- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

Current Chemical Genomics and Translational Medicine
(Discontinued)
ISSN: 2213-9885 ― Volume 12, 2018
A Homogenous Luminescent Proximity Assay for 14-3-3 Interactions with Both Phosphorylated and Nonphosphorylated Client Peptides
Yuhong Du1, 3, Fadlo R Khuri2, Haian Fu*, 1, 2, 3
Abstract
The 14-3-3 proteins are a family of dimeric eukaryotic proteins that mediate both phosphorylation-dependent and -independent protein-protein interactions. Through these interactions, 14-3-3 proteins participate in the regulation of a wide range of cellular processes, including cell proliferation, cell cycle progression, and apoptosis. Because of their fundamental importance, 14-3-3 proteins have also been implicated in a variety of diseases, including cancer and neurodegenerative disorders. In order to monitor 14-3-3/client protein interactions for the discovery of small molecule 14-3-3 modulators, we have designed and optimized 14-3-3 protein binding assays based on the amplified luminescent proximity homogeneous assay (AlphaScreen) technology. Using the interaction of 14-3-3 with a phosphorylated Raf-1 peptide and a nonphosphorylated R18 peptide as model systems, we have established homogenous “add-and-measure” high-throughput screening assays. Both assays achieved robust performance with S/B ratios above 7 and Z’ factors above 0.7. Application of the known antagonistic peptides in our studies further validated the assay for screening of chemical compound libraries to identify small molecules that can modulate 14-3-3 protein-protein interactions.
Article Information
Identifiers and Pagination:
Year: 2008Volume: 2
First Page: 40
Last Page: 47
Publisher Id: CCGTM-2-40
DOI: 10.2174/1875397300802010040
Article History:
Received Date: 30/7/2008Revision Received Date: 01/9/2008
Acceptance Date: 02/9/2008
Electronic publication date: 6/11/2008
Collection year: 2008
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Department of Pharmacology, Emory University School of Medicine and Emory Chemical Biology Discovery Center, Atlanta, GA 30322, USA; Tel: 404-727-0368; Fax: 404-727-0365; E-mail: hfu@emory.edu
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 30-7-2008 |
Original Manuscript | A Homogenous Luminescent Proximity Assay for 14-3-3 Interactions with Both Phosphorylated and Nonphosphorylated Client Peptides | |
INTRODUCTION
The 14-3-3 proteins are a family of eukaryotic regulatory proteins that can recognize phosphoserine/threonine (pSer/Thr)-containing motifs utilized by a variety of signal transduction pathways [1Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation Annu Rev Pharmacol Toxicol 2000; 40: 617-47.-5Mackintosh C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes Biochem J 2004; 381(Pt 2): 329-42.]. 14-3-3 can directly bind pSer or pThr in specific sequence contexts (Fig. 1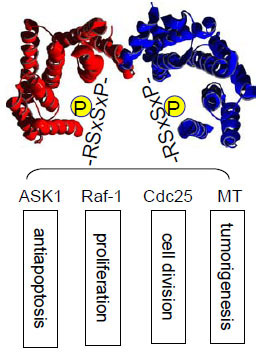 ) [6Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine Cell 1996; 84(6): 889-97., 7Yaffe MB, Rittinger K, Volinia S, et al. The structural basis for 14-3-3: phosphopeptide binding specificity Cell 1997; 91(7): 961-71.]. Sequences around pS259 and pS621 in Raf-1 are well studied examples. This phosphorylation-dictated interaction allows 14-3-3 proteins to bind a multitude of functionally diverse molecules, including kinases, phosphatases, transmembrane receptors, and transcription factors [1Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation Annu Rev Pharmacol Toxicol 2000; 40: 617-47., 5Mackintosh C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes Biochem J 2004; 381(Pt 2): 329-42., 8Dougherty MK, Morrison DK. Unlocking the code of 14-3-3 J Cell Sci 2004; 117(Pt 10): 1875-84.]. In addition, 14-3-3 has been shown to bind to client proteins in a phosphorylation-independent manner. For example, 14-3-3 can activate the Exoenzyme S ADP-ribosyltransferase, a prokaryotic protein factor secreted by Pseudomonas aeruginosa [9Fu H, Coburn J, Collier RJ. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family Proc Natl Acad Sci USA 1993; 90(6): 2320-4.]. The identification of high affinity peptides as 14-3-3 binding sequences, such as R18, by phage display technology further validates this mode of nonphosphorylation-dependent 14-3-3 interaction [10Wang B, Yang H, Liu YC, et al. Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display Biochemistry 1999; 38(38): 12499-504.]. Through these interactions, 14-3-3 proteins participate in the regulation of a wide range of vital cellular processes, including cell proliferation, cell cycle progression, and apoptosis, as well as in a number of pathophysiological conditions, such as cancer and neurodegenerative diseases. Thus, understanding the mechanisms that govern 14-3-3/client protein interactions will provide critical insight into the control of intracellular signal transduction. Availability of 14-3-3 modulators is expected to greatly facilitate our mechanistic investigation of 14-3-3 functions. These 14-3-3 modulators may become leads for potential therapeutic development.
) [6Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine Cell 1996; 84(6): 889-97., 7Yaffe MB, Rittinger K, Volinia S, et al. The structural basis for 14-3-3: phosphopeptide binding specificity Cell 1997; 91(7): 961-71.]. Sequences around pS259 and pS621 in Raf-1 are well studied examples. This phosphorylation-dictated interaction allows 14-3-3 proteins to bind a multitude of functionally diverse molecules, including kinases, phosphatases, transmembrane receptors, and transcription factors [1Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation Annu Rev Pharmacol Toxicol 2000; 40: 617-47., 5Mackintosh C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes Biochem J 2004; 381(Pt 2): 329-42., 8Dougherty MK, Morrison DK. Unlocking the code of 14-3-3 J Cell Sci 2004; 117(Pt 10): 1875-84.]. In addition, 14-3-3 has been shown to bind to client proteins in a phosphorylation-independent manner. For example, 14-3-3 can activate the Exoenzyme S ADP-ribosyltransferase, a prokaryotic protein factor secreted by Pseudomonas aeruginosa [9Fu H, Coburn J, Collier RJ. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family Proc Natl Acad Sci USA 1993; 90(6): 2320-4.]. The identification of high affinity peptides as 14-3-3 binding sequences, such as R18, by phage display technology further validates this mode of nonphosphorylation-dependent 14-3-3 interaction [10Wang B, Yang H, Liu YC, et al. Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display Biochemistry 1999; 38(38): 12499-504.]. Through these interactions, 14-3-3 proteins participate in the regulation of a wide range of vital cellular processes, including cell proliferation, cell cycle progression, and apoptosis, as well as in a number of pathophysiological conditions, such as cancer and neurodegenerative diseases. Thus, understanding the mechanisms that govern 14-3-3/client protein interactions will provide critical insight into the control of intracellular signal transduction. Availability of 14-3-3 modulators is expected to greatly facilitate our mechanistic investigation of 14-3-3 functions. These 14-3-3 modulators may become leads for potential therapeutic development.
