- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Application of MALDI-TOF Mass Spectrometry in Clinical Virology: A Review
Fernando Cobo*
Abstract
MALDI-TOF mass spectrometry is a diagnostic tool of microbial identification and characterization based on the detection of the mass of molecules. In the majority of clinical laboratories, this technology is currently being used mainly for bacterial diagnosis, but several approaches in the field of virology have been investigated. The introduction of this technology in clinical virology will improve the diagnosis of infections produced by viruses but also the discovery of mutations and variants of these microorganisms as well as the detection of antiviral resistance. This review is focused on the main current applications of MALDI-TOF MS techniques in clinical virology showing the state of the art with respect to this exciting new technology.
Article Information
Identifiers and Pagination:
Year: 2013Volume: 7
First Page: 84
Last Page: 90
Publisher Id: TOVJ-7-84
DOI: 10.2174/1874357920130927003
Article History:
Received Date: 3/5/2013Revision Received Date: 1/8/2013
Acceptance Date: 2/8/2013
Electronic publication date: 27 /9/2013
Collection year: 2013
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Section of Microbiology Section (Integrated Biotechnology Area), Hospital de Poniente. Ctra de Almerimar S/N, 04700 El Ejido, Almería, Spain; Tel: +34 950 022 638; Fax: +34 950 022 601; E-mail: fernando.cobo.sspa@juntadeandalucia.es
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 3-5-2013 |
Original Manuscript | Application of MALDI-TOF Mass Spectrometry in Clinical Virology: A Review | |
INTRODUCTION
The matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) is an analytical method of microbial identification and characterization based on the fast and precise assessment of the mass of molecules in a variable range of 100 Da to 100 KDa.
The first description of the use of MS technology for bacterial identification was in 1975 [1Anhalt JP, Fenselau C. Identification of bacteria using mass spectrometry. Anal Chem 1975; 47: 219-5.]. Furthermore, MALDI-TOF MS was used by the mid-1990s for the identification of bacteria in research settings [2Claydon MA, Davey SN, Edwards-Jones V, Gordon DB. The rapid identification of intact microorganisms using mass spectrometry. Nat Biotechnol 1996; 14: 1584-6., 3Holland RD, Wilkes JG, Rafii F. Rapid identification of intact whole bacteria based on spectral patterns using matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 1996; 10: 12227-32.] but it took a long time for the introduction of this technology in routine microbiology. Then, in 2004, the first complete database for bacterial identification was reported [4Keys CJ, Dare DJ, Sutton H. Compilation of a MALDI-TOF mass spectral database for the rapid screening and characterisation of bacteria implicated in human infectious diseases. Infect Genet Evol 2004; 4: 221-42.].
The possibility of MALDI-TOF to quickly characterize a wide variety of microorganisms including bacteria, fungi and viruses [5Giebel R, Worden C, Rust SM. Microbial fingerprinting using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) applications and challenges. Adv Appl Microbiol 2010; 71: 149-84.] increase it potential application in some areas of microbiological diagnosis. Thus, this technology can be used for rapid microbial identification at a relatively low cost and it is an alternative for conventional laboratory diagnosis and molecular identification systems. The major contribution of this method is that has drastically improved the time to identification of a positive sample.
The main use of this technology has been, until now, the identification or typing of bacteria from a positive culture. However, applications in the field of clinical virology have been also performed and are opening up new opportunities, although the introduction in the routine laboratory diagnosis still remains a challenge.
In this review, MALDI-TOF technique is presented including sample preparation as well as data management and quality control, mainly focused in viral research and diagnosis.
MASS SPECTROMETRY PRINCIPLES
The mass spectrometer is composed of three main components: an ion source to ionize and transfer sample molecules ions into a gas phase, a mass analyser device that separate molecules depending to their mass and finally a detector to monitor all separated ions. There are several ionization methods such as chemical ionization, electrospray and MALDI. The choice of method basically depends to the objective of the MS analysis and the nature of the sample, although both MALDI and electrospray are techniques that permit ionization and vaporization of a big quantity of biomolecules [6Emonet S, Shah HN, Cherkaoui A, Schrenzel J. Application and use of various mass spectrometry methods in clinical microbiology. Clin Microbiol Infect 2010; 16: 1604-3.].
