- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Development and Evaluation of an Enzyme-Linked Immunosorbent Assay for Dengue Capsid
Suganya Selvarajah 1, Udayan Chatterji 1, Richard Kuhn 2, Richard Kinney 3, Subhash G Vasudevan 4, Philippe Gallay*, 1
Abstract
The astonishing speed with which Dengue has spread across the world and the severity of its infection make Dengue a prime threat to human life worldwide. Unfortunately, to date there are no effective vaccines or treatments against Dengue. Since only a few assays permit rapid and sensitive detection of Dengue, we developed a specific antigen capture enzyme-linked immunosorbent assay (ELISA) for the abundant structural Dengue-2 capsid protein. We showed that the ELISA allows rapid and sensitive detection of Dengue-2 replication in various cell lines including human and mosquito cells. Using anti-capsid antibodies, we demonstrated that the capsid ELISA is as accurate as other well-characterized Dengue assays such as intracellular FACS staining (IFSA) and fluorescent focus (FFA) assays. The capsid ELISA not only represents a useful tool for in vitro basic research, but it may also represent a valuable diagnostic tool for Dengue infection in patients.
Article Information
Identifiers and Pagination:
Year: 2012Volume: 6
First Page: 29
Last Page: 37
Publisher Id: TOVJ-6-29
DOI: 10.2174/1874357901206010029
Article History:
Received Date: 23/11/2011Revision Received Date: 20/1/2012
Acceptance Date: 20/1/2012
Electronic publication date: 29/3/2012
Collection year: 2012
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Department of Immunology an& Microbial Science, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA; Tel: 858-784-8180; Fax: 858-784-8831; E-mail: gallay@scripps.edu
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 23-11-2011 |
Original Manuscript | Development and Evaluation of an Enzyme-Linked Immunosorbent Assay for Dengue Capsid | |
1. INTRODUCTION
Dengue fever is now found in more than 100 developing countries. Carried by mosquitoes in tropical and sub-tropical areas, dengue virus causes two distinct disease states in humans: a mild disease called dengue fever (DF) and a much more severe disease called dengue hemorrhagic fever (DHF) which is potentially fatal. The World Health Organization has estimated that dengue virus annually causes 100 million new cases of DF, 500,000 cases of DHF, and 25,000 deaths [1Halstead SB. Pathogenesis of dengue: challenges to molecular biology Science 1988; 239: 476-81., 2Putnak JR. Progress in the development of recombinant vaccines against dengue and other arthropod-borne flaviviruses In: E Kurstak , Ed. Modern vaccinology. New York NY: Plenum Press 1994; pp. 231-52.]. In total, 2.5 billion people are considered to be at risk for dengue infection. Originally endemic to Southeast Asia, since WWII dengue has spread around the globe to become endemic to all areas south of the 35th parallel, including Central and South America and Africa. Urbanization in the developing world has created dense living conditions, allowing the disease to spread more easily. At the moment, dengue has not spread significantly to North America, although it has already penetrated Hawaii and Florida. The astonishing speed with which dengue has spread across the rest of the world and the severity of its infection make dengue a prime threat to human life worldwide. Unfortunately, to date there are no effective vaccines or treatments against dengue [3Halstead SB, Deen J. The future of dengue vaccines Lancet 2002; 360: 1243-5.].
Dengue infection presents itself in two modes: a primary and secondary infection. In primary infection, dengue enters target cells via an uncharacterized primary receptor. Secondary infection occurs after inoculation generally with a different dengue serotype. In this case, dengue enters cells as a complex composed of the virus decorated with preexisting non-neutralizing antibodies. These immune complexes may enter host cells by interacting with alternate receptors such as Fc receptors, resulting in antibody-dependent enhancement of infection (ADE). ADE is hypothesized to contribute to the pathogenesis of severe dengue illness, as epidemiological studies have identified secondary infection as a risk factor for DHF and have shown that the presence of pre-existing anti-dengue antibodies correlates with DHF [4Clyde K, Kyle JL, Harris E. Recent advances in deciphering viral and host determinants of dengue virus replication and pathogenesis J Virol 2006; 80: 11418-31.-13Porterfield JS. Antibody-dependent enhancement of viral infectivity Adv Virus Res 1986; 31: 335-55.]. This biphasic manifestation of dengue infection greatly complicates vaccine development.
