- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Effects of Hepatitis B Virus Mutations on its Replication and Liver Disease Severity
Abdulrahim Hakami*, 1, Abdelwahid Ali2, Ahmed Hakami2
Abstract
Hepatitis B virus (HBV), nowadays, is one of the major human pathogens worldwide. Approximately, 400 million people worldwide have chronic HBV infection. Only 5% of persons infected during adulthood develop chronic infection. The reverse is true for those infected at birth or in early childhood, i.e. more than 90% of these persons progress to chronic infection. Currently, eight different genotypes o f HBV have been identified, differing in nucleotide sequence by greater than 8%. In addition, numerous subgenotypes have a l s o been recognized based on the nucleotide sequence variability of 4- 8%. It has invariably been found that these genotypes and mutations play a pivotal role in the liver disease aggravation and virus replication. The precore mutations (G1896A) and the double mutation (T1762/A1764) in the basal core promoter are important mutations that alter expression of the hepatitis B e antigen (HBeAg). The HBeAg is important for establishing viral persistence. The precore G1896A mutation abrogates the expression of HBeAg. Numerous other mutations alter the disease severity and progression. It is predictive that the infected patient has high risk of hepatocellular carcinoma if the genotype C is incriminated or if HBV possesses basal core promoter double mutation. Association of the remaining genotypes have been noted but with less degree than genotype C. Phenotypic assays of the different HBV protein markers with different molecular techniques illustrate the replication efficiency of the virus in cell lines. This review will discuss various mutations into their association with liver disease severity and progression as well as virus replication.
Article Information
Identifiers and Pagination:
Year: 2013Volume: 7
First Page: 12
Last Page: 18
Publisher Id: TOVJ-7-12
DOI: 10.2174/1874357901307010012
Article History:
Received Date: 17/10/2012Revision Received Date: 26/11/2012
Acceptance Date: 27/11/2012
Electronic publication date: 23/1/2013
Collection year: 2013
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Department of Medical Laboratory Sciences, College of Applied Medical Sciences, King Khalid University, P.O. Box 3665, Abha 61481, Saudi Arabia; Tel: +966-72417090 ; Fax: +966-72417746; E-mail: abdu.lightpollution@hotmail.com
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 17-10-2012 |
Original Manuscript | Effects of Hepatitis B Virus Mutations on its Replication and Liver Disease Severity | |
INTRODUCTION
Hepatitis B virus (HBV) has high prevalence worldwide. More than 2 billion people have been infected with around 400 million individuals had chronic HBV infection representing approximately 5% of the world population with more than half million deaths annually due to liver cirrhosis or hepatocellular carcinoma [1 Al-Mahtab M, Rahman S, Khan M, Karim F. Hepatitis B virus genotypes: an overview Hepatobiliary Pancreat Dis Int 2008; 7(5): 457-64., 2 Huang Y, Lok AS. Viral factors and outcomes of chronic hbv infection Am J Gastroenterol 2011; 106: 93-5.]; this indicates that the virus is highly contagious. HBV has four subtypes i.e. ayw, ayr, adw, and adr. At least 8 different HBV genotypes have been identified (A-H) where the nucleotide sequence varies by at least 8% [3 Nakajima A, Usui M, Huy TT, et al. Full-Length Sequence of Hepatitis B virus belong to genotype h identified in a japanese patient with chronic hepatitis Jpn J Infect Dis 2005; 58: 244-6., 4 Devesa M, Rodríguez C, Le??n G, Liprandi F, Pujol FH. Clade analysis and surface antigen polymorphism of hepatitis b virus american genotypes J Med Virol 2004; 72: 377-84.]. Recently, new genotype (I) has also been isolated from Laos, and genotype (J) was identified in Japan [5 Thedja MD, Muljono DH, Nurainy N, Sukowati CH, Verhoef J, Marzuki S. Ethnogeographical structure of hepatitis B virus genotype distribution in Indonesia and discovery of a new subgenotype, B9 Arch Virol 2011; 156 (5): 855-68.]. Many clinical and biological characteristics including transmission, seroconversion and frequency of mutations are completely dependant on HBV genotype (Table 1). These different genotypes generally have restricted geographic distributions, with HBV genotype A is found in North America and Africa, genotype B and C are dominant in Asia, and genotype D present in Mediterranean countries and Europe [6 Ni YH, Chang MH, Wang KJ, et al. Clinical relevance of hepatitis b virus genotype in children with chronic infection and hepatocellular carcinoma Gastroenterology 2004; 127: 1733-8.-9 Lin CL, Kao JH. The clinical implications of hepatitis B virus genotype: Recent advances J Gastroenterol Hepatol 2011; 26: 123-30.] as well as some districts in western Japan [10 Michitaka K, Horiike N, Chen Y, et al. Co-Infection with Hepatitis B Virus Genotypes D and other Genotypes in Western Japan Intervirology 2005; 48: 262-7.]. Numerous subgenotypes that differ in nucleotide sequence by 4-8% have also been identified, and in some cases these too have a geographical limited distribution. For instance, subgenotype Aa (also known as A1) is generally restricted to Africa and Asia, whereas subgenotype Ae (A2) is more frequent in Europe. Subgenotype B1 is restricted to Japan, whereas subgenotypes B2-5 are found throughout China, Indonesia, Vietnam, Philippine respectively. Similarly, HBV genotype C is classified into C1 and C2 subgroups based on preS gene phylogenesis [11 Tanaka Y, Hasegawa I, Kato T, et al. A case-control study for differences among hepatitis b virus infections of genotypes a (subtypes Aa and Ae) and D Hepatology 2004; 40: 747-55.-13 Chan HL, Tsui SK, Ng EY, et al. Epidemiological and virological characteristics of 2 subgroups of hepatitis B virus genotype C J Infect Dis 2005; 191: 2022-32.]. The genotype D subgenotypes have no specified geographic distribution, but D1 was derived from Middle East, D2 from Japan, and a recent subgenotype D6 has been described in Tunisia [14 Schaefer S, Magnius L, Norder H. Under construction: classification of hepatitis B virus genotypes and subgenotypes Intervirology 2009; 52: 323-5., 15 Kramvis A, Arakawa K, Yu MC, Nogueira R, Stram DO, Kew MC. Relationship of serological subtypes, basic core promoter and precore mutations to genotypes/subgenotypes of hepatitis B virus J Med Virol 2008; 80: 27-46.].