 |
Fig. (1) 14-3-3 binds phosphorylated Ser/Thr motifs with RSxpS/TxP as a prototype. It plays an important role in multiple signaling pathways. |
In order to identify small molecule 14-3-3 modulators, fluorescent polarization (FP) assays have been reported [11Du Y, Masters SC, Khuri FR, Fu H. Monitoring 14-3-3 protein interactions with a homogeneous fluorescence polarization assay J Biomol Screen 2006; 11(3): 269-76., 12Wu M, Coblitz B, Shikano S, et al. SWTY--a general peptide probe for homogeneous solution binding assay of 14-3-3 proteins Anal Biochem 2006; 349(2): 186-96.]. The FP-based assays are widely used for high-throughput screening (HTS) due to their simplicity and sensitivity. However, they are prone to interference by fluorescent compounds and are not applicable for protein-protein interactions. Here we describe an alternative HTS assay format for 14-3-3 proteins based on the AlphaScreen (Amplified Luminescent Proximity Homogenous Assay) technology [13Ullman EF, Kirakossian H, Singh S, et al. Luminescent oxygen channeling immunoassay: measurement of particle binding kinetics by chemiluminescence Proc Natl Acad Sci USA 1994; 91(12): 5426-30., 14Eglen RM, Reisine T, Roby P, Rouleau N, Bosse RI, Bielefeld M. The use of AlphaScreen technology in HTS: Current status Curr Chem Genom 2008; 1: 2-10.]. AlphaScreen offers a bead-based, non-radioactive homogenous assay that can be adapted to small volumes in a microplate format. It can be used either as a primary HTS assay or as a confirmative assay and has added value by allowing the detection of interactions between two full length proteins. Using both phosphorylated and nonphosphorylated client targets of 14-3-3 proteins, we have designed and tested AlphaScreen-based 14-3-3 assays for HTS. These assays have been further optimized and validated with known peptide inhibitors for the screening of small molecule 14-3-3 inhibitors.
EXPERIMENTAL PROCEDURES
Purification of Recombinant 14-3-3γ and GST-R18 Proteins
Recombinant 14-3-3γ protein was expressed in Escherichia coli strain BL21 (DE3) as a glutathione S–transferase (GST)-tagged or hexaHistidine (His)-tagged product as previously described [11Du Y, Masters SC, Khuri FR, Fu H. Monitoring 14-3-3 protein interactions with a homogeneous fluorescence polarization assay J Biomol Screen 2006; 11(3): 269-76.]. Similarly, GST-R18 fusion protein was overexpressed in E. Coli. E. coli BL21 (DE3) strains were grown in LB/ampicillin medium for protein auto-induction. After overnight shaking at 37°C, the cells were harvested and the pellets were resuspended in ice-cold PBS buffer supplemented with phenylmethylsulfonyl fluoride (1.0 mM), leupeptin (1μg/ml), and aprotinin (1μg/ml). The resuspended pellets were sonicated and cleared by centrifugation at 4°C (16,000× g, 15 min). His-14-3-3γ protein was purified by a Ni2+-charged His-binding column (Novagen) as previously described [9Fu H, Coburn J, Collier RJ. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family Proc Natl Acad Sci USA 1993; 90(6): 2320-4.], and used for biotinylation. GST-14-3-3γ and GST-R18 proteins were purified using a glutathione-conjugated Sepharose column (GE Healthcare) [11Du Y, Masters SC, Khuri FR, Fu H. Monitoring 14-3-3 protein interactions with a homogeneous fluorescence polarization assay J Biomol Screen 2006; 11(3): 269-76.]. The isolated protein fractions containing 14-3-3γ were desalted using a PD-10 desalting column (GE Healthcare) equilibrated in HEPES buffer. The purified proteins were stored at -20ºC before use.
Basic Assay Development
The AlphaScreen binding assay was carried out in 384-well, white Proxiplates (PerkinElmer, Boston, MA) in a total volume of 20 µl. The AlphaScreen GST detection kit was from PerkinElmer Life Science.
To test the feasibility of the assay platform, we detected the binding of 14-3-3γ to R18 peptide. R18 (PHCVPRDLSWLDLEANMCLP) is an unphosphorylated peptide derived from phage display which exhibits a high affinity for 14-3-3 proteins [10Wang B, Yang H, Liu YC, et al. Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display Biochemistry 1999; 38(38): 12499-504.]. The AlphaScreen donor beads were supplied as streptavidin-coated, and the acceptor beads were conjugated to an anti-GST antibody. Biotinylated 14-3-3γ protein was generate using EZ-LinkTM sulfo-NHS-LC-biotin (Pierce) with purified His-14-3-3γ, while the GST-R18 fusion protein was purified from bacterial cells as described above. To ensure the integrity of the biotinylated His-14-3-3γ, its binding properties were verified using a previously developed FP assay [11Du Y, Masters SC, Khuri FR, Fu H. Monitoring 14-3-3 protein interactions with a homogeneous fluorescence polarization assay J Biomol Screen 2006; 11(3): 269-76.].
Different concentrations of biotin-14-3-3γ protein and GST-R18 were mixed together in 384-well plates (5 μl/well). After a 1 h incubation, acceptor beads (5µl/well) were added and incubated for 30 min. Donor beads (10 μl/well) were subsequently added and incubated for 1.5 h before reading with an EnVision multilabel reader (PerkinElmer life Science). The final concentration of donor and acceptor beads was 20 µg/ml. All dilutions were made in HEPES buffer (10mM HEPES, 150mM NaCl, 0.05% Tween-20, 0.5 mM DTT). Due to the light sensitivity of the donor and acceptor beads, the bead dispensing and reading steps were carried out under dim light conditions.