The identification of primary receptors for dengue would symbolize a major milestone in the development of anti-dengue therapies. Indeed, by targeting the primary dengue receptor, we avoid the ADE biphasic manifestation of dengue infection, which represents a major obstacle in vaccine development. The identification of the primary receptors for dengue entry into a cell remains obscure. Several candidate molecules have been proposed to serve as receptors for dengue including heparan sulfate proteoglycans [14Chen YT, Maguire RE, Hileman JR, et al. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate Nat Med 1997; 3: 866-7.], DC-SIGN [15Tassaneetrithep B, Burgess TH, Granelli-Piperno A, et al. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells J Exp Med 2003; 197: 823-9., 16Navarro-Sanchez E, Altmeyer R, Amara A, et al. Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses EMBO Rep 2003; 4: 723-8.] and unknown cell surface proteins [17Salas-Benito JS, del Angel RM. Identification of two surface proteins from C6/36 cells that bind dengue type 4 virus J Virol 1997; 71: 7246-52., 18Moreno-Altamirano MM, Sanchez-Garcia FJ, Munoz M. Non Fc receptor-mediated infection of human macrophages by dengue virus serotype 2 J Gen Virol 2002; 83: 1123-30.]. However, to date, the true functions of these receptors in dengue infection remain to be further explored. On one hand, these receptors may serve as true entry receptors, which mediate fusion between viral and cellular membranes, a pre-condition for productive infection of enveloped viruses. On the other hand, these receptors may simply serve as attachment receptors, which promote virus adsorption and internalization, either leading to enhanced infection or to a dead-end infection (i.e., virus directed into abortive cellular compartments). Moreover, the identity of the cells, which support robust dengue replication in humans, also remains obscure. Several candidate cell populations have been proposed such as monocytes, B lymphocytes, Langerhans cells, dendritic cells and endothelial cells [4Clyde K, Kyle JL, Harris E. Recent advances in deciphering viral and host determinants of dengue virus replication and pathogenesis J Virol 2006; 80: 11418-31., 5Daughaday CC, Brandt WE, McCown JM, Russell PK. Evidence for two mechanisms of dengue virus infection of adherent human monocytes: trypsin-sensitive virus receptors and trypsin-resistant immune complex receptors Infect Immun 1981; 32: 469-73., 19Andrews BS, Theofilopoulos AN, Peters CJ, Loskutoff DJ, Brandt WE, Dixon FJ. Replication of dengue and junin viruses in cultured rabbit and human endothelial cells Infect Immun 1978; 20: 776-81.-27Wu SJ, Grouard-Vogel G, Sun W, et al. Human skin Langerhans cells are targets of dengue virus infection Nat Med 2000; 6: 816-20.]. However, the conclusive proof that one of these cell populations serves as the main source of virus in the body is still missing. The paucity of information about the identity of the primary receptors and the target cells that dengue exploits in vivo may arise from the paucity of adequate tools to accurately measure dengue attachment, entry and replication in human primary cells. To address this issue, we generated antibodies directed against the abundant structural capsid protein of dengue and used them to develop sensitive and accurate assays, which permit detection of dengue infection: an enzyme-linked immunosorbent assay (ELISA), an intracellular FACS staining assay (IFSA), and a fluorescent focus assay (FFA).
2. MATERIALS AND METHODS
2.1. Cells
African green monkey Vero cells and human 293T cells were grown in DMEM supplemented with 10% FBS, penicillin, streptomycin and glutamine at 37˚C in 10% CO2. Aedes albopictus C6/36 mosquito cells were maintained in EMEM containing Earl’s salts and non-essential amino acids supplemented with 10% FBS, penicillin, streptomycin and glutamine at 28˚C in 5% CO2.
2.2. Viruses
Virus stocks of dengue-2 16681 [28Kinney RM, Butrapet S, Chang GJ, et al. Construction of infectious cDNA clones for dengue 2 virus: strain 16681 and its attenuated vaccine derivative, strain PDK-53 Virology 1997; 230: 300-8.] and TSV01 (accession number AY037116) were produced as described previously [20Diamond MS, Edgil D, Roberts TG, Lu B, Harris E. Infection of human cells by dengue virus is modulated by different cell types and viral strains J Virol 2000; 74: 7814-23.] by inoculating monolayers of Vero or C6/36 cells in 150 cm2 flasks with 100 µl of 1 ( 106 FFU/ml virus stock in 5 ml complete medium. After 2 h, 15 ml of complete medium was added to infected cells. Seven days post-infection, viral supernatants were collected, filtered through a 0.2 micron filter, adjusted to 20% FBS, aliquoted and stored at -80˚C. Viral titers in focus-forming units per milliliter (FFU/ml) were determined by focus-forming assay (FFA) titration in Vero cells.
2.3. Anti-Dengue-2 Capsid Antibody Production
Monoclonal and polyclonal antibodies were produced by immunizing Balb/c mice and New Zealand rabbits with recombinant dengue-2 (PR-159S1 strain) capsid protein [29Jones CT, Ma L, Burgner JW, Groesch TD, Post CB, Kuhn RJ. Flavivirus capsid is a dimeric alpha-helical protein J Virol 2003; 77: 7143-9.] (generous gift from R. Kuhn). Murine and rabbit anti-capsid IgG were purified using protein A beads.