In MS technique [7van Belkum A, Welker M, Erhard M, Chatellier S. Biomedical mass spectrometry in today's and tomorrow's clinical microbiology laboratories. J Clin Microbiol 2012; 50: 1513-7.], samples are prepared by mixing several biomolecules and embedded in a matrix that will crystallize inside. The matrix is composed of small acid molecules that have a strong optical absorption in the range of the laser wavelength used, although the composition is different according to the type of laser used and the biomolecule to be analysed. The molecules are then both desorbed and ionized by charge transfer by absorbing the energy of a short laser pulse. The desorbed and ionized molecules are first accelerated in an electrical field and ejected through a metal flight tube and collide with a detector at the end of the flight tube. This cycle is repeated at a frequency of 50 or more hertz. Thus, biomolecules are separated according to their mass, and create a mass spectrum that is characterized by both the mass and the intensity of the ions. Finally, the result is a spectral signature (spike representation) that is then searched for in the appropriate database for the identification of the microorganism, comparing with values provides by the manufacturer of the database (Fig. 1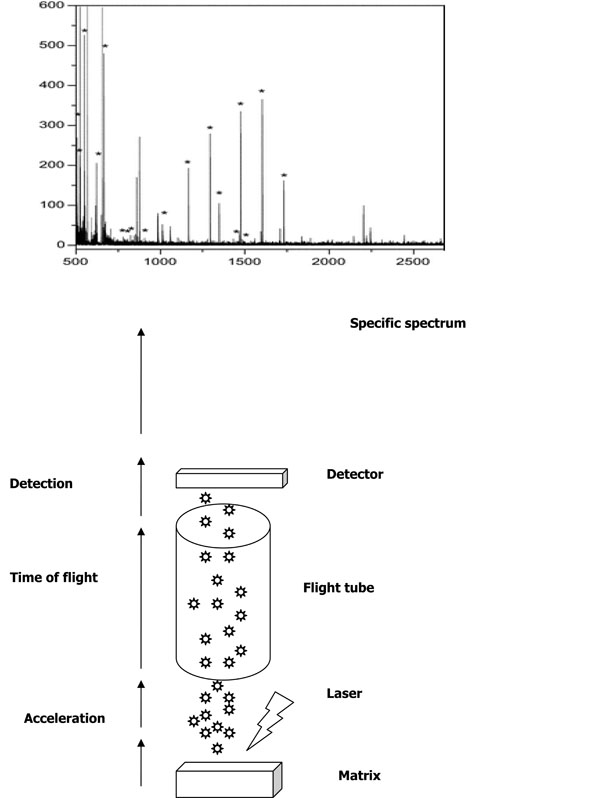 ).
).
On the other hand, electrospray ionization mass spectrometry (ESI-MS) generally uses DNA amplicons that are dissolved in a solvent and then injected in a conductive capillary. Inside this capillary, high voltage is applied resulting in the emission of aerosols of charged drops of the sample. After this, the sample is sprayed through compartments with diminishing pressure, resulting in the formation of gas-phase multiple-charged analyte ions, which are finally detected by the spectrometer. ESI-MS technology permits high resolution analysing the basic composition of amplicons [8Ecker DJ, Sampath R, Massire C. Ibis T5000: a universal biosensor approach for microbiology. Nat Rev Microbiol 2008; 6: 553-8.].
There are also two available methods for typing using this technology. The first method is the MALDI-RE that uses the iSeq method [9Stanssens P, Zabeau M, Meersseman G. High-throughput MALDI-TOF discovery of genomic sequence polymorphisms. Genome Res 2004; 14: 126-33., 10Honisch C, Chen Y, Mortimer C. Automated comparative sequence analysis by base-specific cleavage and mass spectrometry for nucleic acid-based microbial typing. Proc Natl Acad Sci USA 2007; 104: 10649-54.]. In this technique, RNA strands are cleaved and the products of cleavage are analysed by mass spectrometry, and their spectra are compared with the spectra of the reference sequences. Initially, MALDI-RE was used mainly for the detection of single-nucleotide polymorphisms, but currently there are additional applications for this method [11Honisch C, Chen Y, Hillenkamp F. Comparative DNA sequence analysis and typing using mass spectrometry.In: Sha HN.Gharbia SE., eds. Mass spectrometry for microbial proteomics. Chichester Wiley 2010; 443-62.]. The second method for typing is based on the use of ESI technology to analyse specific DNA amplicons (PCR-ESI-MS) [12Ecker DJ, Massire C, Blyn LB. Molecular genotyping of microbes by multilocus PCR and mass spectrometry: a new tool for hospital infection control and public health surveillance. Methods Mol Biol 2009; 551: 71-87.].