For the detection of the 14-3-3 protein binding to a Raf-1-derived phosphopeptide, pS259-Raf-1 [6Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine Cell 1996; 84(6): 889-97.] , biotintylated-pS259-Raf- peptide (Biotin-LSQRSTpSTPNVHM) was custom synthesized (Anaspec, Inc.) and used in an AlphaScreen assay with purified GST-14-3-3γ. The binding experiments were conducted similarly as described above for biotin-14-3-3γ protein and GST-R18.
Peptide Competition Assays
In order to verify the specificity of the 14-3-3 binding assay, two untagged antagonist peptides of 14-3-3 proteins, R18 and a phosphorylated pS967-ASK1- peptide, were used to test their ability to displace the GST-R18 or biotin-pS259-Raf from 14-3-3 proteins. An R18 mutant peptide and a non-phosphorylated ASK1 peptide were used as controls. Peptides were synthesized at the Emory Microchemistry Facility and dissolved in HEPES buffer as stocks. Within each binding experiment, dose-dependent competition was carried out with serial dilutions of peptides, prepared in HEPES buffer. In a 384-well plate, 0.5 μl of each diluted untagged peptide was added to 4.5 µl of the assay mixtures (biotin-14-3-3γ and GST-R18 or GST-14-3-3γ and biotin-pS259-Raf). For each assay, background (one binding component only) and control (without competition peptide) samples were included in each assay plate. The competitive effect of the peptides on binding was expressed as a percentage of control Alpha signal as the following:
Data were plotted against log10 value of peptide concentrations and analyzed using Prism 4.0 software (Graphpad Software, San Diego, CA). IC50 values were determined by non-linear curve fitting as the concentrations of the peptides at which 50% of control Alpha signal was inhibited.
High-Throughput Screening Assay Performance
To monitor assay sensitivity, signal-to-background (S/B) ratios were calculated by the following equation:
S/B = mean signal binding / mean background
To evaluate the quality and suitability of the 14-3-3 binding assay using AlphaScreen for HTS, we determined the Z’ factor [15Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays J Biomol Screen 1999; 4(2): 67-73.]:
Z’ = 1- (3SD of control + 3SD of background) / (mean of control – mean of background).
To monitor the robustness and reproducibility of the 384-well HTS format, the means of both background and control Alpha signal from each assay plate were obtained and analyzed by a scatter plot.
RESULTS AND DISCUSSION
AlphaScreen technology provides a proximity-dependent homogenous assay format for studying molecular interactions [13Ullman EF, Kirakossian H, Singh S, et al. Luminescent oxygen channeling immunoassay: measurement of particle binding kinetics by chemiluminescence Proc Natl Acad Sci USA 1994; 91(12): 5426-30., 14Eglen RM, Reisine T, Roby P, Rouleau N, Bosse RI, Bielefeld M. The use of AlphaScreen technology in HTS: Current status Curr Chem Genom 2008; 1: 2-10.]. This assay system contains two beads, a “donor” bead and an “acceptor” bead. Donor beads contain the photosensitizer phthalocyanine, which upon illumination at 680 nm converts ambient oxygen to an excited form of oxygen, singlet oxygen. If an acceptor bead is within the energy diffusion range (< 200 nm) in solution, energy is transferred from the singlet oxygen to thioxene derivatives within the acceptor bead, resulting in light production at 520-620 nm (Fig. 2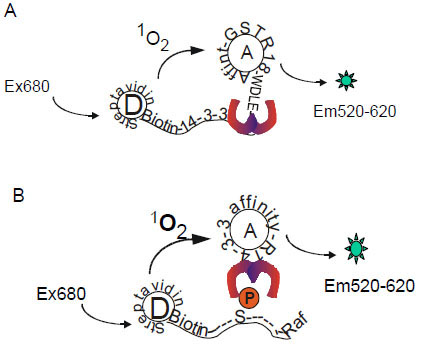 ). The proximity of these two beads can be induced by interaction of two binding components that are coupled to the beads. In the AlphaScreen system employed in our study, the donor beads were coated with streptavidin and acceptor beads were conjugated to anti-GST antibody. Utilizing the AlphaScreen technology, we have designed two assay formats for 14-3-3 binding studies. One assay was used for monitoring the association of 14-3-3 with the non-phosphorylated binding peptide R18 (Fig. 2A
). The proximity of these two beads can be induced by interaction of two binding components that are coupled to the beads. In the AlphaScreen system employed in our study, the donor beads were coated with streptavidin and acceptor beads were conjugated to anti-GST antibody. Utilizing the AlphaScreen technology, we have designed two assay formats for 14-3-3 binding studies. One assay was used for monitoring the association of 14-3-3 with the non-phosphorylated binding peptide R18 (Fig. 2A ), while the other assay was used for detecting the interaction of 14-3-3 with a phosphorylated peptide derived from the well-studied client protein Raf-1 (Fig. 2B
), while the other assay was used for detecting the interaction of 14-3-3 with a phosphorylated peptide derived from the well-studied client protein Raf-1 (Fig. 2B ).
).
Interaction of 14-3-3 with R18
To test the feasibility of using the AlphaScreen technology for monitoring the interaction of 14-3-3 with its client proteins, we utilized the well-established association of 14-3-3 with a high affinity peptide, R18, as a model system. Examinations by mutational and structural approaches have conclusively established that R18 docks in the ligand binding site of 14-3-3 that is also used by natural client proteins [10Wang B, Yang H, Liu YC, et al. Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display Biochemistry 1999; 38(38): 12499-504., 16Petosa C, Masters SC, Bankston LA, et al. 14-3-3zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove J Biol Chem 1998; 273(26): 16305-0.]. Functionally, R18 can effectively compete with other natural proteins for the binding of 14-3-3 proteins [17Masters SC, Fu H. 14-3-3 proteins mediate an essential anti-apoptotic signal J Biol Chem 2001; 276(48): 45193-200.]. Thus, the 14-3-3/R18 interaction was utilized to provide a proof-of-concept for the development of an HTS assay with the AlphaScreen technology platform.
