- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Evaluation of Nucleocapsid and Phosphoprotein P Functionality as Critical Factors During the Early Phase of Paramyxoviral Infection
Sascha Bossow1, a, Sabine Schlecht1, b, Rainer Schubbert2, Matthias Pfeiffer2, Wolfgang J Neubert1, Marian Wiegand*, 1, c
Abstract
In the beginning of a paramyxovirus infection after cell entry viral survival depends on efficient primary (1°) transcription and on the stability of only one input nucleocapsid. Here we examined the influence of the viral polymerase co-factor phosphoprotein P on the very early phase of an infection, i.e. before progeny nucleocapsids are synthesized. We used a novel set-up with Sendai virus (SeV) mutants incapable of genome replication: SeV-ΔP with the entire P ORF deleted, SeV-PΔ2-77 with the deletion of aa 2-77. These mutants allow maintaining the state of the very beginning of an infection when statistically one viral genome is present in the cell. This single genome serves as template for transcription. During SeV-ΔP infections only early 1° transcription takes place at low levels. However, when the truncated P protein is expressed in SeV-PΔ2-77 infections, 1° transcription levels rise significantly up to an 8-fold increased amount of viral mRNA. This shows that the P protein is able to support transcription and thereby mediates the transition from early to late 1° transcription. Importantly, nucleocapsids of both mutants could be shown to remain stable and functional for at least 5 days – even without de novo P protein synthesis. These results describe a novel function of the P protein: enhancing viral gene expression even before genome replication has started. Thus, the since long postulated supportive function of the P protein is not related to stabilization of the nucleocapsid but rather enhances the processivity of the viral polymerase during late 1° and secondary (2°) transcription and genome replication.
Article Information
Identifiers and Pagination:
Year: 2012Volume: 6
First Page: 73
Last Page: 81
Publisher Id: TOVJ-6-73
DOI: 10.2174/1874357901206010073
Article History:
Received Date: 17/2/2012Revision Received Date: 12/4/2012
Acceptance Date: 26/4/2012
Electronic publication date: 14/6/2012
Collection year: 2012
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the AmVac Research GmbH, Lochhamer Str. 29a, D-82152 Martinsried, Germany; Tel: +49 - 89 - 63854363; E-mail: wiegand@amvac-research.de
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 17-2-2012 |
Original Manuscript | Evaluation of Nucleocapsid and Phosphoprotein P Functionality as Critical Factors During the Early Phase of Paramyxoviral Infection | |
INTRODUCTION
After infection of a host cell by a non-segmented negative strand RNA virus (NNSV) most often one viral nucleocapsid (NC) represents the basis for viral reproduction. Transcription of viral genes is the first process that is started even without the support of any de novo synthesized proteins. This early primary transcription (early 1°) accelerates in the presence of de novo synthesized viral nucleocapsid (N) and phospho-(P)-proteins, leading to the state of late primary transcription (late 1°) as shown by using Sendai virus (SeV), a model paramyxovirus [1 Wiegand MA, Bossow S, Schlecht S, Neubert WJ. De novo synthesis of N and P proteins as a key step in Sendai virus gene expression J Virol 2007; 81(24): 13835-44., 2 Curran J, Kolakofsky D. Nonsegmented negative-strand RNA virus RNA synthesis in vivo Virology 2008; 371(2): 227-30.]. During the course of the amount of N, P and polymerase (L) proteins increases and the viral polymerase (vPol) is thought to start genome replication in parallel; newly synthesized genomes serve subsequently as additional templates for secondary transcription (2°), i.e. transcription from NC progeny. A first successful replication of the penetrated NC marks a crucial event in the life cycle of NNSVs. Before this can happen, viral survival inside the host cell depends largely on two factors: NC stability and efficient viral protein expression, based on 1° transcription.
So far the intracellular survival time of NCs was estimated to be 24 h. This result is based on successful replication of defective interfering particles (DIP) in cells super-infected with wild-type SeV [3 Mottet G, Curran J, Roux L. Intracellular stability of nonreplicating paramyxovirus nucleocapsids Virology 1990; 176(1): 1-7.]. Using this set-up, effects of viral de novo protein synthesis on 1° transcription and on template half-life could not be investigated. However, a short NC survival time of only 24 h should hardly be sufficient e.g. for initiation of persistent infections with constant low gene expression levels.
Most of the current understanding of paramyxoviral transcription and replication processes was deduced from in vitro experiments using isolated NC cores (RNP) from DIPs that were incubated with cell extracts containing co-expressed N / P and P / L proteins [4 Curran J, Pelet T, Kolakofsky D. An acidic activation-like domain of the Sendai virus P protein is required for RNA synthesis and encapsidation Virology 1994; 202(2): 875-4., 5 Smallwood S, Ryan KW, Moyer SA. Deletion analysis defines a carboxyl-proximal region of Sendai virus P protein that binds to the polymerase L protein Virology 1994; 202(1): 154-63.]. As large numbers of NC cores were used in those experiments [6 Horikami SM, Curran J, Kolakofsky D, Moyer SA. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro J Virol 1992; 66(8): 4901-8.], the transcriptional activity of a single NC per cell - the situation at the beginning of each natural infection - could not be investigated. In consequence, factors critically involved in early and late 1° transcription could not be determined.
Furthermore, different functions of the vPol co-factor P during RNA synthesis are not completely understood: synthesis of P protein in infected cells is disproportionately high compared to the other viral proteins [7 Lamb RA, Mahy BW, Choppin PW. The synthesis of sendai virus polypeptides in infected cells Virology 1976; 69(1): 116-31., 8 Portner A, Kingsbury DW. Regulatory events in the synthesis of Sendai virus polypeptides and their assembly into virions Virology 1976; 73(1): 79-88.]. In addition, a high turnover rate of P in terms of a time-dependent loss of P interaction with NCs was observed [9 Portner A, Murti KG. Localization of P, NP, and M proteins on Sendai virus nucleocapsid using immunogold labeling Virology 1986; 150(2): 469-78.]. Interestingly, when RNPs were incubated in the presence of elevated levels of P protein, the mRNA synthesis increased 2-fold [10 Curran J. Reexamination of the Sendai virus P protein domains required for RNA synthesis: a possible supplemental role for the P protein Virology 1996; 221(1): 130-40.].