2.4. Western Blot Analysis
Viruses from filtered supernatants of infected Vero cells were purified through a 30% sucrose cushion by ultracentrifugation at 23,000 rpm in an SW28 rotor (Beckman) for 2.5 h at 4˚C. Pelleted viruses were lysed and loaded onto an SDS-gel together with recombinant dengue-2 capsid as a positive control. Western blot analysis was performed according to manufacturer’s instruction for Tris-Glycine precast minigels (Invitrogen). Blots were incubated with primary monoclonal or polyclonal anti-dengue-2 IgG (5 µg/ml) overnight, incubated with secondary horseradish peroxidase (HRP)-conjugated anti-mouse or -rabbit IgG for 2 h at room temperature (Amersham) (1/10,000 dilution) and revealed by ECL (Perkin-Elmer).
2.5. Dengue Replication Detected by Enzyme-Linked Immunosorbent (ELISA)
Cells (2 x 104) were infected with dengue at a multiplicity of infection (MOI) of 0.01 for 2 h at 37˚C, washed twice with medium, and incubated in 2 ml complete medium at 37˚C in 10% CO2. Supernatant aliquots were collected on days 0, 3, 5, 7 and 10 post-infection. Viruses were lysed by mixing supernatant with 0.5% NP-40 in PBS for 10 min at room temperature with vigorous shaking and frozen at -20˚C. 96-well plates coated overnight at 4˚C with rabbit anti-capsid dengue-2 IgG (20 µg/ml) were washed 10 times with H2O, blocked with 3% BSA in PBS for 1 h at room temperature, and washed 4 times with wash buffer (PBS containing 1% BSA and 0.02% Tween). Virus supernatants were added to ELISA plate for 2 h at room temperature. Wells were washed 10 times with washing buffer, incubated for 2 h at 4˚C with biotinylated rabbit or murine anti-capsid IgG (2 µg/ml), washed 10 times, incubated for 30 min at room temperature in the dark with streptavidin-conjugated HRP (Jackson Laboratories and Immunochemicals) (1/1000 dilution) and washed 10 times with washing buffer. The o-phenylenediamine dihydrochloride substrate (Sigma-Aldrich) (100 µl) was added to wells and reaction was stopped after addition of H2SO4 (4 N) (Fisher Scientific) (100 µl). Plates were read at 495 nm on a microplate reader (Molecular Devices).
2.6. Dengue Replication Detected by Intracellular FACS Staining Assay (IFSA)
IFSA was conducted as described previously [30Kao CL, Wu MC, Chiu YH, et al. Flow cytometry compared with indirect immunofluorescence for rapid detection of dengue virus type 1 after amplification in tissue culture J Clin Microbiol 2001; 39: 3672-7., 31Martin NC, Pardo J, Simmons M, et al. An immunocytometric assay based on dengue infection via DC-SIGN permits rapid measurement of anti-dengue neutralizing antibodies J Virol Methods 2006; 134: 74-85.] with minor modifications. Cells (2 x 105) were infected with dengue at an MOI of 0.1 for 2 h at 37˚C, washed twice with medium, and incubated in 2 ml complete medium at 37˚C in 10% CO2. Three days post-infection, cells were washed, trypsinized, resuspended in PBS at 1 x 106/ml and fixed with 0.2% paraformaldehyde in PBS for 30 min on ice. Cells were washed, permeabilized in PBS containing 0.2% Tween for 15 min at 37˚C, washed and resuspended in FACS buffer (PBS containing 2% FBS). For intracellular staining, cells (105) were incubated for 30 min at 4˚C with 10 µg/ml of isotype controls, mouse monoclonal anti-dengue capsid 9A7 IgG, or mouse anti-dengue Env complex IgG (Chemicon). Cell permeabilization was confirmed by staining cells with mouse anti-tubulin IgG (Santa Cruz Biotechnologies). Cells were washed, incubated with secondary phycoerythrin (PE)-conjugated anti-mouse IgG (10 µg/ml) for 30 min at 4˚C, washed again, resuspended in PBS, fixed in 2% paraformaldehyde and stored at 4˚C until FACS analysis.
2.7. Dengue Replication Detected by Fluorescent Focus Assay (FFA)
FFA was conducted as described previously [32Kapadia SB, Brideau-Andersen A, Chisari FV. Interference of hepatitis C virus RNA replication by short interfering RNAs Proc Natl Acad Sci USA 2003; 100: 2014-8., 33Payne AF, Binduga-Gajewska I, Kauffman EB, Kramer LD. Quantitation of flaviviruses by fluorescent focus assay J Virol Methods 2006; 134: 183-9.] with minor modifications. Briefly, Vero cells (1 x 104) seeded overnight in 96-well plates were incubated with supernatants containing viruses (dilutions from 10-1 to 10-6) for 2 h at 37˚C, washed once and incubated for 72 h at 37˚C in 10% CO2. Cells were washed, fixed with 4% formaldehyde in PBS for 20 min at room temperature, washed again and permeabilized with PBS containing 3% BSA, 0.3% TritonX-100 and 10% FBS for 1 h at room temperature. Cells were incubated with mouse monoclonal anti-dengue envelope or anti-capsid 9A7 IgG (1 µg/ml) in 3% BSA and 0.3% Triton X-100 for 1 h at room temperature. Cells were washed 3 times with PBS and incubated with Alexa555-conjugated anti-mouse IgG (Molecular Probes) (1/1000 dilution) for 1 h at room temperature. Cell nuclei were stained with Hoechst dye. Cells were washed 3 times, resuspended in PBS and analyzed for number of infectious foci by fluorescent microscope (Zeiss).