CURRENT CLINICAL VIROLOGY APPLICATIONS
Viral Diagnosis
Common laboratory techniques in viruses’ detection include antibody analysis such as enzyme-linked immunosorbent assay, molecular techniques like polymerase chain reaction (PCR) and dot blot hybridization. However, MALDI-TOF technology has been already used to investigate a wide variety of viruses clinically relevant. There are several approaches of this technology for application in the field of virology.
A recent study performed a multiplex MALDI-TOF MS method that successfully can detect human herpesviruses (HHVs) from a wide variety of archival biological specimens [13Sjöholm MIL, Dillner J, Carlson J. Multiplex detection of human herpesviruses from archival specimens by using matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 2008; 46: 540-.]. The concordance rate between the MALDI-TOF MS technique and reference methods was high and the detection limits of the MS methods were comparable to previous reports of multiplex herpesvirus detection using an oligonucleotide microarray and multiplex PCR techniques. Low viral loads near the detection limit of the method and weak cross-reactivity might contribute to the few discrepant results with reference methods. Thus, this multiplex approach can be very useful for large-scale epidemiological research studies in which broad viruses detection is necessary as well as may also become highly useful for multiplex clinical diagnostic testing.
Other virus like influenza viruses remain a major health problem for human beings. The recent cross-species transmission of avian influenza viruses to humans and the outbreak of the influenza H1N1 variant enhance the need for a rapid and efficient diagnostic tools for these viruses. The detection of influenza viruses is mainly performed using PCR techniques (rRT-PCR) or antibody-based assays to identify the nucleoproteins. Alternative methods have been investigated for viral diagnosis, such as multiplexed flow cytometry [14Yan X, Zhong W, Tang A, Schielke EG, Hang W, Nolan JP. Multiplexed flow cytometry immunoassay for influenza virus detection and differentiation. Anal Chem 2005; 77: 7673-8.], microarrays [15Townsend MB, Dawson ED, Mehlmann M. Experimental evaluation of the FluChip diagnostic microarray for influenza virus surveillance. J Clin Microbiol 2006; 44: 2863-71.] and mass spectrometry [16Downard KM, Morrissey B, Schwahn AB. Mass spectrometry analysis of the influenza virus. Mass Spect Rev 2009; 28: 35-49.]. MS has been considered as one of the best methods for protein analysis because of its low detection limit and high accuracy. It has been also demonstrated that the combined use of antibody-magnetic nanoparticles and MALDI-TOF MS is suitable for sensitive detection of influenza viruses [17Chou TC, Hsu W, Wang CH, Chen YJ, Fang JM. Rapid and specific influenza virus detection by functionalized magnetic nanoparticles and mass spectrometry. J Nanotechnol 2011; 9: 52.]. This method could also be used for the rapid screening of virus subtypes, suggesting a promising application in early and accurate diagnosis of influenza viruses.