Using specific viral deletion mutants, we began investigating the early phase of SeV infections and demonstrated in vivo the need of both N and P proteins for late 1° transcription [1 Wiegand MA, Bossow S, Schlecht S, Neubert WJ. De novo synthesis of N and P proteins as a key step in Sendai virus gene expression J Virol 2007; 81(24): 13835-44.]. More recently Curran and Kolakofsky [2 Curran J, Kolakofsky D. Nonsegmented negative-strand RNA virus RNA synthesis in vivo Virology 2008; 371(2): 227-30.] discovered also in vitro a supporting function of N and P for viral transcription – explained as energy supplier for the vPol in its movement along the NC. These results extend earlier in vitro data where N was described to be necessary only for replication [6 Horikami SM, Curran J, Kolakofsky D, Moyer SA. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro J Virol 1992; 66(8): 4901-8.]. N was described so far to be functional in replication and this only in the form of a complex with P (P:N°) to avoid illegitimate interaction of N with non-viral RNA [11 Curran J, Homann H, Buchholz C, Rochat S, Neubert W, Kolakofsky D. The hypervariable C-terminal tail of the Sendai paramyxovirus nucleocapsid protein is required for template function but not for RNA encapsidation J Virol 1993; 67(7): 4358-64.]. If the N protein also has a function in transcription, the same interaction with P that is responsible for replication, i.e. binding via the N-terminal domain aa 1-77 [4 Curran J, Pelet T, Kolakofsky D. An acidic activation-like domain of the Sendai virus P protein is required for RNA synthesis and encapsidation Virology 1994; 202(2): 875-4.], could be involved.
Here we created a specific virus deletion mutant (SeV-PΔ2-77) which should be incapable of genome replication due to interruption of the P:N° complex necessary for N protein delivery during replication. The influence of a P protein lacking this domain on viral transcription processes should be evaluated. In addition, a mutant with a complete deletion of the P ORF (SeV-ΔP) was employed that has already been shown to be unable to perform genome replication [1 Wiegand MA, Bossow S, Schlecht S, Neubert WJ. De novo synthesis of N and P proteins as a key step in Sendai virus gene expression J Virol 2007; 81(24): 13835-44.]. In the present work, transcriptional processes and nucleocapsid functionality early during infection, before genome replication has started, were analyzed. The obtained results expand the knowledge of viral key factors responsible for survival of NNSVs in the most critical, initial phase of an infection. The intracellular survival of nucleocapsids lasts for several days, even without de novo synthesized proteins. In contrast, for late 1° transcription de novo synthesis of P protein - even without the domain from aa 2 to 77 - is essential and enhances protein expression significantly, thereby securing viral survival.
RESULTS
Generation of SeV P Gene Mutants
In the genome of SeV-ΔP [1 Wiegand MA, Bossow S, Schlecht S, Neubert WJ. De novo
synthesis of N and P proteins as a key step in Sendai virus gene
expression J Virol 2007; 81(24): 13835-44.] the entire P ORF was deleted – leaving behind an “empty” transcription unit. In this way, none of the different P gene-encoded non-structural (NS) proteins (Fig. 1A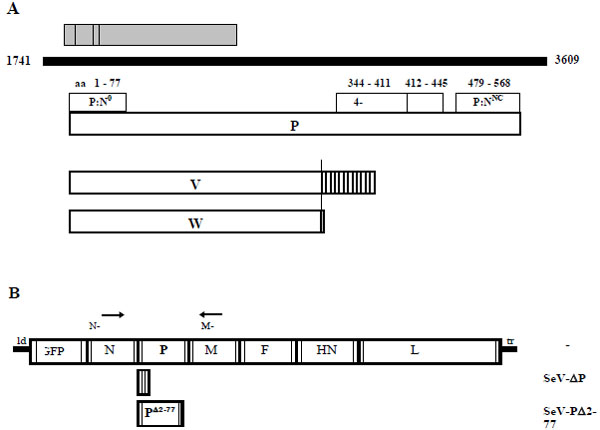 ) nor the P protein can be expressed. This design allowed insertion of a selected alternate P ORF encoding a N-terminally truncated P protein lacking the amino acids 2 to 77. These residues are thought to harbor the domain for formation of the P:N° complex which is essential for genome replication in vitro [4 Curran J, Pelet T, Kolakofsky D. An acidic activation-like domain
of the Sendai virus P protein is required for RNA synthesis and
encapsidation Virology 1994; 202(2): 875-4.] - thus presumably generating a replication-deficient SeV. For monitoring viral gene expression an EGFP ORF was inserted into an additional transcription unit in the genomes of all viral constructs used here (Fig. 1B
) nor the P protein can be expressed. This design allowed insertion of a selected alternate P ORF encoding a N-terminally truncated P protein lacking the amino acids 2 to 77. These residues are thought to harbor the domain for formation of the P:N° complex which is essential for genome replication in vitro [4 Curran J, Pelet T, Kolakofsky D. An acidic activation-like domain
of the Sendai virus P protein is required for RNA synthesis and
encapsidation Virology 1994; 202(2): 875-4.] - thus presumably generating a replication-deficient SeV. For monitoring viral gene expression an EGFP ORF was inserted into an additional transcription unit in the genomes of all viral constructs used here (Fig. 1B ).
).
All recombinant SeV could be rescued successfully from cDNA and amplified using a helper cell line (HCL) expressing the Sendai P protein constitutively [12 Willenbrink W, Neubert WJ. Long-term replication of Sendai virus defective interfering particle nucleocapsids in stable helper cell lines J Virol 1994; 68(12): 8413-7.]. RT-PCR and sequence analyses of recombinant SeV after passage 7 in cell culture confirmed both presence and integrity of the originally cloned variants demonstrating genomic stability during virus propagation.