2.8. RT-PCR
RT-PCR was performed to determine DENV replication by quantifying DENV-2 genomic RNA copies. Viral RNA was extracted using RNA extraction kit (QIAamp Viral RNA mini kit). RT-PCR assay was performed by adding 1 µL of extracted viral RNA to the SensiMix SYBR green reagent, which contained 7.4 μL ddH2O, 10 μL 2x SensiMix One-Step, 0.4 μL 50x SYBR Green solution, 10 U of RNAse Inhibitor, 50 pmoL of forward (DNF) and reverse (D2R) primers. All samples were assayed in triplicate. The amplifications were performed using the DNA Engine Opticon system (MJ Research/Bio-Rad, Hercules, CA) with the following thermal conditions: reverse transcription at 50°C for 30 min, initial denaturation at 95°C for 10 min, followed by 45 cycles of 95°C for 15 sec, 59°C for 30 sec and 72°C for 30 sec. Melting curve analysis was subsequently performed at temperature from 60°C to 98°C to verify the assay specificity. For absolute quantitation of the viral RNA, a standard curve was established with a serially diluted RNA extracted from DEN-2 stock.
2.9. Clinical Specimens
Forty serum samples were obtained from DEN-2 infected patients, who gave a positive result for anti-DEN-2 antibodies. Ten serum samples from blood donors were collected as negative controls.
3. RESULTS
3.1. Production and Characterization of Anti-Dengue Capsid Antibodies
The development of assays, which rapidly and sensitively detect dengue attachment, entry and replication, is essential in order to identify the primary host cells and receptors that support viral replication in the host. To address this issue, we generated antibodies directed against the abundant structural capsid protein of dengue and developed enzyme-linked immunosorbent (ELISA), intracellular FACS staining (IFSA), and fluorescent focus (FFA) assays to detect dengue virus and infection. Balb/c mice and New Zealand rabbits were immunized with recombinant dengue-2 (PR-159S1 strain) capsid core protein purified from E. coli [29Jones CT, Ma L, Burgner JW, Groesch TD, Post CB, Kuhn RJ. Flavivirus capsid is a dimeric alpha-helical protein J Virol 2003; 77: 7143-9.]. We produced mouse monoclonal anti-dengue-2 capsid antibodies and rabbit polyclonal antisera. We analyzed by Western blot the reactivity of the antibodies toward dengue-2 capsid derived either from E. coli or from purified viruses. Recombinant capsid derived from the dengue-2 PR-159S1 strain was purified from E. coli as described previously [29Jones CT, Ma L, Burgner JW, Groesch TD, Post CB, Kuhn RJ. Flavivirus capsid is a dimeric alpha-helical protein J Virol 2003; 77: 7143-9.]. Dengue-2 16681 viruses were purified from supernatant of infected Vero cells by ultracentrifugation over a 30% sucrose cushion. Pelleted viruses were lysed and analyzed by Western blot. We found that both monoclonal (9A7) (Fig. 1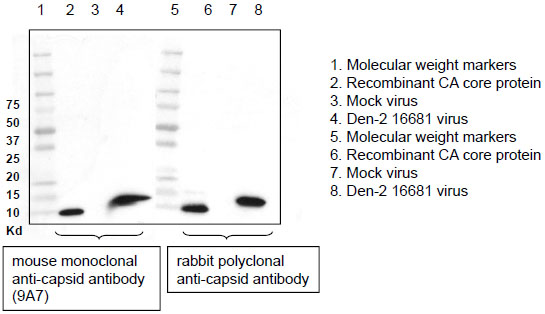 , lane 2) and polyclonal (Fig. 1
, lane 2) and polyclonal (Fig. 1 , lane 6) anti-dengue-2 capsid antibodies efficiently recognize recombinant dengue-2 capsid (at around 10 kDa). Monoclonal (9A7) (Fig. 1
, lane 6) anti-dengue-2 capsid antibodies efficiently recognize recombinant dengue-2 capsid (at around 10 kDa). Monoclonal (9A7) (Fig. 1 , lane 4) and polyclonal (Fig. 1
, lane 4) and polyclonal (Fig. 1 , lane 8) anti-dengue-2 capsid antibodies recognize a single band (around 12 kDa) in viral lysates, corresponding to capsid incorporated into dengue-2 viruses.