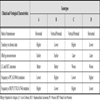
HBV Genotypes and the Comparison of Virological and Clinical Differences [9 Lin CL, Kao JH. The clinical implications of hepatitis B virus genotype: Recent advances J Gastroenterol Hepatol 2011; 26: 123-30.]

Virological Characteristics of PC 1896 and BCP 1762/1764 Mutants [44 Tong MJ, Blatt LM, Kao JH, Cheng JT, Corey WC. Precore/basal core promoter mutants and hepatitis B viral DNA as predictors for liver deaths and hepatocellular carcinoma World J Gastroenterol 2006; 12(41): 6620-.]
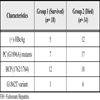
Data of Some Serological Correlates and Disease Outcome and Genomic Mutations from 52 Patients with FH [52 Hou J, Lin Y, Waters J, et al. Detection and significance of a G1862T variant of hepatitis B virus Chinese patients with fulminant hepatitis J Gen Virol 2002; 83: 2291-8.]

Comparison of Three HBV Genotypes and HBeAg Among Different Clinical Patients in Philippines [8 Sakamoto T, Tanaka Y, Orito E, et al. Novel subtypes (subgenotypes) of hepatitis B virus genotypes B and C among chronic liver disease patients in the Philippines J Gen Virol 2006; 87: 1873-82.]
HBV genome is approximately 3.2 kbp. There are four overlapping open reading frames (ORFs) organized in HBV genome. They are C ORF that encodes Hepatitis B core antigen (core or capsid protein; HBcAg) which is a non-structural form, also encodes HBe Ag, P ORF (Polymerase) which has a reverse transcription activity, S ORF (surface protein; HBsAg) and X ORF which was documented to play a role in the development of HBV- related hepatocellular carcinoma, HCC [16 Jalali MV, Alavian SM. Hepatitis B e antigen-negative chronic hepatitis B Hep Mon 2006; 6: 31-5.-19 Hwang GY, Lin CY, Huang LM, et al. Detection of the hepatitis B virus X protein (HBx) antigen and anti-hbx antibodies in cases of human hepatocellular carcinoma J Clin Microbiol 2003; 41(12): 5598-603.]. The basal core promoter (BCP) is located in the X ORF also [20 Yang HI, Yeh SH, Chen PJ, et al. Associations between hepatitis B virus genotype and mutants and risk of hepatocellular carcinoma J Natl Cancer Inst 2008; 100: 1134-43.]. There are two direct repeats, 11 nucleotides each; DR1 (1822-1832) and DR2 (1588-1598) that are involved in the viral replication. DR1 is located in the 3' end of RNA primer. DR2 is the complementary sequence and is close to the 5َ end of the negative DNA strand [21 Haines KM, Loeb DD. The sequence of the RNA primer and the DNA template influence the initiation of plus-strand DNA synthesis in hepatitis B virus J Mol Biol 2007; 370: 471-80.]. Pregenomic RNA (pgRNA) is the major transcription template, as well as the mRNA for core and polymerase. Encapsidation signal (ε) is a hairpin-like structure that consists of 5َ UUC3'. It is the most common cis-element in the pgRNA. The mechanism by which HBV enters the cell is unknown; however, the replication cycle following entry is well characterized. After entering the cell, the viral envelope and protein coats are removed in the cytoplasm and the relaxed circular DNA (rcDNA) is converted to covalently closed circular DNA (cccDNA) in the nucleus [22 Sun D, Nassal M. Stable HepG2- and HuH7-basd human hepatoma cell lines for efficient regulated expression of infectious hepatitis B virus J Hepatol 2006; 45: 636-45.] where it serves as RNA transcription template. Although HBV is a DNA encoding virus, the virus uses reverse transcriptase (RT) as part of its replication cycle. This reverse transcription uses the nuclear cccDNA as a template to produce virions [23 Lentz T, Leob DD. Development of cell cultures that express hepatitis B virus to high levels and accumulate cccDNA J Virol Methods 2010; 169: 52-60.]. After virus infection of hepatocytes, cccDNA is detected within 24 hours [24 Seeger C, Mason W. Hepatitis B Virus Biology Microbiol Mol Biol Rev 2000; 64: 51-68.]. There are around 50 copies of cccDNA in the infected hepatocyte [25 Beck J, Nassal M. Hepatitis B virus replication World J Gastroenterol 2007; 13(1): 48-64.] that can be detected currently by PCR [26 Gao YT, Han T, Li Y, et al. Enhanced specificity of real-time PCR for measurement of hepatitis B virus cccDNA using restriction endonuclease and plasmid-safe ATP-dependent DNase and selective primers J Virol Methods 2010; 169: 181-7.]. Two different RNAs of 3.5 kb (pregenomic RNA and precore RNA) and three other viral RNAs (2.4, 2.1 and 0.8 kb) are transcribed [27 Inoue J, Ueno Y, Nagasaki F, et al. Enhanced intracellular retention of a hepatitis B virus strain associated with fulminant hepatitis Virology 2009; 395: 202-9.]. Transcription of HBV RNA as well as expression of polymerase and core protein is essential to create in vitro replication of HBV DNA and synthesis of its intermediates [28 Hu JL, Cui J, Deng XY, et al. A new strategy for constructing in vitro replication-competent 1.3 copies of hepatitis B virus genome J Virol Methods 2009; 161: 63-9.]. RNA transcription is mediated by host RNA polymerase II and regulated by two enhancers, four promoters and glucocorticoid-responsive element (GRE). Precore RNA and pgRNA are directed by the core promoter [29 Tong S. Mechanism of HBV genome variability and replication of HBV mutants J Clin Virol 2005; S: 134-8.]. The pgRNA is then digested by RNase H activity of viral polymerase but leaving F undigested primer of around 17 nucleotides in the pgRNA 5' end from which (+)-DNA strand is synthesized after template translocation to DR2, then this strand is extended to the 5' end of the (-)-DNA strand. After primer circularization in a subsequent pathway, the rcDNA is made [30 Lewellyn EB, Loeb DD. Base pairing between cis-acting sequences contributes to template switching during plus-strand DNA synthesis in human hepatitis B virus J Virol 2007; 81: 6207-15.]. Polymerase uses 5َ end of internal bulge site in the encapsidation signal (ε) which plays a fundamental role in (-)-strand DNA synthesis and pgRNA encapsidation [31 Abraham TM, Loeb DD. The Topology of Hepatitis B virus pregenomic RNA promotes its replication J Virol 2007; 81: 11577-84.]. The closed-loop of pgRNA is formed by cellular eIF-4G by means of both cap and poly (A) binding proteins. The molecular chaperon heat shock protein 90 (HSP90) is required for RT-(ε) binding [32 Hu J, Flores D, Toft D, Wang X, Nguyen D. Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function J Virol 2004; 78: 13122-31., 33 Hu J, Boyer M. Hepatitis B Virus Reverse Transcriptase and ?? RNA Sequences Required for Specific Interaction In Vitro J Virol 2006; 80: 2141-50.]. The mechanism of capsid formation is unclear but recent results explained that HSP90 is necessary to form the capsid by binding to core protein dimmers [34 Shim HY, Quan X, Yi YS, Jung G. Heat shock protein 90 facilitates formation of the HBV capsid via interacting with the HBV core protein dimmers Virology 2011; 410: 161-9.].