Growth Characteristics of SeV-P Mutants
In order to investigate any influence of the P gene deletions on viral replication, amplification of the SeV P mutants on helper cells was compared to SeV-wt. Besides the full length P protein several non-structural (NS) proteins. which are reported to have an impact on viral replication, are expressed from the P gene (Fig. 1A ): a nested set of four C proteins (C’, C, Y1, Y2) is translated from a +1 frame relative to the P frame by using alternative start codons [13 Latorre P, Kolakofsky D, Curran J. Sendai virus Y proteins are
initiated by a ribosomal shunt Mol Cell Biol 1998; 18(9): 5021-31.]. In addition, V and W proteins result from a co-transcriptional RNA editing process [14 Vidal S, Curran J, Kolakofsky D. A stuttering model for
paramyxovirus P mRNA editing EMBO J 1990; 9(6): 2017-2.], leading to a frame-shift after codon 317. Here, amplification of SeV-P and SeV PΔ2-77 is performed in a HCL which expresses only the full length P protein but no NS proteins. Thus, we have three different settings in this comparative study: (i) SeV-ΔP with expression of P (in trans from HCL) but without any NS proteins; (ii) SeV-PΔ2-77 with expression of P and truncated VΔ2-77 and WΔ2-77 but no C proteins; (iii) SeV-wt with expression of P and all NS proteins.
): a nested set of four C proteins (C’, C, Y1, Y2) is translated from a +1 frame relative to the P frame by using alternative start codons [13 Latorre P, Kolakofsky D, Curran J. Sendai virus Y proteins are
initiated by a ribosomal shunt Mol Cell Biol 1998; 18(9): 5021-31.]. In addition, V and W proteins result from a co-transcriptional RNA editing process [14 Vidal S, Curran J, Kolakofsky D. A stuttering model for
paramyxovirus P mRNA editing EMBO J 1990; 9(6): 2017-2.], leading to a frame-shift after codon 317. Here, amplification of SeV-P and SeV PΔ2-77 is performed in a HCL which expresses only the full length P protein but no NS proteins. Thus, we have three different settings in this comparative study: (i) SeV-ΔP with expression of P (in trans from HCL) but without any NS proteins; (ii) SeV-PΔ2-77 with expression of P and truncated VΔ2-77 and WΔ2-77 but no C proteins; (iii) SeV-wt with expression of P and all NS proteins.
After infection of cells (MOI = 3) virus-containing supernatants of five consecutive days were titrated (Fig. 2 ): SeV-wt infection produced the highest amount (41 x 106 ciu/ml in total) but all cells were already dead after 3 days. SeV-ΔP and SeV-PΔ2-77 provoked a delayed cytopathic effect (CPE; data not shown) and produced titers which were two times lower than the SeV-wt over the period of 5 days. This clearly shows that amplification of the deletion mutants with P protein provided in trans from the HCL allows high titer virus production. In addition no trans-dominant negative effect develops during co-expression of the virus-encoded PΔ2-77 and the full-length P protein, provided by the HCL, as demonstrated by similar titers of SeV-ΔP and SeV-PΔ2-77. Furthermore, the NS proteins expressed in SeV-wt infections but absent in SeV-ΔP or partially absent and truncated in SeV-PΔ2-77 infections had, at best, a minor effect on viral amplification. However, the V and W proteins were reported to have an influence on viral replication and gene expression [15 Horikami SM, Smallwood S, Moyer SA. The Sendai virus V
protein interacts with the NP protein to regulate viral genome RNA
replication Virology 1996; 222(2): 383-90., 16 Yu D, Shioda T, Kato A, Hasan MK, Sakai Y, Nagai Y. Sendai virus-based expression of HIV-1 gp120: reinforcement by the V(-) version Genes Cells Mol Cell Mech 1997; 2(7): 457-66.]. Importantly, both proteins harbor the domain from aa 2 to 77 known to be responsible for binding of P to N (P:N°), essential for viral replication. Comparing propagation of SeV-ΔP and SeV-PΔ2-77 did not reveal any significant difference. This conforms to earlier studies when recombinant viruses with an interrupted expression of the V protein were shown to generally grow well in tissue cultures [17 Delenda C, Hausmann S, Garcin D, Kolakofsky D. Normal cellular
replication of Sendai virus without the trans-frame, nonstructural V
protein Virology 1997; 228(1): 55-62., 18 Kato A, Kiyotani K, Sakai Y, Yoshida T, Nagai Y. The
paramyxovirus, Sendai virus, V protein encodes a luxury function
required for viral pathogenesis EMBO J 1997; 16(3): 578-87.]. Obviously the truncated V and W proteins do not influence replication – possibly due to the deletion which prevents the proteins from interaction with N°.
): SeV-wt infection produced the highest amount (41 x 106 ciu/ml in total) but all cells were already dead after 3 days. SeV-ΔP and SeV-PΔ2-77 provoked a delayed cytopathic effect (CPE; data not shown) and produced titers which were two times lower than the SeV-wt over the period of 5 days. This clearly shows that amplification of the deletion mutants with P protein provided in trans from the HCL allows high titer virus production. In addition no trans-dominant negative effect develops during co-expression of the virus-encoded PΔ2-77 and the full-length P protein, provided by the HCL, as demonstrated by similar titers of SeV-ΔP and SeV-PΔ2-77. Furthermore, the NS proteins expressed in SeV-wt infections but absent in SeV-ΔP or partially absent and truncated in SeV-PΔ2-77 infections had, at best, a minor effect on viral amplification. However, the V and W proteins were reported to have an influence on viral replication and gene expression [15 Horikami SM, Smallwood S, Moyer SA. The Sendai virus V
protein interacts with the NP protein to regulate viral genome RNA
replication Virology 1996; 222(2): 383-90., 16 Yu D, Shioda T, Kato A, Hasan MK, Sakai Y, Nagai Y. Sendai virus-based expression of HIV-1 gp120: reinforcement by the V(-) version Genes Cells Mol Cell Mech 1997; 2(7): 457-66.]. Importantly, both proteins harbor the domain from aa 2 to 77 known to be responsible for binding of P to N (P:N°), essential for viral replication. Comparing propagation of SeV-ΔP and SeV-PΔ2-77 did not reveal any significant difference. This conforms to earlier studies when recombinant viruses with an interrupted expression of the V protein were shown to generally grow well in tissue cultures [17 Delenda C, Hausmann S, Garcin D, Kolakofsky D. Normal cellular
replication of Sendai virus without the trans-frame, nonstructural V
protein Virology 1997; 228(1): 55-62., 18 Kato A, Kiyotani K, Sakai Y, Yoshida T, Nagai Y. The
paramyxovirus, Sendai virus, V protein encodes a luxury function
required for viral pathogenesis EMBO J 1997; 16(3): 578-87.]. Obviously the truncated V and W proteins do not influence replication – possibly due to the deletion which prevents the proteins from interaction with N°.