, lane 8) anti-dengue-2 capsid antibodies recognize a single band (around 12 kDa) in viral lysates, corresponding to capsid incorporated into dengue-2 viruses.
The difference in size between recombinant and viral dengue-2 capsid proteins may result from post-translational modifications such as protein cleavage or phosphorylation. No reactivity was observed in ultracentrifuged supernatant from uninfected Vero cells (mock) (Fig. 1 , lanes 3 and 7). Moreover, no reactivity was observed toward other dengue serotypes such as dengue-1, -3 and -4 (data not shown). This demonstrates that our monoclonal and polyclonal antibodies recognize dengue-2 capsid specifically.
, lanes 3 and 7). Moreover, no reactivity was observed toward other dengue serotypes such as dengue-1, -3 and -4 (data not shown). This demonstrates that our monoclonal and polyclonal antibodies recognize dengue-2 capsid specifically.
3.2. Dengue-2 Replication Detected by Capsid ELISA
Using our monoclonal and polyclonal anti-dengue-2 capsid antibodies, we developed a rapid, reproducible and sensitive ELISA. Briefly, 96-well plates were coated with 20 µg/ml of protein A-purified rabbit anti-capsid dengue-2 IgG, washed and blocked with 3% BSA. Increasing concentrations of recombinant dengue-2 capsid protein diluted in PBS containing 0.5% NP-40 were added to ELISA plates (triplicates). Captured dengue-2 capsid was detected with biotinylated mouse monoclonal anti-dengue-2 capsid 2 µg/ml IgG and streptavidin-conjugated HRP. The threshold of detection of recombinant dengue-2 capsid by the ELISA was 100 to 300 pg/ml (Fig. 2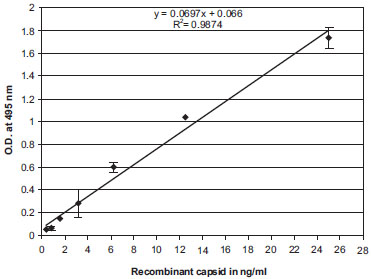 ).
).
We then asked if our ELISA can detect dengue replication over time by measuring levels of capsid released into the supernatant of infected cells. Vero or 293T cells were infected with either 16681 or TSVO1 dengue-2 viruses at an MOI of 0.01. Supernatants from infected cells were collected at different time points and amounts of virus quantified by ELISA. Viral replication can be detected as early as 3 days post-infection and reaches a plateau between day 7 and 10. Note that if we increase the MOI, capsid release into supernatant of infected cells can already be detected 48 h post-infection (data not shown). We found that 16681 replicates more robustly than TSV01 in both Vero and 293T cells (Fig. 2 ). Specifically, 16681 reaches a growth peak of 30-40 ng/ml of capsid in supernatant of infected cells, whereas TSV01 reaches a growth peak of 5-15 ng/ml (Fig. 3
). Specifically, 16681 reaches a growth peak of 30-40 ng/ml of capsid in supernatant of infected cells, whereas TSV01 reaches a growth peak of 5-15 ng/ml (Fig. 3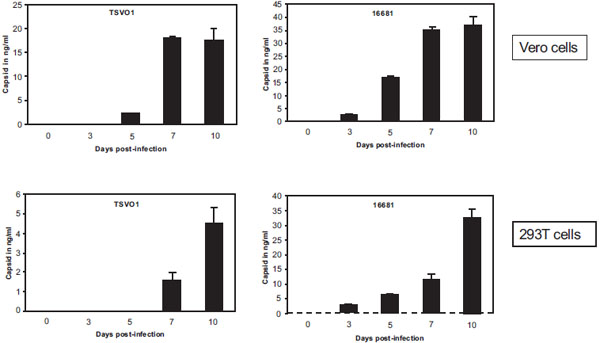 ). We obtained similar growths in human Huh-7 and mosquito C6/36 cells (data not shown). This demonstrates that our capsid ELISA permits rapid and sensitive detection of dengue replication in various cell lines.
). We obtained similar growths in human Huh-7 and mosquito C6/36 cells (data not shown). This demonstrates that our capsid ELISA permits rapid and sensitive detection of dengue replication in various cell lines.