It is laborious to propagate in vitro HBV due to the lack of a robust HBV cell culture and non availability of animal infection model. Cell lines usually lose then tropism to the virus few days after transfection but Huh7 and HepG2 (ATCC HB-8065) cell culture were established as standard HBV cell lines [22 Sun D, Nassal M. Stable HepG2- and HuH7-basd human hepatoma cell lines for efficient regulated expression of infectious hepatitis B virus J Hepatol 2006; 45: 636-45., 35 Lupberger J, Mund A, Kock J, Hildt E. Cultivation of HepG2.15 on Cytodex-3: higher yield of hepatitis B virus and less subviral particles compared to conventional culture methods. J Hepatol 2006; 45(4): 547-2.]. For instance, HBV replication causes HepG2 apoptosis [36 Lu YW, Tan TL, Chan V, Chen WN. The HBSP gene is expressed during HBV replication, and its coded BH3-containing spliced viral protein induces apoptosis in HepG2 cells Biochem Biophys Res Commun 2006; 351: 64-70.], therefore it can be valuable cells in the replication study task. Other animal models exist as hosts for human HBV, among which are Orangutan, Chimpanzee, and Gibbon [37 Dandri M, Voltz TK, Lütgehetmann M, Peterson J. Animal models for the study of HBV replication and its variants J Clin Virol 2007; 152: 1159-73.]. Different HBVs replicate differently in different cell lines; however, hepatic cell lines QSG-7701 showed higher HBV DNA than HepG2 [38 Pan XB, Wei L, Chen HS, Liu F, Gao Y. Liver-derived cell lines QSG7701 and HepG2 support different HBV replication patterns Arch Virol 2007; 152: 1159-73.]. In terms of phenotypic characteristics, the replication products are measured in the cell culture supernatant and then undergo further analyses. Intracellular expression of HBV proteins and the pathogenicity of different genotypes have been compared [39 Sugiyama M, Tanaka Y, Kato T, et al. Influence of hepatitis b virus genotypes on the intra and extracellular expression of viral DNA and antigens Hepatology 2006; 44: 915-24.]. Although a number of old and recent data exist, association of some genotypes with disease outcome is still not conclusive. There are some essential questions to be addressed, for example how do different HBV mutations involve in the severity of the disease?. What is the most virulent genotype and why do HBV variants yield different replication phenotype?. Since the discovery of HBV by Baruch Blumberg in 1965, scientists started to sequence its genome on a regular basis in an attempt to decipher some ambiguity of genomic correlation. The fact that HBV mutate slowly and continuously makes it complicated to fully understand the role of every mutation. This review is an attempt to correlate some mutations in the HBV genome to the virus replication and disease outcome.
HISTORY AND LIVER DISEASE PROGRESSION
The majority of adult-acquired HBV infection is acute and self-limiting. Around 5% of infected patients, during adulthood, will develop chronic hepatitis (CH), and may progress to liver cirrhosis (LC) or hepatocellular carcinoma (HCC). In some clinical cases, HCC can occur without cirrhosis [2 Huang Y, Lok AS. Viral factors and outcomes of chronic hbv infection Am J Gastroenterol 2011; 106: 93-5.]. In chronic carrier patients, HCC has an annual incidence of less than 1% for non- cirrhotic patients and 2-3% for those with cirrhosis [9 Lin CL, Kao JH. The clinical implications of hepatitis B virus genotype: Recent advances J Gastroenterol Hepatol 2011; 26: 123-30.]. Unfortunately, elevated HBV DNA in the serum has been associated with deaths in non-HCC patients [9 Lin CL, Kao JH. The clinical implications of hepatitis B virus genotype: Recent advances J Gastroenterol Hepatol 2011; 26: 123-30.]. Fulminant hepatitis is due to enhanced virus replication and occurs in 1% of infected individuals [27 Inoue J, Ueno Y, Nagasaki F, et al. Enhanced intracellular retention of a hepatitis B virus strain associated with fulminant hepatitis Virology 2009; 395: 202-9.]. Certain HBV excretory proteins are used as markers for disease severity e.g., HBsAg positivity indicates disease chronicity whereas the marker of active viral replication is HBeAg. Elevated alanine transaminase (ALT) enzyme is also used as marker of viral hepatitis or liver damage. It is becoming increasingly apparent that HBV genotype influences HBV natural history and disease pathogenesis. Based on various studies, this genotypic diversity plays a crucial role in the pathogenesis and its prognosis [40 Livezey KW, Negorev D, Simon D. Hepatitis B virus-transfected Hep G2 cells demonstrate genetic alterations and de novo viral integration in cells replication HBV Mutat Res 2000; 452: 163-78.]. HBV genotype A is predominant in asymptomatic individuals and the genotype most frequently associated with severe liver disease, including HCC is genotype C [41 Toan NL, Song LH, Kremsner PG, et al. Impact of the hepatitis b virus genotype and genotype mixtures on the course of liver disease in Vietnam Hepatology 2006; 43: 1375-84.], which is associated with more serious liver illnesses compared to genotype B [42 Tanaka Y, Orito E, Yuen MF, et al. Two subtypes (subgenotypes) of hepatitis B virus genotype C: a novel subtyping assay based on restriction fragment length polymorphism Hepatol Res 2005; 33: 216-4.].