Enteroviruses are also a major cause of illness in children causing gastroenteritis, hand, foot and mouth disease and severe neurological complications such as aseptic meningitis, encephalitis and acute neurogenic pulmonary edema. Classical detection of enteric virus is mostly based on conventional approaches such as indirect immunofluorescence assay, neutralizing testing, and cell culture and immunohistochemical detection. However, some clinical samples remain undetectable because of the absence of antibodies against viral proteins and cell culture and neutralization tests are time-consuming and laborious. Multiplex RT-PCR and real-time PCR have shown good sensitivity and specificity [18Thao NTT, Ngoc NTK, Tu PV. Develoment of a multiplex polymerase chain reaction assay for simultaneous identification of human enterovirus 71 and coxsackievirus A16. J Viral Methods 2010; 170: 134-9.]. For these viruses, the MALDI-TOF MS technology has demonstrated high capacity to diagnosis with high throughput and short turnaround time. MALDI-TOF MS measures the intrinsic physical properties of molecules directly. A PCR mass assay combining multiplex PCR with MALDI-TOF MS technology has shown simultaneous detection of eight human enteric viruses such as hepatitis E virus, coxsackievirus, poliovirus, astrovirus, norovirus, echovirus and reovirus [19Piao J, Jiang J, Xu B. Simultaneous detection and identification of enteric viruses by PCR-mass assay. PLoS ONE 2012; 7: e42251.]. This method has a sensitivity ranging from 100 to 1000 copies/reaction and shows that this assay could be used for the simultaneous detection of co-infections with several viruses.
MALDI-TOF MS has been also used for the diagnosis of hand, foot, and mouth disease that is caused by acute enterovirus infections such as poliovirus, coxsackievirus A and B and echovirus. These infections are clinically indistinguishable but infection with echovirus 71 could be complicated with aseptic meningitis, encephalitis and acute flaccid paralysis. Recently, a study has developed a MALDI-TOF MS technique for the rapid diagnosis of these viruses [20Peng J, Yang F, Xiong Z. Sensitive and rapid detection of viruses associated with hand foot and mouth disease using multiplexed MALDI-TOF analysis. J Clin Virol 2013; 56: 170-4.]. The diagnosis of these pathogens can help to a more comprehensive surveillance of enteroviruses to understand the molecular epidemiology of enteroviruses detecting multiple genotypes simultaneously. The main advantage of this strategy is the identification of several genotypes at the same time, opening up new possibilities for the diagnosis and discovering pathological and epidemiological characteristics.
On the other hand, infection with high-risk human papillomaviruses (HPV) has been closely associated with cervical cancer as well as other types of cancer such as anus, vagina, penis and head and neck. The diagnosis of HPV infection is based on several methods, but HPV DNA detection is necessary for the following-up of the patients. Currently, there are three HPV assays approved for clinical use by the United States Food and Drug Administration. Two non-PCR-based assays (e.g. Cervista, Hologic, Marlborough, MA; Hybrid Capture 2, Qiagen, Valencia, CA) and a PCR-based method such as the Cobas HPV assay. The majority of these assays use GP5+/GP6+ consensus primers, MY09/11 primers and their multiprimer derivates PGMY09/11 primers and the SPF10 system. However, a new method based on the amplification of a fragment of the L1 gene region using GP5+/GP6+ consensus primers using mass array technique based on the MALDI-TOF MS technology has been recently developed to identify 14 high-risk HPVs [21Yi X, Li J, Yu S. A new PCR-based mass spectrometry system for high-risk HPV. Part I Am J Clin Pathol 2011; 136: 913-., 22Duu H, Yi J, Wu R. A new PCR-based mass spectrometry system for high risk HPV. Part II Am J Clin Pathol 2011; 136: 920-3.]. This MS assay was found to be non-cross reactive with common low risk HPV genotypes affecting the genital tract and more sensitive (General sensitivity results ranging from 10 to 100 copies) than the Hybrid Capture 2 and conventional PCR methods. An advantage of this method is that it is possible to extend the HPV genotype spectrum by adding new type-specific primers. The use of this technique together with automated DNA extraction and PCR pipetting procedures could increase the viral detection which a decrease in the cost of the overall assay.
Diagnosis of Mutations and Identification of Viral Genotypes
Hepatitis B virus (HBV) and hepatitis C virus (HCV) infections are still a major public health problem due to their worldwide morbidity and mortality. For this reason, screening for these infections is an important part of routine laboratory activity. Serological and molecular markers are key elements for the diagnosis, prognosis and monitoring of treatment in HBV and HCV infections. Currently, chemiluminiscence assays are widely being used for viral diagnosis. Molecular techniques are routinely used for detecting, quantifying viral genomes and analyzing their sequence, in order to determine their genotype and detect resistance to antiviral drugs. However, MassARRAY systems based on nucleic acid analysis by MALDI-TOF MS technology provides an alternative approach to HBV and HCV genotyping [23Ganova-Raeva L, Ramachandran S, Honisch C. Robust hepatitis B virus genotyping by mass spectrometry. J Clin Microbiol 2010; 48: 4161-8.], being accurate to identify all eight HBV genotypes. MassARRAY system generates MS patterns that are compared to sequences derived from known HBV genotypes. The main advantages of this assay are its rapidity, cost-effectively and versatility. The assay produces automatic data reports and does not require any additional data interpretation.