The AlphaScreen requires the coupling of test protein components onto specific donor and acceptor beads. Accordingly, we generated biotinylated His-14-3-3γ protein for coupling to the streptavidin-coated donor beads (Fig. 2 ). To ensure that the biotinylation did not compromise the binding activity of 14-3-3 proteins, we examined the interaction of the biotin-14-3-3γ protein with a Raf-1 peptide using a FP assay [11Du Y, Masters SC, Khuri FR, Fu H. Monitoring 14-3-3 protein interactions with a homogeneous fluorescence polarization assay J Biomol Screen 2006; 11(3): 269-76.]. Biotinylation appeared to have no obvious effect on the binding properties of 14-3-3γ, with a KD (0.76 μM) comparable to nonbiotinylated 14-3-3 (KD of 0.42 μM [11Du Y, Masters SC, Khuri FR, Fu H. Monitoring 14-3-3 protein interactions with a homogeneous fluorescence polarization assay J Biomol Screen 2006; 11(3): 269-76.]; Fig. 3A
). To ensure that the biotinylation did not compromise the binding activity of 14-3-3 proteins, we examined the interaction of the biotin-14-3-3γ protein with a Raf-1 peptide using a FP assay [11Du Y, Masters SC, Khuri FR, Fu H. Monitoring 14-3-3 protein interactions with a homogeneous fluorescence polarization assay J Biomol Screen 2006; 11(3): 269-76.]. Biotinylation appeared to have no obvious effect on the binding properties of 14-3-3γ, with a KD (0.76 μM) comparable to nonbiotinylated 14-3-3 (KD of 0.42 μM [11Du Y, Masters SC, Khuri FR, Fu H. Monitoring 14-3-3 protein interactions with a homogeneous fluorescence polarization assay J Biomol Screen 2006; 11(3): 269-76.]; Fig. 3A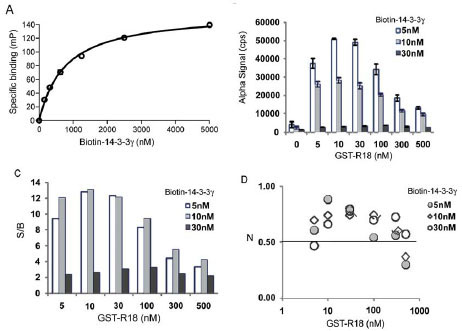 ). For generating the corresponding acceptor beads, a GST-tagged R18 protein-peptide fusion was purified for coupling to the anti-GST antibody-coated beads. We used the R18 fusion protein in order to place the 14-3-3 recognition sequence within the context of a full length protein to mimic the situation in natural 14-3-3 client proteins. Our previous studies demonstrated that the fusion of GST had no detectable effect on the efficacy of R18 for 14-3-3 binding [10Wang B, Yang H, Liu YC, et al. Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display Biochemistry 1999; 38(38): 12499-504.]. These reagents were then added to the reaction in different sequences or combinations before the Alpha luminescence signal was recorded. As shown in Fig. (3B
). For generating the corresponding acceptor beads, a GST-tagged R18 protein-peptide fusion was purified for coupling to the anti-GST antibody-coated beads. We used the R18 fusion protein in order to place the 14-3-3 recognition sequence within the context of a full length protein to mimic the situation in natural 14-3-3 client proteins. Our previous studies demonstrated that the fusion of GST had no detectable effect on the efficacy of R18 for 14-3-3 binding [10Wang B, Yang H, Liu YC, et al. Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display Biochemistry 1999; 38(38): 12499-504.]. These reagents were then added to the reaction in different sequences or combinations before the Alpha luminescence signal was recorded. As shown in Fig. (3B ), when biotin-14-3-3γ proteins (5 nM, 10 nM, or 30 nM) were incubated with the donor and acceptor beads in the absence of GST-R18, only a low background Alpha signal (~2,000 counts) was generated. However, the addition of increasing concentrations of GST-R18 (5 nM to 500 nM) to the biotin-14-3-3γ solutions (5 nM or 10 nM) induced the robust production of Alpha signal in a dose-dependent manner. The specific Alpha signal, calculated by subtracting the background counts, approached approximately 46,000 counts with 5 nM of biotin-14-3-3γ and 10 nM of GST-R18. The S/B ratio of the assay reached beyond 10 under the conditions tested, which defines an excellent assay window (Fig. 3C
), when biotin-14-3-3γ proteins (5 nM, 10 nM, or 30 nM) were incubated with the donor and acceptor beads in the absence of GST-R18, only a low background Alpha signal (~2,000 counts) was generated. However, the addition of increasing concentrations of GST-R18 (5 nM to 500 nM) to the biotin-14-3-3γ solutions (5 nM or 10 nM) induced the robust production of Alpha signal in a dose-dependent manner. The specific Alpha signal, calculated by subtracting the background counts, approached approximately 46,000 counts with 5 nM of biotin-14-3-3γ and 10 nM of GST-R18. The S/B ratio of the assay reached beyond 10 under the conditions tested, which defines an excellent assay window (Fig. 3C ). Further increases in the GST-R18 concentration beyond 100 nM or the biotin-14-3-3 protein concentration beyond 30 nM resulted in a decrease in Alpha signal and S/B ratio. This “hooking effect”, an effect common to many immunoassays relying on antibody recognition and binding of target molecules, is likely due to the saturation of available binding sites on one or both of the AlphaScreen beads [18PerkinElmer LaAS. A practical guide to working with AlphaScreen 2004 [cited; Available from: http: //las.perkinelmer.com/ Content/Manuals/GDE_AlphaScreenPracticalGuide.pdf] ]. It has been shown that Alpha signal increases with increasing target molecule concentrations up to a point, after which the target molecules will become inhibitory to the production of Alpha signals [19Sehr P, Pawlita M, Lewis J. Evaluation of different glutathione S-transferase-tagged protein captures for screening E6/E6AP interaction inhibitors using AlphaScreen J Biomol Screen 2007; 12(4): 560-7., 20Mills NL, Shelat AA, Guy RK. Assay optimization and screening of RNA-Protein interactions by AlphaScreen J Biomol Screen 2007; 12(7): 946-55.]. These studies suggest that the AlphaScreen assay platform can be used to monitor the interaction of 14-3-3 with its binding partners.