SOME INSIGHTS OF HBV MUTATIONS RELATEDNESS TO THE VIRUS REPLICATION
Variability of HBV genome is basically attributed to lack of proofreading during the replication process and the high copy number of the virus [43 Sheldon J, Rodès B, Zoulim F, Bartholomeusz A, Soriano V. Mutations affecting the replication capacity of the hepatitis B virus J Viral Hepatitis 2006; 13: 427-34.]. Characteristically, single mutation has different effects on viral replication and/or disease severity than double mutation. Virus mutation was predictive for liver disease but the role of precore (PC) mutation (A1896) in HCC is still not clear [44 Tong MJ, Blatt LM, Kao JH, Cheng JT, Corey WC. Precore/basal core promoter mutants and hepatitis B viral DNA as predictors for liver deaths and hepatocellular carcinoma World J Gastroenterol 2006; 12(41): 6620-.]. Furthermore, association of 1896 mutation with fulminant hepatitis has been documented [27 Inoue J, Ueno Y, Nagasaki F, et al. Enhanced intracellular retention of a hepatitis B virus strain associated with fulminant hepatitis Virology 2009; 395: 202-9.]. The G1896A precore stop codon mutation at the codon 28 decreased HBV replication if paired with T1858 mutation [43 Sheldon J, Rodès B, Zoulim F, Bartholomeusz A, Soriano V. Mutations affecting the replication capacity of the hepatitis B virus J Viral Hepatitis 2006; 13: 427-34.]. Replicative intermediates were higher intracellularly for the PC mutant compared to wild type (WT) virus [45 Heipertz RA, Miller TG, Kelley CM, Delaney WE, Locarnini SA, Isom HC. In vitro study of the effects of precore and lamivudineresistant mutations on hepatitis B virus replication J Virol 2007; 81: 3068-76.]. Intermittent and persistent replication in vivo is associated with dual PC/BCP mutation [46 Biswas A, Banerjee A, Chandra PK, et al. Variations in the functional domain of basal core promoter of hepatitis B virus among eastern indian patients with prevalence of genotypes A, C, and D among the same ethnic population J Med Virol 2011; 83: 253-60.]. Furthermore, triple BCP mutation; T1762/A1764, and 1766 increases transcription of pgRNA whereas the double mutation 1762/1764 is less effective [47 Günther S, Piwon N, Jung A, Iwanska A, Schmitz H, Will H. Enhanced replication contributes to enrichment of hepatitis B virus with a deletion in the core gene Virology 2000; 273: 286-99., 48 Tsai A, Kawai S, Kwei K, et al. Chimeric constructs between two hepatitis B virus genomes confirm transcriptional impact of core promoter mutations and reveal multiple effects of core gene mutations Virology 2009; 387: 364-72.]. This double mutation though increased the genome replication in vitro but decreased HBeAg synthesis [20 Yang HI, Yeh SH, Chen PJ, et al. Associations between hepatitis B virus genotype and mutants and risk of hepatocellular carcinoma J Natl Cancer Inst 2008; 100: 1134-43.], and contributed to HBV pathogenesis in vivo [8 Sakamoto T, Tanaka Y, Orito E, et al. Novel subtypes (subgenotypes) of hepatitis B virus genotypes B and C among chronic liver disease patients in the Philippines J Gen Virol 2006; 87: 1873-82.]. PC mutation (G1896A) prevents HBeAg precursor translation whereas T1762/A1764 core mutation interfered with core mRNAs transcription leading to suppressed HBeAg synthesis [49 Kobayashi M, Arase Y, Ikeda K, et al. Precore wild-type hepatitis B virus with G1896 in the resolution of persistent hepatitis B virus infection Intervirology 2003; 46: 157-63.] and was associated with high replication phenotype [50 Scaglioni PP, Melegari M, Wands JP. Biologic properties of hepatitis B viral genomes with mutations in the precore promoter and precore open reading frame Virology 1997; 233: 374-81.]. The prevalence of PC 1896 and BCP 1762/1764 mutations in hyperendemic districts is examined for the first time by Yang et al. 2008 [20 Yang HI, Yeh SH, Chen PJ, et al. Associations between hepatitis B virus genotype and mutants and risk of hepatocellular carcinoma J Natl Cancer Inst 2008; 100: 1134-43.]. A1899 mutation is reported in higher proportion in liver cirrhosis patients as well [46 Biswas A, Banerjee A, Chandra PK, et al. Variations in the functional domain of basal core promoter of hepatitis B virus among eastern indian patients with prevalence of genotypes A, C, and D among the same ethnic population J Med Virol 2011; 83: 253-60.]. In relation to the association of genotypes and these common mutations, 1762/1764 is significantly higher in genotype A but lower in the genotype C. Genotype D has the highest A1896 mutation proportion [51 Kramvis A, Kew MC. Relationship of genotypes of hepatitis B virus to mutations, disease progression and response to antiviral therapy J Viral Hepatitis 2005; 12: 456-64.]. Although PC and BCP were extensively analyzed, other mutations have been correlated with various effects, e.g. the bulge of ε signal is affected by G1862T mutation. These recent findings suggest the noticeable influence of genotypes on virus replication.
In a study of in vitro translation, G1862T mutation abolished precore/core proteins synthesis but not entirely impaired [52 Hou J, Lin Y, Waters J, et al. Detection and significance of a G1862T variant of hepatitis B virus Chinese patients with fulminant hepatitis J Gen Virol 2002; 83: 2291-8.]. Inuoe et al. 2009 [27 Inoue J, Ueno Y, Nagasaki F, et al. Enhanced intracellular retention of a hepatitis B virus strain associated with fulminant hepatitis Virology 2009; 395: 202-9.] found that G1862T has no effect on HBeAg expression. G1862T was also implicated in reducing DNA replication severely [53 Guarnieri M, Kim KH, Bang G, et al. Point mutations upstream of hepatitis B virus core gene affect DNA replication at the step of core protein expression J Virol 2006; 80(2): 587-95.]. G1899A mutations increased DNA replication but the double mutation G1899A/G1862T was associated with considerably more replication compared to G1862T alone. Interestingly, U:G pair is converted into U:A in the lower stem (ε) in the two positions 1855/1899 if HBV strain possesses G1899A mutation. Using western blot analysis, core protein expression was reduced by G1862T mutation.