Moreover, this method can discriminate single-point mutations and is capable of detecting wild-type and mutant alleles [24Hong SP, Kim NK, Hwang SG. Detection of hepatitis B virus YMDD variants using mass spectrometry analysis of oligonucleotide fragments. J Hepatol 2004; 40: 837-44., 25Luan J, Yuan J, Li X. Multiplex detection of 60 hepatitis B virus variants by MALDI-TOF mass spectrometry. Clin Chem 2009; 55: 1503-9.]. Variations in the HBV genome are biologically and clinically important and might confer drug resistance or affect virus replication capacity, resulting in failure of antiviral therapy. Some MALDI-TOF MS platforms are capable to detect 60 HBV variants with accuracy and low detection limits [25Luan J, Yuan J, Li X. Multiplex detection of 60 hepatitis B virus variants by MALDI-TOF mass spectrometry. Clin Chem 2009; 55: 1503-9.]. The main disadvantage of this method is that only HBV genotypes B and C were tested.
Some characteristically mutations in HBV could cause lamivudine resistance due to prolonged treatment with this antiviral agent. Current methods for detecting such variants (e.g. sequencing analysis, RFLP and hybridization-based assays) are time-consuming and unsuitable for screening large numbers of samples. For these reasons, it has been developed a MALDI-TOF MS assay suitable for detecting YMDD mutants [24Hong SP, Kim NK, Hwang SG. Detection of hepatitis B virus YMDD variants using mass spectrometry analysis of oligonucleotide fragments. J Hepatol 2004; 40: 837-44.]. This technique provides high rates of accuracy for YMDD mutations and has advantages over RFLP and direct DNA sequencing, including the ability to more sensitively and specifically detects mixed populations as well as being a simpler procedure without need for cloning or multiple PCR steps producing earlier detection of HBV mutations. Moreover, this platform is adaptable for the detection of other mutations or polymorphisms. This method will be very useful to evaluate the emergence of mutant viruses and to monitoring of antiviral treatment of chronic hepatitis B, correlating them to clinical features with regard to response to drug therapy in these patients.
On the other hand, the identification of HCV genotypes has become important in order to determine the clinical course and the outcome of antiviral therapy. Many HCV genotyping methods have been developed, including genotype-specific PCR, RFLP, hybridization techniques and heteroduplex motility assays. Currently, the method of choice for this purpose is the nucleotide sequencing of a subgenomic region, but all these methods are complex to standardize, labor-intensive or non-useful for determining multiple HCV genotypes. However, it is frequent heterogeneity during the course of infection, producing mixed-genotypes infections in some patients. Thus, researchers have developed some MALDI-TOF MS-based assays for HCV genotyping during last years [26Kim YJ, Kim SO, Chung HJ. Population genotyping of hepatitis C virus by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry analysis of short DNA fragments. Clin Chem 2005; 51: 1123-31.-28Oh HB, Kim SO, Cha CH. Identification of hepatitis C virus genotype 6 in Korean patients by analysis of 5' untranslated region using a matrix assisted laser desorption/ionization time of flight-based assay. restriction fragment mass polymorphism. J Med Virol 2008; 80: 1712-9.]. For genotyping purposes, MS provides accurate information on the molecular mass of the analyte and the procedure can be fully automated, as well as the direct analysis of molecules with no labels to be introduced and no separation steps followed by the image processing.