3.3. Dengue-2 Replication Detected by Intracellular FACS Staining Assay (IFSA) Using Anti-Capsid Antibodies
We then asked whether our ELISA is as accurate as other previously established assays, which measure dengue infection and replication. Specifically, we developed an intracellular FACS staining assay (IFSA) as well as a fluorescent focus assay (FFA) using anti-capsid antibodies. We then compared ELISA, IFSA and FFA for their capacities to monitor dengue infection in various cell lines. First, we developed an IFSA using anti-capsid antibodies. Vero cells (2 x 105) were infected with 16681 dengue-2 virus at an MOI of 0.1 for 2 h at 37˚C, washed twice and incubated in complete medium for three days. Cells were then washed, trypsinized, fixed and permeabilized. Permeabilized infected cells were incubated with the monoclonal anti-dengue capsid 9A7 antibody followed by a secondary phycoerythrin (PE)-conjugated anti-mouse antibody. Cells were then washed, fixed in paraformaldehyde and analyzed by FACS. Efficiency of permeabilization was verified by anti-tubulin staining. Ninety-five to 99% of the cells were positive for tubulin staining after permeabilization (Fig. 4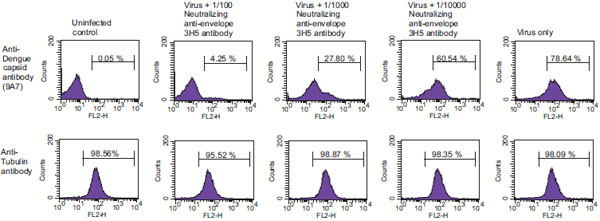 , bottom panels), whereas less than 0.01% of the cells were positive for tubulin staining without permeabilization (data not shown). Importantly, we found that 75 to 80% of infected cells were positive after anti-dengue capsid 9A7 antibody staining (Fig. 4
, bottom panels), whereas less than 0.01% of the cells were positive for tubulin staining without permeabilization (data not shown). Importantly, we found that 75 to 80% of infected cells were positive after anti-dengue capsid 9A7 antibody staining (Fig. 4 , top right panel) compared to 0.03 to 0.07% of non-infected cells (Fig. 4
, top right panel) compared to 0.03 to 0.07% of non-infected cells (Fig. 4 , top left panel). We obtained similar results using mouse anti-dengue envelope complex antibodies (Chemicon) (data not shown). Our data indicate that anti-dengue-2 capsid antibodies can be used to measure dengue replication by ELISA (Fig. 3
, top left panel). We obtained similar results using mouse anti-dengue envelope complex antibodies (Chemicon) (data not shown). Our data indicate that anti-dengue-2 capsid antibodies can be used to measure dengue replication by ELISA (Fig. 3 ) as well as by IFSA (Fig. 4
) as well as by IFSA (Fig. 4 ).
).
3.4. Inhibition of Dengue-2 Replication Detected by ELISA and IFSA Using Anti-Capsid Antibodies
In order to compare the accuracies of ELISA and IFSA, we tested them for their abilities to measure the potency of anti-dengue agents. Specifically, we compared side-by-side ELISA and IFSA capacities to detect inhibition of dengue-2 infection by neutralizing antibodies. Vero cells were exposed to dengue-2 16681 virus at an MOI of 0.1 in the presence or absence of the neutralizing anti-dengue-2 envelope 3H5 antibody (hybridoma supernatant diluted at 1/100, 1/1000 and 1/10,000) (kindly provided by R. Kinney). Three days post-infection, infection was monitored either by IFSA, by measuring amounts of intracellular capsid, or by ELISA, by measuring amounts of capsid released in the supernatant. By IFSA, we found that the neutralizing 3H5 antibody efficiently reduces dengue-2 infection. The percentage of capsid positive Vero cells drastically decreases (60.5%, 27.8% and 4.25% of positive cells) in the presence of increasing concentration of 3H5 antibody (Fig. 4 ). Specifically, we found that dengue-2 infectivity goes from 100 down to 76, 16 and 5.6% in the presence of 3H5 hybridoma diluted at 1/10000, 1/1000 and 1/100, respectively. Importantly, we observed a very similar pattern of inhibition of dengue-2 replication by 3H5 when we quantified amounts of capsid released in the supernatant of infected cells by ELISA (Table 1, right column). Dengue-2 infectivity monitored by ELISA is gradually reduced from 100 to 75, 36 and 5.4% when 3H5 is added at a dilution of 1/10000, 1/1000 and 1/100, respectively. These data indicate that ELISA and IFSA assays using anti-capsid antibodies represent accurate tools to measure dengue-2 replication as well as to evaluate the potency of anti-dengue agent candidates.
). Specifically, we found that dengue-2 infectivity goes from 100 down to 76, 16 and 5.6% in the presence of 3H5 hybridoma diluted at 1/10000, 1/1000 and 1/100, respectively. Importantly, we observed a very similar pattern of inhibition of dengue-2 replication by 3H5 when we quantified amounts of capsid released in the supernatant of infected cells by ELISA (Table 1, right column). Dengue-2 infectivity monitored by ELISA is gradually reduced from 100 to 75, 36 and 5.4% when 3H5 is added at a dilution of 1/10000, 1/1000 and 1/100, respectively. These data indicate that ELISA and IFSA assays using anti-capsid antibodies represent accurate tools to measure dengue-2 replication as well as to evaluate the potency of anti-dengue agent candidates.