ASSOCIATION OF HBV MUTATION TO THE DISEASE OUTCOME
I. . Precore (PC) and Double Basal Core Promoter (BCP) Mutations
Kobayashi et al. 2003 [49 Kobayashi M, Arase Y, Ikeda K, et al. Precore wild-type hepatitis B virus with G1896 in the resolution of persistent hepatitis B virus infection Intervirology 2003; 46: 157-63.] showed that PC WT virus (G1896) was significantly more prevalent in patients who cleared the virus (negative HBeAg) compared to the G1896A precore mutant which persisted in patients who developed HCC. There were no discrepancies in the prevalence of T1762/A1764 double mutation in the BCP among the two groups of infected patients. Tsai et al. 2009 [48 Tsai A, Kawai S, Kwei K, et al. Chimeric constructs between two hepatitis B virus genomes confirm transcriptional impact of core promoter mutations and reveal multiple effects of core gene mutations Virology 2009; 387: 364-72.] confirmed that triple BCP mutation (1762/1764/1766) was associated with the highest increase in the transcription of pgRNA whereas double mutation was least effective. Biswas et al. 2011 [46 Biswas A, Banerjee A, Chandra PK, et al. Variations in the functional domain of basal core promoter of hepatitis B virus among eastern indian patients with prevalence of genotypes A, C, and D among the same ethnic population J Med Virol 2011; 83: 253-60.] found that the double BCP mutation (1762/1764) was lower in genotype A than C but higher in D. Some patients with BCP mutants developed HCC [44 Tong MJ, Blatt LM, Kao JH, Cheng JT, Corey WC. Precore/basal core promoter mutants and hepatitis B viral DNA as predictors for liver deaths and hepatocellular carcinoma World J Gastroenterol 2006; 12(41): 6620-.]. The triple mutation (1753/1762/1764) was associated with severe liver illness [46 Biswas A, Banerjee A, Chandra PK, et al. Variations in the functional domain of basal core promoter of hepatitis B virus among eastern indian patients with prevalence of genotypes A, C, and D among the same ethnic population J Med Virol 2011; 83: 253-60.]. C1753 mutation was associated with the previous BCP mutation in 42 patients whereas 38 patients with the BCP mutation did not have C1753, and 50 patients have had WT BCP [46 Biswas A, Banerjee A, Chandra PK, et al. Variations in the functional domain of basal core promoter of hepatitis B virus among eastern indian patients with prevalence of genotypes A, C, and D among the same ethnic population J Med Virol 2011; 83: 253-60.]. Inactive carriers were associated with WT BCP; HBeAg status in these clinical cases did not play any role. 1762/1764 in chronic liver disease was only slightly significant. T1766/A1768 was not taken into account because of its very low prevalence. Patients with PC and BCP mutants showed consistent results as expected [44 Tong MJ, Blatt LM, Kao JH, Cheng JT, Corey WC. Precore/basal core promoter mutants and hepatitis B viral DNA as predictors for liver deaths and hepatocellular carcinoma World J Gastroenterol 2006; 12(41): 6620-.]. To illustrate this, 60% of (+)-HBeAg was detected in PC WT and 30% in PC mutants. On the other hand, 66% of patients with BCP WT were (+)-HBeAg compared to 55% for BCP mutants (Table 2).
II. . G1862T and G1896A Mutations
G1862 mutation was 5 times more common in fulminant hepatitis (FH) patients compared to chronic carriers [52 Hou J, Lin Y, Waters J, et al. Detection and significance of a G1862T variant of hepatitis B virus Chinese patients with fulminant hepatitis J Gen Virol 2002; 83: 2291-8.]. Among 52 FH patients, G1862T was detected in 7 patients (Table 3).
ESSENTIAL TECHNIQUES FOR MUTATIONS DETECTIONS
There are fundamental techniques to study viral replication and further to assess protein expression utilized by most scientific papers, among which are cell culture (mainly Huh7 and HepG2 cells) to transfect infectious cDNA clones of the virus, DNA purification, Southern blotting, determination of genotype, polymerase chain reaction (PCR), real time PCR, cloning, and sequencing. The HBV genome is often cloned into pCR-XL-TOPO, introduced into competent cells such as E. coli and kept at -70˚C for later studies. Site directed mutagenesis can be used to generate a specific virus mutant to be utilized to meet the criteria of the study being investigated. RT-PCR detects HBV virions in the cell culture supernatant. Günther et al. 1995 [54 Günther S, Li BC, Miska S, Krüger DH, Meisel H, Will H. A novel method for amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients J Virol 1995; 69: 5437-44.] introduced a novel technique to be able to analyse full HBV genome length in which genome is released from the clone by SapI restriction endonuclease digestion, then transfected into HuH7 cell lines and this can generate an entire replication cycle with efficient viral protein expression. The technique was considered as a breakthrough in HBV molecular research. Clinical trials are valuable and may provide better prediction for the role of genotypes and mutations.
RELATIONSHIP OF HBV GENOTYPES TO REPLICATION DISEASE OUTCOME
According to Sakamoto et al. 2006 [8 Sakamoto T, Tanaka Y, Orito E, et al. Novel subtypes (subgenotypes) of hepatitis B virus genotypes B and C among chronic liver disease patients in the Philippines J Gen Virol 2006; 87: 1873-82.], HBV/A prevalence was higher in CH patients compared to LC or HCC patients. HBV/C was significantly higher in LC and HCC. CH patients had higher HBeAg prevalence and they were mostly younger than LC patients. Frequency of PC mutation (A1896) did not show interesting difference among HBV/A. In HCC patients, HBeAg positivity, age and the frequency of PC mutation did not represent any significant differences. T1653 mutation was observed only in HBV/C (in Sakamoto et al. study) but not in HBV/A, and was higher in HCC than CH patients with HBV/C. 100 patients with different clinical status have been studied to show the relationship of different genotypes with the severity of liver disease (Table 4).