). Further increases in the GST-R18 concentration beyond 100 nM or the biotin-14-3-3 protein concentration beyond 30 nM resulted in a decrease in Alpha signal and S/B ratio. This “hooking effect”, an effect common to many immunoassays relying on antibody recognition and binding of target molecules, is likely due to the saturation of available binding sites on one or both of the AlphaScreen beads [18PerkinElmer LaAS. A practical guide to working with AlphaScreen 2004 [cited; Available from: http: //las.perkinelmer.com/ Content/Manuals/GDE_AlphaScreenPracticalGuide.pdf] ]. It has been shown that Alpha signal increases with increasing target molecule concentrations up to a point, after which the target molecules will become inhibitory to the production of Alpha signals [19Sehr P, Pawlita M, Lewis J. Evaluation of different glutathione S-transferase-tagged protein captures for screening E6/E6AP interaction inhibitors using AlphaScreen J Biomol Screen 2007; 12(4): 560-7., 20Mills NL, Shelat AA, Guy RK. Assay optimization and screening of RNA-Protein interactions by AlphaScreen J Biomol Screen 2007; 12(7): 946-55.]. These studies suggest that the AlphaScreen assay platform can be used to monitor the interaction of 14-3-3 with its binding partners.
 |
Fig. (3) AlphaScreen assay for measuring the interaction of 14-3-3γ with R18. (A) Biotinylation has no effect on the ligand binding capability of His-14-3-3γ proteins. Binding of biotin-14-3-3γ to pS259-Raf-1 peptide was determined by FP assay [11Du Y, Masters SC, Khuri FR, Fu H. Monitoring 14-3-3 protein interactions with a homogeneous fluorescence polarization assay J Biomol Screen 2006; 11(3): 269-76.]. Biotin-14-3-3 proteins were incubated with a TMR-pS259-Raf peptide (1 nM) at RT for 30 min and the FP signal [expressed in millipolarization units (mP)] was read with an Analyst HT reader (Molecular Devices). Specific binding (mP) was obtained by subtracting free peptide tracer values from values recorded in the presence of 14-3-3 proteins. (B) Interaction of biotin-14-3-3 with GST-R18 generates an Alpha luminescence signal. Increasing concentrations of GST-R18 protein were incubated with biotin-14-3-3 protein at RT for 30 min. Donor and acceptor beads were added with an interval of 30 min. The plate was assayed using an EnVision multilabel plate reader after 1.5 hr incubation at room temperature. Data are expressed as mean ± SD from triplicate samples. (C) S/B ratios of the 14-3-3γ/R18 AlphaScreen assay. (D) Z’ values of the assay. |
To evaluate the quality and suitability of the AlphaScreen assay for HTS in a 384-well microplate format, the Z’ factors were determined. The Z’ factor incorporates the dynamic assay range and the assay variability into a single metric, and is a parameter for the quality of the assay itself without test compounds [21Zhang L, Chen J, Fu H. Suppression of apoptosis signal-regulating kinase 1-induced cell death by 14-3-3 proteins Proc Natl Acad Sci USA 1999; 96(15): 8511-5.]. The Z’-factor is widely utilized for quality assessment in assay development and optimization. Assays with Z’ factors between 0.5 and 1 (the maximum) are considered of high quality and suitable for automation and the subsequent screening. As shown in Fig. (3D ), when biotin-14-3-3γ at a concentration of 5 nM or 10 nM was incubated with GST-R18 (5 nM, 10 nM, or 30 nM), the calculated Z’ values were all above 0.7, supporting a robust assay format. Thus, this AlphaScreen assay for the 14-3-3 and R18 interaction can be further developed for HTS of 14-3-3 modulators.
), when biotin-14-3-3γ at a concentration of 5 nM or 10 nM was incubated with GST-R18 (5 nM, 10 nM, or 30 nM), the calculated Z’ values were all above 0.7, supporting a robust assay format. Thus, this AlphaScreen assay for the 14-3-3 and R18 interaction can be further developed for HTS of 14-3-3 modulators.
To validate the AlphaScreen assay for the discovery of 14-3-3 inhibitors, we tested inhibition of the interaction of 14-3-3 with the R18 by two untagged antagonist peptides: an R18 peptide and an ASK1 peptide, pS967-ASK1. The untagged R18 and pS967-ASK1 peptides are expected to compete with GST-R18 for the binding of 14-3-3 proteins. Indeed, incubation of increasing concentrations of untagged R18 peptide with biotin-14-3-3γ and GST-R18 led to a dose-dependent decrease in Alpha signal with an IC50 value of 28 ± 1.1 nM (Fig. 4A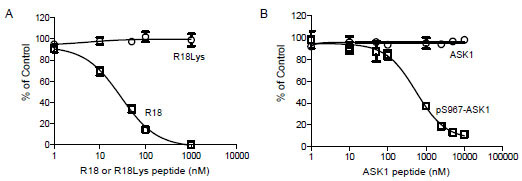 ). On the other hand, untagged R18Lys, a R18 peptide with a mutated 14-3-3 binding residue, failed to displace GST-R18 from the 14-3-3 protein, supporting a specific effect of the R18 competitor. Many 14-3-3 client proteins, including ASK1, require phosphorylation to mediate the binding to 14-3-3 proteins. The pS967-ASK1 peptide is a 14-3-3 binding peptide that contains phosphorylated Ser-967 and its surrounding sequence within the ASK1 protein [21Zhang L, Chen J, Fu H. Suppression of apoptosis signal-regulating kinase 1-induced cell death by 14-3-3 proteins Proc Natl Acad Sci USA 1999; 96(15): 8511-5., 22Subramanian RR, Zhang H, Wang H, Ichijo H, Miyashita T, Fu H. Interaction of apoptosis signal-regulating kinase 1 with isoforms of 14-3-3 proteins Exp Cell Res 2004; 294(2): 581-91.]. As shown in Fig. (4B
). On the other hand, untagged R18Lys, a R18 peptide with a mutated 14-3-3 binding residue, failed to displace GST-R18 from the 14-3-3 protein, supporting a specific effect of the R18 competitor. Many 14-3-3 client proteins, including ASK1, require phosphorylation to mediate the binding to 14-3-3 proteins. The pS967-ASK1 peptide is a 14-3-3 binding peptide that contains phosphorylated Ser-967 and its surrounding sequence within the ASK1 protein [21Zhang L, Chen J, Fu H. Suppression of apoptosis signal-regulating kinase 1-induced cell death by 14-3-3 proteins Proc Natl Acad Sci USA 1999; 96(15): 8511-5., 22Subramanian RR, Zhang H, Wang H, Ichijo H, Miyashita T, Fu H. Interaction of apoptosis signal-regulating kinase 1 with isoforms of 14-3-3 proteins Exp Cell Res 2004; 294(2): 581-91.]. As shown in Fig. (4B ), the free pS967-ASK1 peptide competed with the binding of GST-R18 to 14-3-3γ, resulting in a dose-dependent decrease in Alpha signal with an IC50 value of 549.4 ± 1.1 nM. The same ASK1 peptide without phosphorylation, however, had no effect on the Alpha signal generated from the 14-3-3 binding to R18 (Fig. 4
), the free pS967-ASK1 peptide competed with the binding of GST-R18 to 14-3-3γ, resulting in a dose-dependent decrease in Alpha signal with an IC50 value of 549.4 ± 1.1 nM. The same ASK1 peptide without phosphorylation, however, had no effect on the Alpha signal generated from the 14-3-3 binding to R18 (Fig. 4 ). These results are consistent with reported interactions of 14-3-3 with phosphorylated client peptides [6Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine Cell 1996; 84(6): 889-97., 7Yaffe MB, Rittinger K, Volinia S, et al. The structural basis for 14-3-3: phosphopeptide binding specificity Cell 1997; 91(7): 961-71.]. Thus, small molecules that compete for the binding of R18 to 14-3-3 proteins can be revealed in the AlphaScreen-based assay platform.
