice for each cell lysate (40 µl) with an Anthos Lucy 3 luminometer (Bio Advance). LUC activities were normalized based on β-galactosidase expression. Transfections were done in triplicates. As shown on Fig. (3A ), maximum levels of LUC activity were obtained when 250 ng of wt L expressing plasmid was used for the transfection. The three constructs HA-L, mCherry-L and eGFP-L presented an RNA polymerase activity, as demonstrated by LUC activity, but with relative activities of about 30% of the wt L. No activity was detected for LN812Q and LN812D although both were still able to co-localize with P and N in cells (Fig. 3B
), maximum levels of LUC activity were obtained when 250 ng of wt L expressing plasmid was used for the transfection. The three constructs HA-L, mCherry-L and eGFP-L presented an RNA polymerase activity, as demonstrated by LUC activity, but with relative activities of about 30% of the wt L. No activity was detected for LN812Q and LN812D although both were still able to co-localize with P and N in cells (Fig. 3B ). These results demonstrate that, as has been shown for morbilliviruses [19Duprex WP, Collins FM, Rima BK. Modulating the function of the measles virus RNA-dependent RNA polymerase by insertion of green fluorescent protein into the open reading frame J Virol 2002; 76(14): 7322-8., 20Brown DD, Rima BK, Allen IV, et al. Rational attenuation of a morbillivirus by modulating the activity of the RNA-dependent RNA polymerase J Virol 2005; 79(22): 14330-8.] and rhabdoviruses [28Ruedas JB, Perrault J. Insertion of enhanced green fluorescent protein in a hinge region of vesicular stomatitis virus L polymerase protein creates a temperature-sensitive virus that displays no virion-associated polymerase activity in vitro J Virol 2009; 83(23): 12241-52.], the pneumovirus RSV L can tolerate a large insertion of 11% within its coding sequence.
). These results demonstrate that, as has been shown for morbilliviruses [19Duprex WP, Collins FM, Rima BK. Modulating the function of the measles virus RNA-dependent RNA polymerase by insertion of green fluorescent protein into the open reading frame J Virol 2002; 76(14): 7322-8., 20Brown DD, Rima BK, Allen IV, et al. Rational attenuation of a morbillivirus by modulating the activity of the RNA-dependent RNA polymerase J Virol 2005; 79(22): 14330-8.] and rhabdoviruses [28Ruedas JB, Perrault J. Insertion of enhanced green fluorescent protein in a hinge region of vesicular stomatitis virus L polymerase protein creates a temperature-sensitive virus that displays no virion-associated polymerase activity in vitro J Virol 2009; 83(23): 12241-52.], the pneumovirus RSV L can tolerate a large insertion of 11% within its coding sequence.
In conclusion, we have demonstrated that i) the GDNQ motif (residues 810-813) of the pneumovirus RSV L protein is critical for its activity, and ii) the L polymerase contains a variable region (VR2) that can be used to tag the L protein in order to track it in living cells. The VR2 region is localized between CRs V and VI of L and is equivalent to the H2 hinge region previously identified for paramyxovirus and rhabdovirus L proteins. Insertion of short (HA) or longer sequences (eGFP and mCherry) within this region creates functional RdRp but with reduced activities (roughly 30% of the unmodified L). These tagged L proteins constitute a useful and valuable tool and open up a number of avenues for studies of RSV L trafficking and function in living cells. Furthermore, using reverse genetics to insert these mutations into virus may prove to be an effective means to rationally attenuate RSV with the potential to produce RSV strains suitable for use as a vaccine.
ACKNOWLEDGEMENTS
We thank Nathalie Castagné for technical help, Michel Brémont, Stéphane Biacchesi, and David Bhella for critical reading of the manuscript.