3.5. Detection of Dengue-2 Replication by Fluorescent Focus Assay (FFA) Using Anti-Capsid Antibodies
We then developed an FFA using anti-capsid antibodies to compare its potential to measure dengue infection with that of ELISA and IFSA described above. FFA was conducted as described previously [32Kapadia SB, Brideau-Andersen A, Chisari FV. Interference of hepatitis C virus RNA replication by short interfering RNAs Proc Natl Acad Sci USA 2003; 100: 2014-8.-33Payne AF, Binduga-Gajewska I, Kauffman EB, Kramer LD. Quantitation of flaviviruses by fluorescent focus assay J Virol Methods 2006; 134: 183-9.] with minor modifications. Briefly, supernatants from Vero and 293T cells infected with 16681 dengue-2 virus were harvested on days 0, 3, 5, 7 and 10 and immediately used to infect naïve Vero cells. Infected Vero cells were washed, fixed with formaldehyde and permeabilized with TritonX-100. Cells were then incubated with monoclonal anti-dengue-2 capsid or anti-envelope antibodies. Cells were washed and incubated with Alexa555-conjugated anti-mouse antibodies. Infectious foci were visualized using a fluorescent microscope.
We found that the subcellular localization of capsid and envelope in infected Vero cells greatly differs (Fig. 5A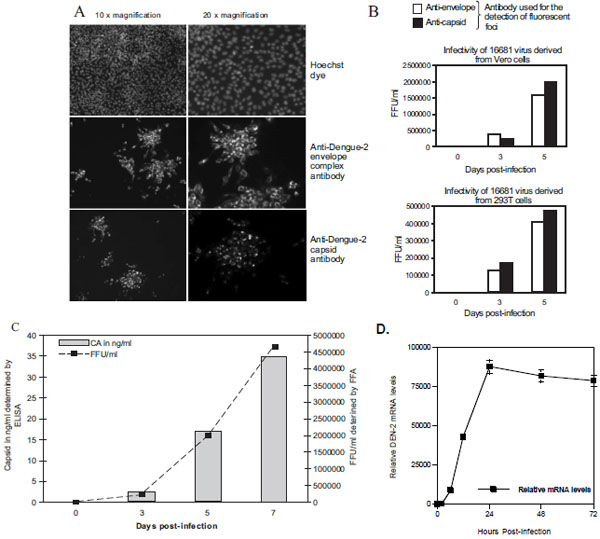 ). The anti-capsid antibody mainly stains the nucleus and nucleolus (Fig. 5A
). The anti-capsid antibody mainly stains the nucleus and nucleolus (Fig. 5A , bottom panels), whereas the anti-envelope complex antibody stains the cytoplasm (Fig. 5A
, bottom panels), whereas the anti-envelope complex antibody stains the cytoplasm (Fig. 5A , top panel). This is in accordance with previous studies, which showed that capsid localizes in the nuclear compartment [34Bulich R, Aaskov JG. Nuclear localization of dengue 2 virus core protein detected with monoclonal antibodies J Gen Virol 1992; 73: 2999-3003., 35Wang SH, Syu WJ, Huang KJ, et al. Intracellular localization and determination of a nuclear localization signal of the core protein of dengue virus J Gen Virol 2002; 83: 3093-102.]. Importantly, dengue-2 replication monitored by FFA using the anti-capsid antibody perfectly correlates with that using the anti-envelope antibody (Fig. 5B
, top panel). This is in accordance with previous studies, which showed that capsid localizes in the nuclear compartment [34Bulich R, Aaskov JG. Nuclear localization of dengue 2 virus core protein detected with monoclonal antibodies J Gen Virol 1992; 73: 2999-3003., 35Wang SH, Syu WJ, Huang KJ, et al. Intracellular localization and determination of a nuclear localization signal of the core protein of dengue virus J Gen Virol 2002; 83: 3093-102.]. Importantly, dengue-2 replication monitored by FFA using the anti-capsid antibody perfectly correlates with that using the anti-envelope antibody (Fig. 5B ). More importantly, dengue-2 replication monitored by FFA using anti-capsid antibodies perfectly correlates with that using our capsid ELISA and FFA (Fig. 5C
). More importantly, dengue-2 replication monitored by FFA using anti-capsid antibodies perfectly correlates with that using our capsid ELISA and FFA (Fig. 5C ). RT-PCR analysis of dengue-2 replication match well the capsid ELISA data, although there is a small window period between when viral RNA is detectable by the sensitive RT-PCR and when capsid is detected by ELISA (Fig. 5D
). RT-PCR analysis of dengue-2 replication match well the capsid ELISA data, although there is a small window period between when viral RNA is detectable by the sensitive RT-PCR and when capsid is detected by ELISA (Fig. 5D ). Altogether these data indicate that antibodies directed against the structural capsid protein of dengue-2 represent ideal tools to measure dengue-2 replication in various detection assays.