In Sugiyama et al. 2003 study [39 Sugiyama M, Tanaka Y, Kato T, et al. Influence of hepatitis b virus genotypes on the intra and extracellular expression of viral DNA and antigens Hepatology 2006; 44: 915-24.], HBV/C had the highest HBV DNA expression followed by Bj/Ba and then by genotypes A and D. In terms of phenotypic analysis, the genotypes Bj had the highest intracellular expression of core protein followed by HBV/C and subgenotype Ba. Extracellular expression of HBsAg was the highest for subgenotype Ae, then Aa and Ba using Northern Blot analysis. PreS/S mRNA expressed intracellularly in high level for Ae. Level of precore mRNA was high for Bj and C but lower in Ba. Intracellular DNA expression was significantly high for HBV/C but was very low for genotypes A and D, and lowest for Ae. HBV/A was significantly higher in asymptomatic patients whereas HBV/C is higher in HCC. Replication level in transfected HepG2 cells was high in genotype A than C and D [41 Toan NL, Song LH, Kremsner PG, et al. Impact of the hepatitis b virus genotype and genotype mixtures on the course of liver disease in Vietnam Hepatology 2006; 43: 1375-84.]. Vietnamese LC patients were associated with genotype D.
DISCUSSION
HBV genotypes may induce varied liver disease but the virological differences are still to be defined and evidence behind this is becoming stronger nowadays. HBeAg secretion is affected by PC (A1896) mutation; this is why it is usually detected in HBeAg negative patients [20 Yang HI, Yeh SH, Chen PJ, et al. Associations between hepatitis B virus genotype and mutants and risk of hepatocellular carcinoma J Natl Cancer Inst 2008; 100: 1134-43.]. PC mutation prevalence was low and did not show any significant association with clinical status [46 Biswas A, Banerjee A, Chandra PK, et al. Variations in the functional domain of basal core promoter of hepatitis B virus among eastern indian patients with prevalence of genotypes A, C, and D among the same ethnic population J Med Virol 2011; 83: 253-60.]; this is confirmed by Sakamoto et al. results [8 Sakamoto T, Tanaka Y, Orito E, et al. Novel subtypes (subgenotypes) of hepatitis B virus genotypes B and C among chronic liver disease patients in the Philippines J Gen Virol 2006; 87: 1873-82.]. It was also strongly evident that virus replication was not altered by PC mutation because HBeAg is not required for virus replication. Kobayashi et al. showed that patients infected with HBV PC WT are prone to clear the virus [49 Kobayashi M, Arase Y, Ikeda K, et al. Precore wild-type hepatitis B virus with G1896 in the resolution of persistent hepatitis B virus infection Intervirology 2003; 46: 157-63.]. I perceive, the advantage of this is the large sample size (1077 patients) and long study duration (15 years). According to Tong et al. 2006 [44 Tong MJ, Blatt LM, Kao JH, Cheng JT, Corey WC. Precore/basal core promoter mutants and hepatitis B viral DNA as predictors for liver deaths and hepatocellular carcinoma World J Gastroenterol 2006; 12(41): 6620-.], 42% of the cirrhotic patients had PC 1896 mutant virus whereas the percentage of patients who developed HCC with PC 1896 mutation was 45% which is an indication of high risk to HCC. Significance of this mutation is still not obvious despite all related studies and further investigation is required. Table 2 elucidates that PC 1896 mutation is not 100% associated with negative HBeAg expression but its possible association with HBeAg negative status is verified. (+)-HBeAg was higher in PC mutants than PC WTs whereas (-)-HBeAg did not show significant variation among BCP mutants and WTs. The prevalence of PC WTs was higher in HBV/A. HBV/B on the other hand was associated with high PC mutant prevalence (Table 2). Anti-HBe antibodies and (-)-HBeAg have given understanding of HBV biology. In (+)-HBeAg WT strain, anti-HBe can destroy hepatocytes and stop persistent infection; however, HBV can mutate into (-)-HBeAg to escape anti-HBe antibodies. Table 2 did not mention genotype D in its association with the prevalence of either PC (1896) or BCP (1762/1764), but another study showed that BCP double mutation was less in HBV/D whereas A1896 was significantly higher in HBV/D compared to subgenotypes Aa and Ae [11 Tanaka Y, Hasegawa I, Kato T, et al. A case-control study for differences among hepatitis b virus infections of genotypes a (subtypes Aa and Ae) and D Hepatology 2004; 40: 747-55.]. All these data confirm the association of HBV mutations to disease severity and outcome.
BCP 1762/1764 mutants have strong evidence of its relation with HCC progression. This double mutation can be a bad indication to the infected individual due to its relation with HCC as presented in some previous studies [44 Tong MJ, Blatt LM, Kao JH, Cheng JT, Corey WC. Precore/basal core promoter mutants and hepatitis B viral DNA as predictors for liver deaths and hepatocellular carcinoma World J Gastroenterol 2006; 12(41): 6620-.]. Other reports showed that PC mutant is correlated with high viral load in (-)-HBeAg, and lower with BCP mutants [56 Fang ZL, Sabin CA, Dong BQ, et al. The association of HBV core promoter double mutations (A1762T and G1764A) with viral load differs between HBeAg positive and anti-HBe positive individuals: A longitudinal analysis J Hepatol 2009; 50: 273-80.]. Accordingly, Xu et al. 2011 exhibited opposite results in an investigation of 793 patients to understand the role of PC/BCP mutation [57 Xu Z, Ren X, Liu Y, et al. Association of hepatitis B virus mutations in basal core promoter and precore regions with severity of liver disease: an investigation of 793 Chinese patients with mild and severe chronic hepatitis B and acute-on-chronic liver failure J Gastroenterol 2011; 46: 391-400.]. Tong et al. 2006 [44 Tong MJ, Blatt LM, Kao JH, Cheng JT, Corey WC. Precore/basal core promoter mutants and hepatitis B viral DNA as predictors for liver deaths and hepatocellular carcinoma World J Gastroenterol 2006; 12(41): 6620-.] limitation is that BCP 1762/1764 analysis was not possible in some patients due to the low level of the virus DNA, but this does not weaken the evidence. As previously shown, data may not be agreeable. It is becoming apparent that triple BCP mutation is associated with higher replication efficiency and more severe illness than double mutation. This indicates the critical role of different patterns of mutations.