). These results are consistent with reported interactions of 14-3-3 with phosphorylated client peptides [6Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine Cell 1996; 84(6): 889-97., 7Yaffe MB, Rittinger K, Volinia S, et al. The structural basis for 14-3-3: phosphopeptide binding specificity Cell 1997; 91(7): 961-71.]. Thus, small molecules that compete for the binding of R18 to 14-3-3 proteins can be revealed in the AlphaScreen-based assay platform.
Interaction of 14-3-3 with a Raf-1 Peptide
With the establishment of a general platform for monitoring 14-3-3 interactions, we extended our study to examine the utility of the AlphaScreen technology for the binding of 14-3-3 to a phosphorylated peptide derived from Raf-1, a mitogen-activated protein kinase regulator. It has been well-established that phosphorylation of Raf-1 at Ser-259 induces its association with the 14-3-3 proteins, which plays a critical role in Raf-1-mediated cell signaling [6Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine Cell 1996; 84(6): 889-97., 8Dougherty MK, Morrison DK. Unlocking the code of 14-3-3 J Cell Sci 2004; 117(Pt 10): 1875-84., 23Fantl WJ, Muslin AJ, Kikuchi A, et al. Activation of Raf-1 by 14-3-3 proteins Nature 1994; 371(6498): 612-4.-25Fu H, Xia K, Pallas DC, et al. Interaction of the protein kinase Raf-1 with 14-3-3 proteins Science 1994; 266(5182): 126-9.]. To test the 14-3-3/Raf-1 interaction in the AlphaScreen system, we generated a biotinylated Raf peptide, pS259-Raf-1, for coupling to streptavadin-coated donor beads. 14-3-3γ was produced and purified as a GST-tagged fusion protein for coupling to the GST-antibody coated acceptor beads. The interaction of biotin-pS259-Raf-1 peptide with GST-14-3-3γ would bring donor and acceptor beads together and induce the production of Alpha signal (Fig. 2 ). Indeed, mixing increasing concentrations of GST-14-3-3γ and biotin-pS259-Raf-1 with both beads resulted in a dose-dependent increase in Alpha signal (Fig. 5A
). Indeed, mixing increasing concentrations of GST-14-3-3γ and biotin-pS259-Raf-1 with both beads resulted in a dose-dependent increase in Alpha signal (Fig. 5A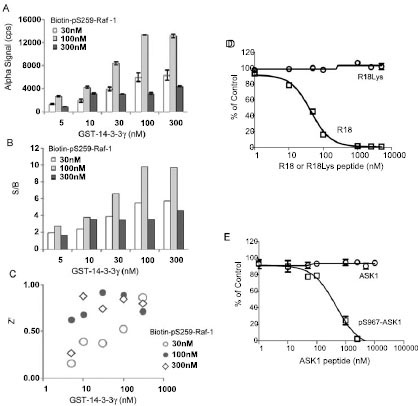 ). When incubated with 30 or 100 nM of biotin-pS259-Raf-1, the S/B ratio significantly increased with increasing concentration of 14-3-3 proteins (Fig. 5B
). When incubated with 30 or 100 nM of biotin-pS259-Raf-1, the S/B ratio significantly increased with increasing concentration of 14-3-3 proteins (Fig. 5B ). The calculated Z’ factors were above 0.6 when 100 nM of biotin-pS259-Raf-1 was incubated with increasing concentrations of GST-14-3-3 protein (Fig. 5C
). The calculated Z’ factors were above 0.6 when 100 nM of biotin-pS259-Raf-1 was incubated with increasing concentrations of GST-14-3-3 protein (Fig. 5C ). Z’ factors reached 0.9 when 30 or 100 nM of 14-3-3 was used, indicating a robust assay performance.