Genome Replication Deficiency of the SeV P Mutants
In order to validate our experimental set-up which should particularly reflect the situation at the beginning of an infection when only one initial NC is present in the cell, we examined the potential replication deficiency of our SeV P mutants. Checking viral amplification in a cell line not expressing the viral P protein is the optimal set-up. Therefore, Vero cells infected with SeV-ΔP or SeV-PΔ2-77 (MOI = 1) were monitored for a period of 10 days for any viral spread – detectable via GFP expression: in SeV-ΔP infection no EGFP expression could be observed, in SeV-PΔ2-77 infections only single green cells (comparable to Fig. 5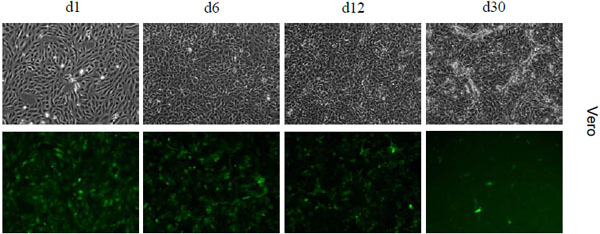 ). No spread of green fluorescence to neighboring cells could be found. In contrast, SeV-wt infections spread rapidly and destroyed all cells until day 3 post infection (p.i.). As further and more sensitive control, aliquots of the supernatants of infected Vero cells were taken at day 3 and day 10 and transferred onto fresh Vero cells and HCLs. Over a period of 7 days only SeV-wt progeny but no progeny of the SeV-P mutants could be detected.
). No spread of green fluorescence to neighboring cells could be found. In contrast, SeV-wt infections spread rapidly and destroyed all cells until day 3 post infection (p.i.). As further and more sensitive control, aliquots of the supernatants of infected Vero cells were taken at day 3 and day 10 and transferred onto fresh Vero cells and HCLs. Over a period of 7 days only SeV-wt progeny but no progeny of the SeV-P mutants could be detected.
In addition to this sensitive biological test setting above, replication-deficiency of SeV-ΔP and SeV-PΔ2-77 was verified directly via RT-PCR. Total cellular RNA was isolated from infected Vero cells (MOI = 1) at 24 h and 48 h p.i. A 3 h incubation time served as reference (initial amount of 5 x 105 input genomes), ensuring that virus adsorption and internalization is completed [19 Yonemitsu Y, Kitson C, Ferrari S, et al. Efficient gene transfer to
airway epithelium using recombinant Sendai virus Nat Biotechnol 2000; 18(9): 970-3.] and primary transcription is initialized [20 Plumet S, Duprex WP, Gerlier D. Dynamics of viral RNA
synthesis during measles virus infection J Virol 2005; 79(11): 6900-8.]. Equal amounts of RNA were subjected to selective reverse transcription of only the negative strand genomic viral RNA by employment of a SeV leader-specific positive-sense primer. Identical aliquots of the RT samples of each virus were used for a semi-quantitative endpoint PCR (Fig. 3A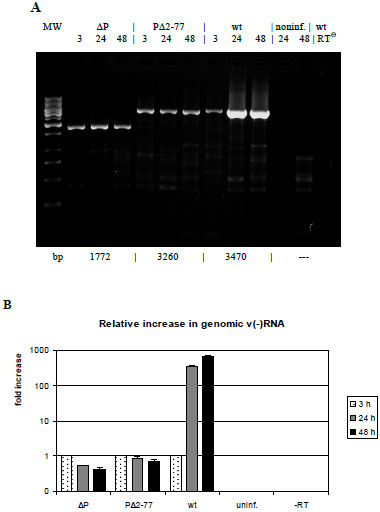 ) and a quantitative PCR analysis (Fig. 3B
) and a quantitative PCR analysis (Fig. 3B ). Both attempts reveal a constant (SeV-PΔ2-77) or marginally decreasing (SeV-ΔP) template number during the course of infection, in contrast to SeV-wt infections with a 366-fold (24 h) and 704-fold increase (48 h), respectively. In summary, our data demonstrate the incapability of both mutants SeV-ΔP and SeV-PΔ2-77 to replicate genomes in cells not providing the viral P protein. Thus, deletion the N-terminal 76 aa suffices to create a replication-deficient phenotype.
). Both attempts reveal a constant (SeV-PΔ2-77) or marginally decreasing (SeV-ΔP) template number during the course of infection, in contrast to SeV-wt infections with a 366-fold (24 h) and 704-fold increase (48 h), respectively. In summary, our data demonstrate the incapability of both mutants SeV-ΔP and SeV-PΔ2-77 to replicate genomes in cells not providing the viral P protein. Thus, deletion the N-terminal 76 aa suffices to create a replication-deficient phenotype.