). Altogether these data indicate that antibodies directed against the structural capsid protein of dengue-2 represent ideal tools to measure dengue-2 replication in various detection assays.
3.6. Detection of Dengue-2 Capsid in Patient Sera by ELISA
We then asked whether this capsid ELISA can be used as a diagnostic tool to identify infected patients. Specifically, we collected 10 donor sera and 50 dengue-2 patient sera and tested them for capsid content by ELISA. Importantly, we constantly found that patient sera gave capsid values superior to those of donor sera (Table 2). This demonstrates that the capsid ELISA assay could be of a great utility for screening Dengue 2 infected patients. It also suggests that similar assays can be developed when similar antibodies will be generated for the detection of the 3 other serotypes.
DISCUSSION
In this study, we produced monoclonal and polyclonal antibodies directed against the structural capsid protein of dengue-2. The antibodies recognize capsid specifically since no cross-reactivity was observed by Western blot toward other viral or cellular proteins. Moreover, no cross-reactivity was observed toward capsids derived from other serotypes such as dengue-1, -3 and -4. We then developed an ELISA for dengue-2 capsid. We showed that the ELISA allows rapid and sensitive (ng/mL range) detection of dengue-2 (i.e., 16681 and TSVO1) replication in various cell lines including human and mosquito cells. We then compared the accuracy of the ELISA to measure dengue-2 replication with that of other previously established assays. Specifically, we developed an IFSA as well as an FFA using anti-capsid antibodies. Importantly, we found that kinetic and peak amplitude of dengue-2 replication detected by ELISA correspond perfectly with those measured by IFSA or FFA. Although a couple of studies also generated anti-dengue capsid antibodies [34Bulich R, Aaskov JG. Nuclear localization of dengue 2 virus core protein detected with monoclonal antibodies J Gen Virol 1992; 73: 2999-3003.-35Wang SH, Syu WJ, Huang KJ, et al. Intracellular localization and determination of a nuclear localization signal of the core protein of dengue virus J Gen Virol 2002; 83: 3093-102.], to our knowledge, this is the first demonstration that anti-capsid antibodies represent valuable tools to measure dengue virus infection using various assays including ELISA, IFSA and FFA.
There are several advantages for using a capsid ELISA to detect dengue infection. The ELISA is a rapid test that can be completed in a few hours. It thus represents an ideal alternative assay for the time-consuming standard plaque assay. The 96-well format ELISA test is a very qualitative tool that permits the analysis of a large number of samples in duplicate or triplicate. The capsid ELISA is thus extremely convenient to measure growth of numerous dengue-2 strains in several cell types over a long period of time (2-3 weeks) or to screen a large panel of anti-dengue compound candidates. In contrast to the plaque assay, the ELISA does not rely on virus-induced cytopathic effects and thus can be used in a broader range of target cells or mutant viruses that do not form plaques. Another advantage of using a capsid ELISA to detect dengue infection is that accumulation of capsid into the supernatant of infected cells (peak growth) represents a reliable proof of replication. In contrast, positive intracellular stainings of viral proteins by IFSA or FFA may result from dengue capture and internalization by attachment receptors (i.e., heparan sulfate proteoglycans or lectins) in the absence of true productive infection (without fusion between the viral and the cellular membrane). Moreover, IFSA and FFA procedures are more time-consuming than ELISA procedures, mainly when a large number of samples need to be analyzed. A major advantage of the capsid ELISA is that it detects viruses independently of their infectious capacities. This is extremely important for studies proposing to identify regions or residues of the viral envelope necessary for infection. Indeed, envelope mutant viruses released into the supernatant of transfected cells can be standardized for capsid content by ELISA and subsequently tested for infection by IFSA or FFA. Similarly, the capsid ELISA will be extremely useful to map the domains or residues of the structural capsid protein necessary for dengue-2 infection. The capsid ELISA also represents an ideal tool to analyze several steps of the dengue life cycle because it permits the quantitative measure of amounts of viruses, which are released from infected cells (viral budding), which are attached onto the surface of a cell (viral attachment), or which entered into the cell (viral internalization).
In conclusion, we developed an ELISA for capsid that allows rapid, sensitive and specific detection of dengue-2 replication in human and mosquito cells. We demonstrated that the capsid ELISA is as accurate as other established infection assays such as IFSA and FFA. The capsid ELISA may not only represent a useful tool for in vitro basic research, but it may also represent a valuable diagnostic tool for dengue detection in clinical patients in epidemic conditions. Prospective studies should be carried out to confirm the clinical value of this new test.
CONFLICT OF INTEREST
Declared none.
ACKNOWLEDGEMENT
We thank J. Kuhns for secretarial assistance. This is publication no. 18697 from the Department of Immunology & Microbial Science, The Scripps Research Institute, La Jolla, CA.