). Z’ factors reached 0.9 when 30 or 100 nM of 14-3-3 was used, indicating a robust assay performance.
 |
Fig. (5) AlphaScreen assay for measuring the interaction of 14-3-3γ with pS259-Raf-1. Experiments were performed as described in the legend to Fig. (4 |
To validate this assay for the screening of 14-3-3 inhibitors, two untagged 14-3-3 peptide antagonists were tested for inhibition of the interaction of 14-3-3 with Raf-1. R18 or pS967-ASK1 antagonist peptides were incubated with GST-14-3-3γ (100 nM) and biotin-pS259-Raf-1 peptide (100 nM), and Alpha signals were recorded. These two 14-3-3 antagonists effectively competed with the 14-3-3/Raf-1 binding as shown by decreased Alpha signal with the increasing concentrations of R18 or pS967-ASK1 peptide (Figs. 5D and 4E
and 4E ). Importantly, the R18Lys or non-phosphorylated ASK1 control peptides showed no effect in this assay. The calculated IC50 values for the R18 peptide was 35.7 ± 1.1 nM and for the pS967-ASK1 peptide was 534.9 ± 1.1 nM, which is consistent with the results obtained in the 14-3-3/R18 interaction assay (Fig. 4
). Importantly, the R18Lys or non-phosphorylated ASK1 control peptides showed no effect in this assay. The calculated IC50 values for the R18 peptide was 35.7 ± 1.1 nM and for the pS967-ASK1 peptide was 534.9 ± 1.1 nM, which is consistent with the results obtained in the 14-3-3/R18 interaction assay (Fig. 4 ).
).
HTS Assay Validation
One potential use of a homogeneous assay for 14-3-3 binding is to carry out HTS of chemical libraries for the identification of small molecule 14-3-3 inhibitors. In order to evaluate the suitability of the AlphaScreen-based 14-3-3 assays for HTS, a series of experiments were performed.
First, the dimethyl sulfoxide (DMSO) tolerance level of the assay was evaluated. DMSO is a common solvent used in dissolving chemical compounds. To determine the effect of DMSO on the AlphaScreen based 14-3-3 assay, experiments were performed in the presence of varying amounts of DMSO. As shown in Fig. (6A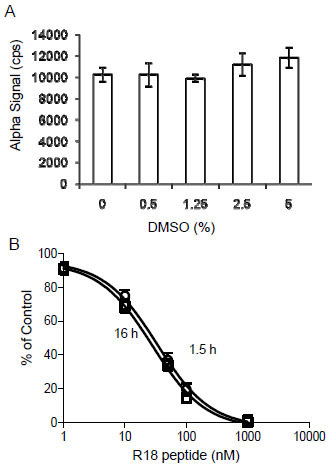 ), the Alpha signals for the binding of biotin-14-3-3γ (5 nM) to GST-R18 (15 nM) were not significantly altered in the presence of up to 5% DMSO. Then, we determined the assay stability by performing competition experiments and recording the AlphaScreen luminescence signal over time. As shown in Fig. (6B
), the Alpha signals for the binding of biotin-14-3-3γ (5 nM) to GST-R18 (15 nM) were not significantly altered in the presence of up to 5% DMSO. Then, we determined the assay stability by performing competition experiments and recording the AlphaScreen luminescence signal over time. As shown in Fig. (6B ), the addition of untagged R18 peptide to the reaction containing biotin-14-3-3γ, GST-R18, and the donor/acceptor beads, decreased the Alpha luminescence signals in a dose-dependent manner. No significant difference was noticed when the result was recorded either at 1.5 hr or 16 hr after mixing the reaction components. Similar IC50 values were obtained with these two time points (28.0 ± 1.1 nM for 1.5 hr and 33.6 ± 1.1 nM for 16 hr). These results suggest that the AlphaScreen assay for HTS of 14-3-3 inhibitors is stable over a time period of at least 16 hr, which allows the reading of more than 120 microplates (384-well) with the Envision plate reader. Finally, we evaluated parameters for HTS performance and assay consistency by testing well-to-well and plate-to-plate variation of the binding assay in a total volume of 20 μl. Each reaction contained 5 nM of biotin-14-3-3 and 15 nM of GST-R18 as in Fig. (3
), the addition of untagged R18 peptide to the reaction containing biotin-14-3-3γ, GST-R18, and the donor/acceptor beads, decreased the Alpha luminescence signals in a dose-dependent manner. No significant difference was noticed when the result was recorded either at 1.5 hr or 16 hr after mixing the reaction components. Similar IC50 values were obtained with these two time points (28.0 ± 1.1 nM for 1.5 hr and 33.6 ± 1.1 nM for 16 hr). These results suggest that the AlphaScreen assay for HTS of 14-3-3 inhibitors is stable over a time period of at least 16 hr, which allows the reading of more than 120 microplates (384-well) with the Envision plate reader. Finally, we evaluated parameters for HTS performance and assay consistency by testing well-to-well and plate-to-plate variation of the binding assay in a total volume of 20 μl. Each reaction contained 5 nM of biotin-14-3-3 and 15 nM of GST-R18 as in Fig. (3 ). The Z’ factor and S/B ratio of reactions from ten individual plates were recorded and compared (Figs.7A
). The Z’ factor and S/B ratio of reactions from ten individual plates were recorded and compared (Figs.7A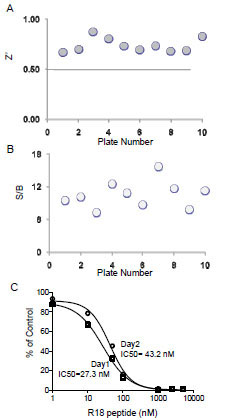 and 7B
and 7B ). Among these 10 plates, the average value for Z’ was 0.73 with a standard deviation of 0.06 and the average S/B ratio was 10.6 with a standard deviation of 2.4, indicating the robustness and suitability of this assay for automation. We further evaluated the assay consistency by performing the R18 peptide competition assay with the biotin-14-3-3γ/GST-R18 interaction pair on different days. As shown in Fig. (7C
). Among these 10 plates, the average value for Z’ was 0.73 with a standard deviation of 0.06 and the average S/B ratio was 10.6 with a standard deviation of 2.4, indicating the robustness and suitability of this assay for automation. We further evaluated the assay consistency by performing the R18 peptide competition assay with the biotin-14-3-3γ/GST-R18 interaction pair on different days. As shown in Fig. (7C ), the dose-response curves obtained from measurements in two different plates at two different days gave rise to similar IC50 values ( 27.3 nM and 43.2 nM, respectively), demonstrating high consistency of the AlphaScreen assays.