Viral Transcription and Gene Expression with or without De Novo Synthesized P Protein
After validation of our test system which guarantees a constant minimal template number during infection experiments, we could next evaluate the influence of P gene products solely on viral transcription. First we looked at the immediate products of transcription, i.e. mRNA. Total RNA from infected Vero cells was isolated at 3 h (reference), 24 h and 48 h p.i. with SeV ΔP, PΔ2-77 and wt. After use of an oligo-dT primer for RT in order to transcribe only mRNA, a quantitative PCR analysis was performed amplifying a part of the N ORF (Fig. 4A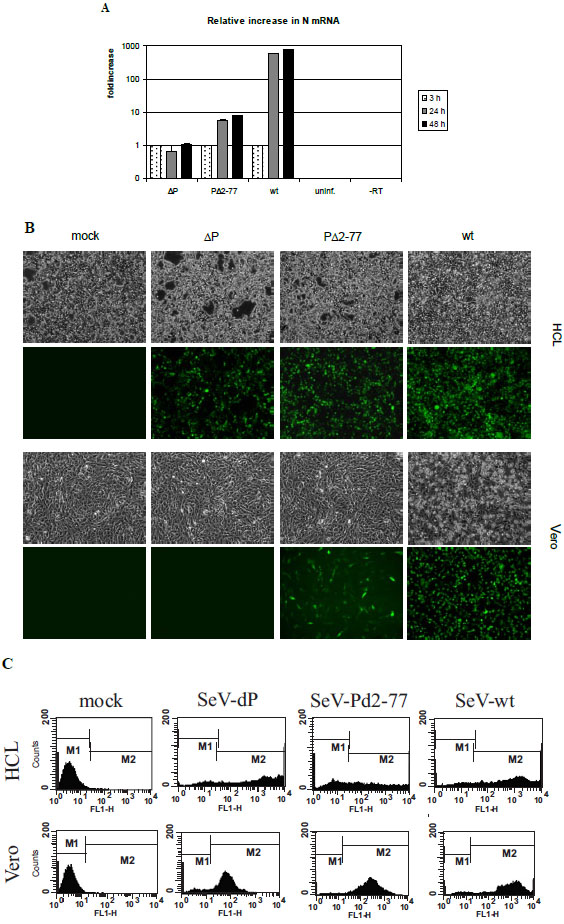 ). SeV-ΔP infected cells show no detectable increase from 3 h to 48 h p.i. Interestingly, SeV PΔ2-77 very clearly is able to transcribe in an enhanced mode – without being able to replicate viral genomes - showing a 5.7-fold (24 h) to 8.2-fold (48 h) increase in mRNA. Compared to SeV-wt (increase 596-fold at 24 h and 806-fold at 48 h p.i.), the rise in mRNA levels shows a parallel progression but in total at a 100-fold reduced amount. The 8.2-fold increase in 1° transcription during SeV-PΔ2-77 infections resulted statistically from only one template NC per infected cell. SeV-wt, as expected, transcribes at high rates, reaching the 800-fold amount of mRNA 48 h p.i. This high number is probably mainly based on 2° transcription after replication with 704-fold more genomic templates compared to 3 h (Fig. 3B
). SeV-ΔP infected cells show no detectable increase from 3 h to 48 h p.i. Interestingly, SeV PΔ2-77 very clearly is able to transcribe in an enhanced mode – without being able to replicate viral genomes - showing a 5.7-fold (24 h) to 8.2-fold (48 h) increase in mRNA. Compared to SeV-wt (increase 596-fold at 24 h and 806-fold at 48 h p.i.), the rise in mRNA levels shows a parallel progression but in total at a 100-fold reduced amount. The 8.2-fold increase in 1° transcription during SeV-PΔ2-77 infections resulted statistically from only one template NC per infected cell. SeV-wt, as expected, transcribes at high rates, reaching the 800-fold amount of mRNA 48 h p.i. This high number is probably mainly based on 2° transcription after replication with 704-fold more genomic templates compared to 3 h (Fig. 3B ).
).
Based on these surprising data that SeV-PΔ2-77 is able to efficiently support viral transcription at constant template number – even reaching SeV-wt performance when the effects from 2° transcription are deducted – we wanted to analyze next which consequences at the protein level could be detected. Here, we even have a special situation, because SeV-PΔ2-77 is unable to produce progeny virus in cells not trans-complementing P. This means synthesized proteins cannot be drained off the cell in form of released viral particles. Therefore, we compared protein expression levels from infected HCL and Vero cells. EGFP expression was monitored by fluorescence microscopy for a rough estimation (Fig. 4B ) and by FACS analysis to determine the number and the intensity of fluorescent cells (Fig. 4C
) and by FACS analysis to determine the number and the intensity of fluorescent cells (Fig. 4C ). In HCL, providing the full-length P protein in trans, both mutants induced a strong EGFP fluorescence at 48 h p.i. comparable to infections with SeV-wt (Fig. 4B
). In HCL, providing the full-length P protein in trans, both mutants induced a strong EGFP fluorescence at 48 h p.i. comparable to infections with SeV-wt (Fig. 4B , upper panel). In Vero cells, however, SeV-ΔP could not produce any visible fluorescence, while SeV-PΔ2-77 infected cells still showed a strong but slightly reduced expression compared to SeV-wt 48 h p.i. (Fig. 4B
, upper panel). In Vero cells, however, SeV-ΔP could not produce any visible fluorescence, while SeV-PΔ2-77 infected cells still showed a strong but slightly reduced expression compared to SeV-wt 48 h p.i. (Fig. 4B , lower panel).
, lower panel).
In the FACS analyses of the same setting quantification of gene expression revealed some interesting numbers. In the HCL infected with SeV-ΔP or SeV-PΔ2-77 a continuous distribution with fluorescence intensities comparable to wt infections could be observed (Fig. 4C , upper panel). In Vero cells, however, EGFP expression in SeV-ΔP infections was only weak, whereas SeV-PΔ2-77 induced a significant signal of medium intensity (peak at 300 FL-1H units) (Fig. 4C
, upper panel). In Vero cells, however, EGFP expression in SeV-ΔP infections was only weak, whereas SeV-PΔ2-77 induced a significant signal of medium intensity (peak at 300 FL-1H units) (Fig. 4C , lower panel). Strikingly, the EGFP level was only 7-10 times reduced compared to SeV-wt infections (peak at 2000 FL-1H units). This is a surprising result because it shows that gene expression from only one template on average can lead to a significant amount of protein, especially when taking the drastically differing levels of expressed mRNA into consideration (Fig. 4A
, lower panel). Strikingly, the EGFP level was only 7-10 times reduced compared to SeV-wt infections (peak at 2000 FL-1H units). This is a surprising result because it shows that gene expression from only one template on average can lead to a significant amount of protein, especially when taking the drastically differing levels of expressed mRNA into consideration (Fig. 4A ).