Other methods based on MALDI-TOF MS technology for viral diagnosis purposes have been also recently developed. A method based on MALDI-TOF MS technology studied the possibility of genotyping JC virus using urine samples in patients without clinical evidence of progressive multifocal leukoencephalopathy [29Bayliss J, Moser R, Bowden S, McLean CA. Characterisation of single nucleotide polymorphisms in the genome of JC polyomavirus using MALDI TOF mass spectrometry. J Virol Methods 2010; 164: 63-7.]. MALDI-TOF showed successfully genotype of these viruses and identifies sequence variations in the target genome. Other study used this technique in order to identify the possible mutations of influenza A (H5) viruses for monitoring purposes [30Yea C, McCorrister S, Westmacott G, Petric M, Tellier R. Early detection of influenza A (H5) viruses with affinity for the human sialic acid receptor by MALDI-TOF mass spectrometry based mutation detection. J Virol Methods 2011; 172: 72-.].
Identification of Drug Resistance
Finally, the application of MALDI-TOF MS technique could be useful for detecting drug resistance against some antivirals. A recent study has developed a sensitive and rapid method for the detection of ganciclovir resistance by PCR based MALDI-TOF analysis [31Zürcher S, Mooser C, Lüthi AU. Sensitive and rapid detection of ganciclovir resistance by PCR based MALDI-TOF analysis. J Clin Virol 012 ; 54: 359-63.].
Use of MALDI-TOF Techniques in Epidemiology
Other potential application of MALDI-TOF MS technology is to provide critical epidemiological data about viral infections. From an epidemiological point of view, one of the main applications of these methods is determining specific data of clinical isolates for viral diseases’ outbreaks. Because of their complexity, some viral typing techniques require specialized test methods and additional diagnostic devices. For this reason, the typing is not routinely performed in every laboratory, so these samples are being sent out of the hospital and this fact could delay the investigation of these outbreaks and hospital-associated infections. The introduction of MALDI-TOF MS technology in the laboratories can facilitate the performing of tests leading to improve the information to the infection control staff. The introduction of MS techniques into the clinical laboratory can provide accurate and rapid epidemiological data and it could have a critical role in hospital infection control and surveillance of viral outbreaks.
Table 1 summarizes the main applications of MALDI-TOF MS to clinical virology.
ADVANTAGES OF MALDI-TOF MS TECHNOLOGY AND COST-EFFECTIVENESS
With regard to viral diagnosis, there are few studies comparing MALDI-TOF MS technology with other conventional based methods (e.g. viral, culture, nucleic acid based techniques). MALDI-TOF method successfully detects viruses in a wide variety of biological specimens, and the concordance rate between MALDI-TOF and these techniques is high. PCR based identification strategies have some limitations such as much longer turnaround time than MS, reagent and labour costs and some technical problems. On the other hand, MALDI-TOF technology can also be used to identify viruses in co-infected samples. These methods are capable of simultaneous detection of multiple pathogens in a single assay and can then prevent misdiagnosis and delays in treatment without increasing costs or adding a new step to the process.
With regard to the technical problems, PCR methods could have inhibitory particles and issues of contamination; thus, separate areas for sample preparation, amplification and analysis are needed. Furthermore, nucleic acid based techniques are expensive and time-consuming and appear in most cases less convenient than MS for routine laboratory identifications. For MALDI-TOF there must be significant progress made in closing the upfront of PCR processing to include DNA extraction and the PCR process itself.
MALDI-TOF MS methods also allow large-scale research studies not only for fresh samples but also on archival samples from several biological specimens.
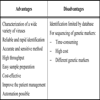
The main advantages and disadvantages of MALDI-TOF MS technology are shown in Table 2.
The most remarkable differences between MALDI-TOF method and other conventional techniques are the estimated time and costs required for sample identification. There are no studies of cost-effectiveness in viruses, but it is estimated that the cost of bacterial identification represent only 17-32% of the costs of conventional identification methods [32Seng P, Drancourt M, Gouriet F. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 2009; 49: 543-1.]. Other studies have shown that the cost of reagents required for phenotypic and nucleic acid identification is higher than for MS techniques [33Cherkaoui A, Hibbs J, Emonet S. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J Clin Microbiol 2010; 48: 1169-75.]. The expensive cost of MALDI-TOF devices is comparable with other laboratory equipment. Moreover, MALDI-TOF is a rapid technique if compared with other methods, so this technology improves the working time (e.g. pre-analytical procedure) as well as the turnaround time (e.g. analytical proceduce). The time needed for microorganism identification is < 15 minutes for MS method, whereas conventional techniques need 5-48 hours.