), the dose-response curves obtained from measurements in two different plates at two different days gave rise to similar IC50 values ( 27.3 nM and 43.2 nM, respectively), demonstrating high consistency of the AlphaScreen assays.
 |
Fig. (6) Evaluation of the DMSO tolerance and stability of the 14-3-3 AlphaScreen assay. (A) Effect of DMSO on Alpha signal. The biotin-14-3-3γ (5 nM) and GST-R18 (15 nM) were incubated with increasing amounts of DMSO, and reactions were carried out as described in the legend to Fig. (4 |
 |
Fig. (7) Evaluation of the 14-3-3 AlphaScreening assay for HTS. The binding assay was performed essentially as described in Fig. (4 |
CONCLUSIONS
We have developed and optimized a HTS assay in a 384-well format by using the AlphaScreen technology for the detection of 14-3-3 protein interaction with both phosphorylated and nonphosphorylated client peptides. Experimental conditions for both 14-3-3/R18 and 14-3-3/pS259-Raf-1 pairs have been established that gave rise to a robust assay performance with S/B ratio above 7 and Z’ factors above 0.7. Further validation experiments provided evidence that the competitive 14-3-3 binding assays using AlphaScreen can be used for HTS for the screening of 14-3-3/client peptide interaction inhibitors. These assays utilize a small amount of test proteins (low nM) in a low volume (20 μl), which makes them particularly suited for HTS. It is expected that this assay is also applicable for monitoring the interaction of 14-3-3 with its full length client proteins in future HTS campaigns.
ACKNOWLEDGEMENTS
We are grateful to Jeff Pfohl and Jeff Selander from PerkinElmer for valuable suggestions and to Lisa M. Cockrell for critical reading of the manuscript. We also thank members of the Fu lab for assistance. This work was supported in part by National Institutes of Health Grant GM60033, R03MH076385, U54HG003918 and P01CA116676. Y.D. is a recipient of the Emory Drug Development and Pharmacogenomics Academy Research Fellowship and an Emory Head and Neck Cancer SPORE career development award (P50 CA128613). F.R.K. and H.F. are Georgia Cancer Coalition Distinguished Cancer Scholars and H.F. is a Georgia Research Alliance Distinguished Investigator.
REFERENCES
| [1] | Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation Annu Rev Pharmacol Toxicol 2000; 40: 617-47. |
| [2] | Muslin AJ, Xing H. 14-3-3 proteins: regulation of subcellular localization by molecular interference Cell Signal 2000; 12(11-12): 703-9. |
| [3] | Yaffe MB. How do 14-3-3 proteins work?-- Gatekeeper phosphorylation and the molecular anvil hypothesis FEBS Lett 2002; 513(1): 53-7. |
| [4] | Aitken A. 14-3-3 proteins: a historic overview Semin Cancer Biol 2006; 16(3): 162-72. |
| [5] | Mackintosh C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes Biochem J 2004; 381(Pt 2): 329-42. |
| [6] | Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine Cell 1996; 84(6): 889-97. |
| [7] | Yaffe MB, Rittinger K, Volinia S, et al. The structural basis for 14-3-3: phosphopeptide binding specificity Cell 1997; 91(7): 961-71. |
| [8] | Dougherty MK, Morrison DK. Unlocking the code of 14-3-3 J Cell Sci 2004; 117(Pt 10): 1875-84. |
| [9] | Fu H, Coburn J, Collier RJ. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family Proc Natl Acad Sci USA 1993; 90(6): 2320-4. |
| [10] | Wang B, Yang H, Liu YC, et al. Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display Biochemistry 1999; 38(38): 12499-504. |
| [11] | Du Y, Masters SC, Khuri FR, Fu H. Monitoring 14-3-3 protein interactions with a homogeneous fluorescence polarization assay J Biomol Screen 2006; 11(3): 269-76. |
| [12] | Wu M, Coblitz B, Shikano S, et al. SWTY--a general peptide probe for homogeneous solution binding assay of 14-3-3 proteins Anal Biochem 2006; 349(2): 186-96. |
| [13] | Ullman EF, Kirakossian H, Singh S, et al. Luminescent oxygen channeling immunoassay: measurement of particle binding kinetics by chemiluminescence Proc Natl Acad Sci USA 1994; 91(12): 5426-30. |
| [14] | Eglen RM, Reisine T, Roby P, Rouleau N, Bosse RI, Bielefeld M. The use of AlphaScreen technology in HTS: Current status Curr Chem Genom 2008; 1: 2-10. |
| [15] | Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays J Biomol Screen 1999; 4(2): 67-73. |
| [16] | Petosa C, Masters SC, Bankston LA, et al. 14-3-3zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove J Biol Chem 1998; 273(26): 16305-0. |
| [17] | Masters SC, Fu H. 14-3-3 proteins mediate an essential anti-apoptotic signal J Biol Chem 2001; 276(48): 45193-200. |
| [18] | PerkinElmer LaAS. A practical guide to working with AlphaScreen 2004 [cited; Available from: http: //las.perkinelmer.com/ Content/Manuals/GDE_AlphaScreenPracticalGuide.pdf] |
| [19] | Sehr P, Pawlita M, Lewis J. Evaluation of different glutathione S-transferase-tagged protein captures for screening E6/E6AP interaction inhibitors using AlphaScreen J Biomol Screen 2007; 12(4): 560-7. |
| [20] | Mills NL, Shelat AA, Guy RK. Assay optimization and screening of RNA-Protein interactions by AlphaScreen J Biomol Screen 2007; 12(7): 946-55. |
| [21] | Zhang L, Chen J, Fu H. Suppression of apoptosis signal-regulating kinase 1-induced cell death by 14-3-3 proteins Proc Natl Acad Sci USA 1999; 96(15): 8511-5. |
| [22] | Subramanian RR, Zhang H, Wang H, Ichijo H, Miyashita T, Fu H. Interaction of apoptosis signal-regulating kinase 1 with isoforms of 14-3-3 proteins Exp Cell Res 2004; 294(2): 581-91. |
| [23] | Fantl WJ, Muslin AJ, Kikuchi A, et al. Activation of Raf-1 by 14-3-3 proteins Nature 1994; 371(6498): 612-4. |
| [24] | Freed E, McCormick F, Ruggieri R. Proteins of the 14-3-3 family associate with Raf and contribute to its activation Cold Spring Harb Symp Quant Biol 1994; 59: 187-93. |
| [25] | Fu H, Xia K, Pallas DC, et al. Interaction of the protein kinase Raf-1 with 14-3-3 proteins Science 1994; 266(5182): 126-9. |