).
Taking all results of the different experiments together, we could clearly show that mutant SeV-PΔ2-77, but not SeV-ΔP, is able to actively transcribe and to express proteins to significant levels. In conclusion, domain aa 2 to 77 of the Sendai P protein seems to be dispensable for transition from early 1° transcription to late 1° transcription at the beginning of an infection.
Intracellular Survival Time of Replication-Deficient Nucleocapsids
Besides mRNA and protein expression levels during 1° transcription we wanted to evaluate NC stability as another critical parameter at the beginning of an infection. The longer stability and functionality of the infiltrated NC lasts, the higher the likelihood of viral survival and the establishment of a productive infection. We applied our replication-deficient SeV-PΔ2-77 and simulated infections of Vero cells (Fig. 5 ) with statistically calculated only one NC per cell. Only from day 6 p.i. on a fading EGFP signal could be observed. To exclude that this fading fluorescence was the result of a reduced viability and arrested cell division, we split infected Vero cells twice (at day 7 and day 16 p.i.). However, the same observation was made: EGFP expression decreased from day 6 p.i. on. Interestingly, this decline progressed slowly. As long as until day 30 p.i. fluorescent cells were still detectable. SeV-ΔP infected cells exhibited also a constantly moderate fluorescence for a similar period of time (data not shown) when infected with a MOI of 15. A presumed CPE resulting from SeV-PΔ2-77 infection was not visible - indicating that the decline in EGFP expression was rather due to inactivation of the NC inside the cell than the consequence of cellular break-down due to the viral infection. In conclusion, with respect to the half-life of EGFP of 26 h [21 Corish P, Tyler-Smith C. Attenuation of green fluorescent protein
half-life in mammalian cells Protein Eng 1999; 12(12): 1035-40.], we discovered that NCs remain functional during an infection for at least 5 days but maybe even much longer (> 15 days). This clearly extends existing knowledge that describes NC activity of SeV to last only 24 h. The documented period of at least 5 days seems to be a comfortable time frame for the virus to establish a productive infection.
) with statistically calculated only one NC per cell. Only from day 6 p.i. on a fading EGFP signal could be observed. To exclude that this fading fluorescence was the result of a reduced viability and arrested cell division, we split infected Vero cells twice (at day 7 and day 16 p.i.). However, the same observation was made: EGFP expression decreased from day 6 p.i. on. Interestingly, this decline progressed slowly. As long as until day 30 p.i. fluorescent cells were still detectable. SeV-ΔP infected cells exhibited also a constantly moderate fluorescence for a similar period of time (data not shown) when infected with a MOI of 15. A presumed CPE resulting from SeV-PΔ2-77 infection was not visible - indicating that the decline in EGFP expression was rather due to inactivation of the NC inside the cell than the consequence of cellular break-down due to the viral infection. In conclusion, with respect to the half-life of EGFP of 26 h [21 Corish P, Tyler-Smith C. Attenuation of green fluorescent protein
half-life in mammalian cells Protein Eng 1999; 12(12): 1035-40.], we discovered that NCs remain functional during an infection for at least 5 days but maybe even much longer (> 15 days). This clearly extends existing knowledge that describes NC activity of SeV to last only 24 h. The documented period of at least 5 days seems to be a comfortable time frame for the virus to establish a productive infection.
DISCUSSION
After release of a nucleocapsid (NC) into the host cell at the beginning of infection, two main factors are crucial for survival of the ongoing NNSV life cycle: Stability of the NC - until progeny templates are synthesized - and in parallel efficient primary (1°) transcription. To allow insights into early stages of viral RNA synthesis we created a new test system that provides a constant minimal template number in virus infected cells (i.e. statistically one input NC per cell on average) by using specific viral mutants (SeV-ΔP and SeV-PΔ2-77) with which viral transcription could be uncoupled from genome replication.
The survival time of NCs - a crucial factor in early phase of an infection - and the processivity of the viral polymerase (vPol) during 1° transcription could now be investigated at constant template number, without being affected by NC progeny. Upon entry of the NC - containing 30 to 50 vPol molecules - into the host cell, its stability is a prerequisite for sufficient primary transcription. Using SeV-PΔ2-77 we were able to monitor viral gene expression kinetically based on only one template in the infected cell. Interestingly, a constant EGFP expression could be detected for up to 6 days. With respect to the EGFP half-life of ca. 26 h [21 Corish P, Tyler-Smith C. Attenuation of green fluorescent protein half-life in mammalian cells Protein Eng 1999; 12(12): 1035-40.], a NC activity of at least 5 days can be concluded. The readout of our test system for NC stability is coupled to the activity of early 1° transcription. Therefore, we cannot exclude that early 1° de novo synthesized viral proteins are influencing NC stability. However, due to the fact that even NCs of the SeV-ΔP mutant mediated constant EGFP expression, we conclude no relevant influence of de novo synthesized P protein on NC stability. If any of the remaining viral gene products affect NC stability positively could not be determined here. Surprisingly, in previous studies an intracellular survival time of NCs of only one day was described [3 Mottet G, Curran J, Roux L. Intracellular stability of nonreplicating paramyxovirus nucleocapsids Virology 1990; 176(1): 1-7.]. However, these studies were performed using DIPs that do not express any viral genes which might causes the underestimation of the NC half-life. Furthermore, such a short survival time of NCs would not correspond well with the development of persistent infections during which RNA synthesis is carried out at very low levels [22 Cattaneo R, Schmid A, Rebmann G, et al. Accumulated measles virus mutations in a case of subacute sclerosing panencephalitis: interrupted matrix protein reading frame and transcription alteration Virology 1986; 154(1): 97-107., 23 Homann HE, Hofschneider PH, Neubert WJ. Sendai virus gene expression in lytically and persistently infected cells Virology 1990; 177(1): 131-40.].