One of the main weaknesses of MALDI-TOF is that the identification is limited by database. The mass profile is used as a mass spectrum to compare with those of well-characterized organisms in a database. The spectrum typically includes specific peaks, so that with a comprehensive library of spectra, the identification might be performed by using bioinformatics.
Currently, there are a wide number of references in the database from the available commercial platforms for MALDI-TOF techniques. These mass spectra can establish the comparison and further identification of microorganisms, but until now these spectra are limited. The database will expand due to the collaboration between commercial companies and the hospitals of several countries, so the available databases should be improved in the near future. It will be very important the comparison of data from different companies’ instruments, but this fact is currently not feasible.
DATA MANAGEMENT AND QUALITY CONTROL
Sample preparation by MALDI-TOF MS remains laborious today, although some robots are being developed in order to improve the deposit of the sample and the matrix on the target. Moreover, some aspects of MALDI-TOF MS technology are being also investigated; for example, the laser pulse frequency, disposable target slides and the mass spectrometers as well as the software used for the communication between laboratory equipment and laboratory information systems.
Data management primarily depends on the interpretation of crude data. Currently, some methods for data interpretation have been investigated [34Welker M. Proteomics for routine identification of microorganisms. Proteomics 2011; 11: 3143-53.]. An important fact is the integration of information communication systems to physicians that requires hospital or laboratory information systems (HIS, LIS).
Reference databases require updating if new information is obtained. The addition of new isolates or mutations could improve this tool and the final diagnosis of the diseases. A robust system of information for regional prevalence of infectious diseases, emerging and re-emerging infectious pathogens, the discovery of new mutations and local or pandemic changes in epidemiology is necessary.
The MALDI-TOF MS system is increasingly used in clinical diagnostic laboratory for viral identification, mutation analysis, genotyping and antiviral resistance studies. However, there are still some problems arising during sample extraction affecting to the extraction protocol and to the adequate reagent conservation for the assay. These problems could be avoided by the introduction of an adequate quality control program. The extraction step should be daily checked by training technicians as well as the introduction of routinely internal quality controls.
Other aspect of this technique that must be submitted to an adequate quality control is the deposit of the sample on the microplate and the cleaning of this microplate. It is necessary implement a control about these issues in order to the correct identification of the samples.
Finally, it is a good practice for these devices the adequate maintenance of the MALDI-TOF based on a global quality control program. In this sense, the calibration control is a tool useful to recalibrate the spectrometer device and to identify technical problems. Appropriate maintenance of these devices is essential to carry out an adequate and accurate viral identification.
It is essential both to continuously improve the quality of the database and to introduce a quality control program for the MS device. The implementation of these programs can help to improve the quality of the overall procedure, so it will be the basis to improve the usefulness and the accuracy of MALDI-TOF.
CONCLUDING REMARKS
MALDI-TOF MS is a relatively recent innovation for the identification of microorganisms and is being used in many clinical microbiology laboratories. The main application of this technology is, at the moment, the identification of bacteria to species level in less than a minute, although this method is about to become the standard of identification of microorganisms in most laboratories.
MALDI-TOF MS technology is also being introduced in virology assays. The main applications in this field are the identification of viruses from clinical samples, the discovery of mutations for these viruses, the rapid screening of virus subtypes, its use in molecular epidemiology and the detection of resistance to several antiviral agents.
Future applications of MALDI-TOF MS in clinical virology will be developed in order to provide more accurate identification on clinical specimens and will include the detection of resistance protein markers and specific virulence for several viruses. However, the introduction of this technology routinely for virology diagnosis would enable a change in working practices. Moreover, this fact will require a complete integration into laboratory workflow and several technical improvements such as to detection sensitivity. The continuous updating of databases for microorganisms will be also an important issue, as well as the possibility for the diagnosis directly from clinical samples.
MS methods can be used currently for viral diagnosis but refinement of technology will be an important approach for the introduction of MALDI-TOF in routine viral diagnosis.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
ACKNOWLEDGEMENTS
Declared none.