G1862T showed no effects on HBeAg expression [27 Inoue J, Ueno Y, Nagasaki F, et al. Enhanced intracellular retention of a hepatitis B virus strain associated with fulminant hepatitis Virology 2009; 395: 202-9., 53 Guarnieri M, Kim KH, Bang G, et al. Point mutations upstream of hepatitis B virus core gene affect DNA replication at the step of core protein expression J Virol 2006; 80(2): 587-95.]. In Guarnieri et al. study [53 Guarnieri M, Kim KH, Bang G, et al. Point mutations upstream of hepatitis B virus core gene affect DNA replication at the step of core protein expression J Virol 2006; 80(2): 587-95.] there was a reduction in the core protein expression. Conversely, they presented enhancement of the same protein for G1899A. Both 1862/1896 mutations reduced expression of HBeAg but the difference is that G1862T reduced virus replication; however, G1896A did not. Interestingly, association of 1899 mutation with G1862A greatly enhanced the replication. G1862T mutation should motivate researchers due to its vague effect by its own or by association with other mutations and because there are quite a few research on G1862T. This mutation can be associated with fulminant hepatitis (Table 3). G1899A mutation has an interesting effect on the pair of 1855 position. These results suggest that investigation of the nucleotide position 1899 becomes mandatory in HBV molecular research due to its unfathomable influence on replication. Extracellular and intracellular expression among genotypes were not consistent according to Sugiyama et al. [39 Sugiyama M, Tanaka Y, Kato T, et al. Influence of hepatitis b virus genotypes on the intra and extracellular expression of viral DNA and antigens Hepatology 2006; 44: 915-24.] and some observations may indicate that it is somewhat troublesome to link a particular genotype or mutation with certain degree of liver disease, but other mutations are confirmed by agreement and consensus though some mutations may show significant association, for instance T1653 mutation observed by Sakamoto et al. [8 Sakamoto T, Tanaka Y, Orito E, et al. Novel subtypes (subgenotypes) of hepatitis B virus genotypes B and C among chronic liver disease patients in the Philippines J Gen Virol 2006; 87: 1873-82.] is a predictive factor for HCC in patients infected with HBV/C. This factor can be added with genotype C as risk to HCC. Although pre-S mutation studies are limited, it is expected as a factor of infection chronicity [51 Kramvis A, Kew MC. Relationship of genotypes of hepatitis B virus to mutations, disease progression and response to antiviral therapy J Viral Hepatitis 2005; 12: 456-64.]. For example, T3098C in the preS1 region and T53C were associated with HCC but further investigation is essential [40 Livezey KW, Negorev D, Simon D. Hepatitis B virus-transfected Hep G2 cells demonstrate genetic alterations and de novo viral integration in cells replication HBV Mutat Res 2000; 452: 163-78.]. C1753 with 1762/1764 is indicative of severe liver disease [46 Biswas A, Banerjee A, Chandra PK, et al. Variations in the functional domain of basal core promoter of hepatitis B virus among eastern indian patients with prevalence of genotypes A, C, and D among the same ethnic population J Med Virol 2011; 83: 253-60.]. Other stressors may negatively exacerbate the clinical status like the presence of C1753 with BCP double mutation. According to Biswas et al. 2011 observation, C1753 is becoming more common among newly infected patients. To analyze the difference between T1858 and C1858 mutations and their association with unanticipated factors, G1896 strengthens T1858 pairing [49 Kobayashi M, Arase Y, Ikeda K, et al. Precore wild-type hepatitis B virus with G1896 in the resolution of persistent hepatitis B virus infection Intervirology 2003; 46: 157-63.] but replication capacity was equal in C1858 and T1858 [58 Alestig E, Hannoun C, Horal P, Lindh M. Phylogenetic origin of hepatitis B virus strains with precore C-1858 variants J Clin Microbiol 2001; 39(9): 3200-.]. The type of precore mutation detected is influenced by HBV genotype. G1896 is less common in HBV/A because the nucleotide at 1858 is a C. If 1858 is a T, the nucleotide at 1896 is able to mutate to an A, and maintain stability of the epsilon stem loop [55 Revill P, Yuen L, Walsh R, Perrault M, Locarnini S, Kramvis A. Bioinformatic analysis of the hepadnavirus e-antigen and its precursor identifies remarkable sequence conservation in all orthohepadnaviruses J Med Virol 2010; 82(1): 104-5.]. Interestingly, T1858 prevalence was 100% in patients infected with genotype D (n= 61) compared to 0% for subgenotypes Aa and Ae (n = 54; n= 57) respectively [11 Tanaka Y, Hasegawa I, Kato T, et al. A case-control study for differences among hepatitis b virus infections of genotypes a (subtypes Aa and Ae) and D Hepatology 2004; 40: 747-55.].