Another key factor in viral life cycle is the phase of 1° transcription during which sufficient viral gene products have to be synthesized in order to ensure amplification of the initial NC and thereby secure viral survival. At the very beginning early 1° transcription functions without support of any de novo synthesized viral proteins. In SeV-ΔP infections we could confirm a moderate but constant mRNA level through early 1° transcription compared to SeV-wt infections, measured by quantitative RT-PCR. In SeV-PΔ2-77 infections a further support of transcription was not expected, since complex formation between N and P without domain aa 1-77 was described to be unstable [24 Curran J, Marq JB, Kolakofsky D. An N-terminal domain of the Sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication J Virol 1995; 69(2): 849-55.]. However, when PΔ2-77 proteins were synthesized during infections, mRNA levels rose towards late 1o transcription. So far, N and P proteins have been shown to be essential for late 1° transcription [1 Wiegand MA, Bossow S, Schlecht S, Neubert WJ. De novo synthesis of N and P proteins as a key step in Sendai virus gene expression J Virol 2007; 81(24): 13835-44.]. Here we demonstrate that also truncated PΔ2-77 is sufficient for an 8-fold increase of mRNA levels even though it carries a deletion which could influence P protein function. Thus, this N-terminal domain is not essential for functionality of P in viral transcription - unlike in genome replication. During SeV-wt infections, the overall amount of mRNA is 100-fold higher compared to SeV-PΔ2-77 infections due to 2° transcription from newly synthesized progeny genomes. But when looking at the protein level instead of mRNA, gene expression by PΔ2-77 reaches 7-10% of the wild type protein amount. This might be the result of a reduced burden for the cell due to a reduced viral CPE and the fact that no progeny particles drain off viral products due to abrogated genome replication.
Early 1° transcription runs without de novo synthesized P and N proteins but late 1° transcription necessitates the expression of both N and P proteins. For subsequent viral replication N protein has to be bound in a complex to P (P:N°) to be prevented from illegitimate binding to RNA [11 Curran J, Homann H, Buchholz C, Rochat S, Neubert W, Kolakofsky D. The hypervariable C-terminal tail of the Sendai paramyxovirus nucleocapsid protein is required for template function but not for RNA encapsidation J Virol 1993; 67(7): 4358-64.] and only this P:N° is a substrate for encapsidation of the nascent RNA chain. Interestingly, also truncated P protein supports late 1° transcription and consequently a P:N° with P as PΔ2-77 should be involved. Taking all this into account one can postulate that in P:N° two domains of P that are responsible for the interaction with N are present at the same time. One serves the process of RNA synthesis in transcription and replication - by mediating the interaction of the viral polymerase with the RNP template. The other functions in encapsidation of the nascent genomic RNA strand. Besides aa 1-77 relevant for nascent RNA encapsidation, another more C-terminal domain supports vPol processivity. That domain may be located within aa 78-144 as concluded earlier from in vitro experiments [4 Curran J, Pelet T, Kolakofsky D. An acidic activation-like domain of the Sendai virus P protein is required for RNA synthesis and encapsidation Virology 1994; 202(2): 875-4.]. Domain 78-144 was suggested to be able to compensate for the supportive function of aa 1-77 but complexes of P and N without domain aa 1-77 were described to be unstable [24 Curran J, Marq JB, Kolakofsky D. An N-terminal domain of the Sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication J Virol 1995; 69(2): 849-55.]. However, in our opinion, the existence of two alternative domains both dedicated to the same function, i.e. genome replication, does not make sense from an evolutionary point of view. We therefore postulate the existence of a separate domain in addition to aa 1-77 that supports vPol processivity during transcription but probably also during replication.
In order to further define this supportive activity of P, the influence of additionally expressed V and W proteins during SeV-ΔP infections on transcription was evaluated (data not shown). Both proteins – formerly described as inhibiting genome replication [4 Curran J, Pelet T, Kolakofsky D. An acidic activation-like domain of the Sendai virus P protein is required for RNA synthesis and encapsidation Virology 1994; 202(2): 875-4.] – contain the domains aa 1-77 and 78-144; however, in our approach they did not support late 1° transcription. This indicates that the supportive function of P for vPol processivity is not only based on two domains within the N-terminal part of P (aa 1-77 and 78-144) but at the same time on domains within the C-terminal part (aa 318-568). Suitable domains within P would be: (i) aa 412-445, responsible for the interaction with L in the vPol complex or (ii) aa 479-568, responsible for binding to N as part of the RNP template during RNA synthesis [24 Curran J, Marq JB, Kolakofsky D. An N-terminal domain of the Sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication J Virol 1995; 69(2): 849-55.]. Furthermore, the vPol is able to transcribe with the help of PΔ2-77:N° complexes, as shown here by late 1° transcription during SeV-PΔ2-77 infections. Thus, these C-terminal domains of P seem not to be inhibited by the N-terminal deletion.
In conclusion, the SeV P protein seems to contain two different domains for binding monomeric N protein. The one located C-terminally beyond aa 78 supports vPol processivity during all RNA synthesis activities, whereas the N-terminal domain (aa 2-77) is necessary specifically for the process of genome replication. In this way, de novo synthesized P protein early in infection plays an important role in securing viral survival by supporting vPol activity but not NC stabilization. The nucleocapsid remains stable for several days even without de novo synthesized P protein.
MATERIAL & METHODS
Cells and Viruses
Vero cells were purchased from ATCC (CCL-81). The helper cell line (HCL; derived from HEK 293 cells) expresses constitutively SeV P protein [12 Willenbrink W, Neubert WJ. Long-term replication of Sendai virus defective interfering particle nucleocapsids in stable helper cell lines J Virol 1994; 68(12): 8413-7.]. BSR-T7 - BHK cells expressing T7 RNA polymerase [25 Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter J Virol 1999; 73(1): 251-9.] - were a kind gift from K.-K. Conzelmann (Munich). Cell lines were maintained in DMEM (Invitrogen) supplemented with 10% heat-inactivated FCS (Invitrogen). Recombinant SeV derived from strain Fushimi (murine PIV1, ATCC VR-105). All incubation steps of infected cells took place at 33 °C.