HBV/A expression was low in Sugiyama et al. study; on the other hand, replication level in HepG2 cells was high for genotype A according to Toan et al. [41 Toan NL, Song LH, Kremsner PG, et al. Impact of the hepatitis b virus genotype and genotype mixtures on the course of liver disease in Vietnam Hepatology 2006; 43: 1375-84.]. The corresponding results for the same genotype were contradicted. Genotype A is becoming common in Vietnam [41 Toan NL, Song LH, Kremsner PG, et al. Impact of the hepatitis b virus genotype and genotype mixtures on the course of liver disease in Vietnam Hepatology 2006; 43: 1375-84.] compared to previous studies showed that genotypes B and C are the most common in Asia including Vietnam. The limitations of Sugiyama et al. study [39 Sugiyama M, Tanaka Y, Kato T, et al. Influence of hepatitis b virus genotypes on the intra and extracellular expression of viral DNA and antigens Hepatology 2006; 44: 915-24.] is the low sample size and did not use PC/BCP mutations, but they studied in vivo and in vitro experiments and it was a rare comparative study of many genotypes. Toan et al. [41 Toan NL, Song LH, Kremsner PG, et al. Impact of the hepatitis b virus genotype and genotype mixtures on the course of liver disease in Vietnam Hepatology 2006; 43: 1375-84.] paper was significant because 460 patients were studied from different countries (Vietnam, Europe, and Africa). Although HBV/B encompassed the majority of participants in Yang et al. paper [20 Yang HI, Yeh SH, Chen PJ, et al. Associations between hepatitis B virus genotype and mutants and risk of hepatocellular carcinoma J Natl Cancer Inst 2008; 100: 1134-43.], they argued that HBV/C was associated with high risk of HCC. Yang et al. [20 Yang HI, Yeh SH, Chen PJ, et al. Associations between hepatitis B virus genotype and mutants and risk of hepatocellular carcinoma J Natl Cancer Inst 2008; 100: 1134-43.] discussed other factors like cigarette smoking and alcohol drinking. Their large sample size is another advantage of the study. Sakamoto et al. [8 Sakamoto T, Tanaka Y, Orito E, et al. Novel subtypes (subgenotypes) of hepatitis B virus genotypes B and C among chronic liver disease patients in the Philippines J Gen Virol 2006; 87: 1873-82.] exhibited that HBV/A and B were associated with 35% of HCC cases compared to 30% to genotype C in Philippines (Table 4). There is a consensus that HBV/C is the most common genotype involved in HCC [19 Hwang GY, Lin CY, Huang LM, et al. Detection of the hepatitis B virus X protein (HBx) antigen and anti-hbx antibodies in cases of human hepatocellular carcinoma J Clin Microbiol 2003; 41(12): 5598-603., 35 Lupberger J, Mund A, Kock J, Hildt E. Cultivation of HepG2.15 on Cytodex-3: higher yield of hepatitis B virus and less subviral particles compared to conventional culture methods. J Hepatol 2006; 45(4): 547-2., 50 Scaglioni PP, Melegari M, Wands JP. Biologic properties of hepatitis B viral genomes with mutations in the precore promoter and precore open reading frame Virology 1997; 233: 374-81.], thus, it is recommended to investigate genotype C especially in the field of antiviral research to prevent the fatal sequelae.
The association of HBV infection with other common viruses that are known to infect the liver such as cytomegalovirus (CMV) and Epstein-Barr virus (EBV) could have deceitful effect. It is very unlikely and seldom discussed in scientific HBV research albeit the role of some mutations alone is identified. The presence of other hepatitis viruses with HBV for long period of time might have a role in the replication and disease progression. One difference between in vivo and in vitro studies is the immune response in the infected individuals, and this should be taken into account. Another drawback is the infection duration difference i.e. the long period of infection inside human hepatocytes compared to few days of the in vitro infection model. The effect of viral load should be considered as well. Alcohol abuse, iron level, history of toxic or drug usage and impaired metabolisms must also be considered in the interpretation as factors for chronic liver disease. Some previous studies did not discuss the cell lines they are using as an interfering factor, for example, different cell lines produce different HBV DNA level for the same HBV variant, and thus it is preferred to compare the type of cell line in the replication studies. The subgenotypes are not discussed in some original reports; these subgenotypes within the same genotype (like Bj and Ba) have sometimes immense disparity in their effect on either replication or progression of infection. Skeptical observations lead to controversialism in some mechanism of the replication and the biologic function of HBeAg.
Comparative studies between distinct HBV genotypes are difficult due to environmental and host differences [55 Revill P, Yuen L, Walsh R, Perrault M, Locarnini S, Kramvis A. Bioinformatic analysis of the hepadnavirus e-antigen and its precursor identifies remarkable sequence conservation in all orthohepadnaviruses J Med Virol 2010; 82(1): 104-5.]. Another factor that makes these studies difficult is the various geographical distributions [50 Scaglioni PP, Melegari M, Wands JP. Biologic properties of hepatitis B viral genomes with mutations in the precore promoter and precore open reading frame Virology 1997; 233: 374-81.], the considerable mutations being analyzed and the emerging mutations such as A1899 and C1753. Inevitably, the very low prevalence of some newly emerging mutations is one of the interfering factors in HBV molecular studies. As can be seen, the exact effect of certain mutations could be perplexing, but future studies will provide full understanding. Classification is one significant issue in this context. HBV mutations change this classification, for example C1 and C2 subgenotypes classification were reported interchangeably by two studies [7 Norder H, Couroucé AM, Coursaget P, et al. Genetic diversity of hepatitis b virus strains derived worldwide: genotypes, subgenotypes, and hbsag subtypes Intervirology 2004; 47: 289., 32 Hu J, Flores D, Toft D, Wang X, Nguyen D. Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function J Virol 2004; 78: 13122-31.] and this will lead to confusion [12 Ahn SH, Yuen L, Revill P. Clarification required for the definition of hepatitis b virus subgenotypes C1 and C2 Intervirology 2009; 52: 321-2.]. In addition, Norder et al. reported four subgenotypes for HBV/C, two years later; Sakamoto et al. showed five subgenotypes (C1- C5), therefore precise HBV designation is very significant [12 Ahn SH, Yuen L, Revill P. Clarification required for the definition of hepatitis b virus subgenotypes C1 and C2 Intervirology 2009; 52: 321-2.].
In summary, HBV resolution can be facilitated by the presence of WT virus G1896 in CH patients with (+)-HBeAg. It can be concluded that BCP mutation (1762/1764) and genotype C are associated with high risk of HCC. As mentioned in the introduction, new genotypes and subgenotypes are being discovered, thus, it makes these studies difficult to accurately link specific genotypes and mutations with certain liver disease or in vitro replication study. HBeAg negativity might be associated with severe liver damage as in mutations affecting HBeAg expression. Future studies are valuable to determine the pathogenic outcome. More research will be needed considering more than one factor with more genotypes to reach agreement and to decipher uncertainties about the precise role of certain mutations and genotypes on either replication cycle or liver pathogenesis.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
ACKNOWLEDGEMENTS
I would like to express my appreciation and gratitude to Dr. Peter Revill (research scientist at VIDRL research center, Australia) and Dr. Peter Roche (the program coordinator at RMIT University, Melbourne, Australia) for their technical advice.