Construction of Modified Genomic SeV Plasmids
Numbering of positions given below refers to GenBank submission M30202, sequences are denoted in the positive antigenomic direction. The plasmid pRS3Gg, containing the SeV wt genome [26 Leyrer S, Neubert WJ, Sedlmeier R. Rapid and efficient recovery of Sendai virus from cDNA: factors influencing recombinant virus rescue J Virol Methods 1998; 75(1): 47-58.], was used as a template to create fragments harboring an alternate P gene product by PCR (“Expand High Fidelity PCR system”, Roche). The fragment encoding the N-terminally truncated protein PΔ2-77 was amplified with the primer pair PΔ2-77 Xho for (5’-CCCCCTTTTTCTCGAGATGTCGACCCAAGATAATCGATCAGGTGAGG-3’) and PΔ2-77 Xho rev (5’-TTTTT CCCCCCTCGAGTTACTAGTTGGTCAGTGACTCTATGTCCTC-3’), fusing the P start codon directly to the codon for amino acid 78 Ser and introducing XhoI restriction sites (underlined) flanking the ORF. Construction of the genomic plasmids containing the SeV wt genome or the mutant SeV ΔP lacking the entire P ORF (corresponding to original position 1844-3550) was described previously [1 Wiegand MA, Bossow S, Schlecht S, Neubert WJ. De novo synthesis of N and P proteins as a key step in Sendai virus gene expression J Virol 2007; 81(24): 13835-44.]. The plasmid of SeV PΔ2-77 was obtained by inserting the PCR product into the “empty” transcription cassette of SeV ΔP via XhoI. All genomic constructs contain an additional transcription unit with the EGFP ORF between the leader and the N gene [27 Bernloehr C, Bossow S, Ungerechts G, et al. Efficient propagation of single gene deleted recombinant Sendai virus vectors Virus Res 2004; 99(2): 193-7.] as well as the oligobasic F cleavage site Fmut and were confirmed by sequencing.
Rescue of Recombinant SeV and Propagation
Recombinant viruses were recovered from transfected BSR-T7 cells as described in Wiegand et al. [1 Wiegand MA, Bossow S, Schlecht S, Neubert WJ. De novo synthesis of N and P proteins as a key step in Sendai virus gene expression J Virol 2007; 81(24): 13835-44.] with a modification of the rescue procedure for the P mutants. After three days, the cells were transfected a second time with 1.0 µg pTM-P/C- and incubated for 2-3 more days. The recovered viruses were propagated on the P-expressing helper cell line (HCL) several times using DMEM + 5% FCS. Virus stocks were prepared from the 5th passage and used for all following experiments.
Replication Kinetics, Titration and Fluorescent Micrographs
Confluent HCL monolayers in 12-well plates (5 × 105 per well) were inoculated with virus (MOI = 3). After adsorption (1 h at 33 °C), the inoculum was removed and cells were washed twice before adding 1 ml medium supplemented with 5% FCS. Supernatants were isolated at the indicated time points and replaced by 1 ml fresh medium. Released progeny viruses were titrated in duplicate on HCL in a dilution series, counting EGFP-positive cells. Cells were monitored by live cell fluorescence microscopy (488 nm excitation) using a Leica DM IL microscope (Leica Microsystems), a F-View II CCD camera and AnalySIS software (Soft Imaging Systems).
Reverse Transcription, PCR and Quantitative PCR
Total cellular RNA was isolated (RNeasy; Qiagen) from infected cells at the indicated time points. Applying 1 µg RNA each, viral (-)genomes were selectively reverse transcribed (Superscript III; Invitrogen) using the primer “ld-25” 5’-TGGAATATATAATGAAGTCAGA-3’ (nt 25 to 46). An aliquot of the resulting viral genome-specific cDNA (1/10th) was employed in a standard PCR with the oligonucleotides “N-1280” 5’-GGATACAGCCAAGGAGA GGC-3’ (nt 1280 to 1299) and “M-4749+” 5’-GGGGCATT GTCGCAGGTC-3’ (nt 4749 to 4732). To amplify viral mRNAs, 1 µg of each total RNA was reverse transcribed with an oligo(dT)20 primer. The conditions for the RT and PCR were as in Wiegand et al. [1 Wiegand MA, Bossow S, Schlecht S, Neubert WJ. De novo synthesis of N and P proteins as a key step in Sendai virus gene expression J Virol 2007; 81(24): 13835-44.].
For real-time quantitative PCR, different sets of primers were designed using the ABI Software Primer Express (Applied Biosystems). The primer pair “SendaiNF” 5’-GCCAG AGGAGCACAGTCTCAGT-3’/“SendaiNR” 5’-GTCCAAT GAGTGAGCTAGGAAGG-3’ was applied for amplification of a 101 nt long region within the N gene. Aliquots (1/20th) of viral (-)genome-specific and mRNA-specific RT samples were amplified using “SYBR Green Taqman Universal Mastermix” (25 µl volume) with activation (50 °C, 2 min and 95 °C, 10 min), followed by 40 cycles of 95 °C, 15 sec and 60 °C, 1 min. The reactions were performed and detected in MicroAmp Optical 96-well Reaction Plates on an ABI PRISM 7900 Sequence Detection System (9600 emulation mode). At completion, the threshold cycles were determined and the data were analyzed via the Sequence Detection Software 2.1 (Applied Biosystems).
Flow Cytometry
Infected cells were trypsinized, washed and resuspended in 200 µl PBS (approximately 5 × 105 cells). The number of EGFP-expressing cells was immediately analyzed by flow cytometry (FACScalibur; Becton Dickinson) using CellQuest software.
ACKNOWLEDGEMENTS
We thank Michael Schurig for establishing and optimizing the rescue procedure based on BSR-T7 cells. This work was supported by grants from the Bundesministerium für Bildung und Forschung (AZ: 0312193) and from the European Commission (QLK2-2002-01722).
CONFLICT OF INTEREST
All authors declare no conflict of interest.


