- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Clinical Cancer Journal
(Discontinued)
ISSN: 1874-1894 ― Volume 5, 2011
Individualized Survival and Treatment Response Predictions in Breast Cancer Patients: Involvements of Phospho-EGFR and Phospho-Her2/Neu Proteins
Lan Guo*, 1, Jame Abraham2, Daniel C. Flynn3, Vincent Castranova4, Xianglin Shi4 , Yong Qian*, 4
Abstract
Our robust prediction system for individual breast cancer patients combines three well-known machinelearning classifiers to provide stable and accurate clinical outcome prediction (N=269). The average performance of the selected classifiers is used as the evaluation criterion in breast cancer outcome predictions. A profile (incorporating histology, lymph node status, tumor grade, tumor stage, ER, PR, Her2/neu, patient’s age and smoking status) generated over 95% accuracy in individualized disease-free survival and treatment response predictions. Furthermore, our analysis demonstrated that the measurement of phospho-EGFR and phospho-Her2/neu is more powerful in breast cancer survival prediction than that of total EGFR and total Her2/neu (p < 0.05). The incorporation of hormone receptor status, Her2/neu, patient’s age and smoking status into the traditional pathologic markers creates a powerful standard to perform individualized survival and treatment outcome predictions for breast cancer patients
Article Information
Identifiers and Pagination:
Year: 2008Volume: 2
First Page: 18
Last Page: 31
Publisher Id: TOCCJ-2-18
DOI: 10.2174/1874189400802010018
Article History:
Electronic publication date: 27/05/2008Collection year: 2008
open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.5/), which permits unrestrictive use, distribution, and reproduction in any medium, provided the original work is properly cited.
* Address correspondence to these authors at the MBR Cancer Cen- ter/Department of Community Medicine, School of Medicine, West Virginia University, Morgantown, WV 26506-9300, USA; Fax: (304) 293-4667; Tel: (304) 293-6455; E-mail: lguo@hsc.wvu.edu
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on |
Original Manuscript | Individualized Survival and Treatment Response Predictions in Breast Cancer Patients: Involvements of Phospho-EGFR and Phospho-Her2/Neu Proteins | |
INTRODUCTION
Breast cancer is the most common cancer among women in the United States. There was an estimated 212,600 new cases diagnosed as breast cancer in the United States in 2003. Among them, 39,800 women were expected to die of metastatic diseases [1]. For the past decade, the overall risk of mortality due to breast cancer has been declining with the development of advanced therapies as well as the early detection [2]. However, the survival rate has not been substantially improved for patients with recurrent or metastatic breast cancer [1]. One of the main obstacles to improve the survival rate is to accurately classify breast cancer patients into subgroups of good prognosis and poor prognosis, reflecting a different probability of disease recurrence and survival after therapy. Following this, another unsolved problem is to decide which combination of treatment is most suitable for each individual patient. Accurate survival and treatment response prediction for a given breast cancer patient will guide the selection of the optimal therapy.
Current research in breast cancer outcome prediction based on the pathologic information is not optimized. In addition, efforts have been focused on predictions for populations, rather than for individuals. Most predictions produce a relative chance of survival for an individual based on the experience with a historical population of patients, as approximated by a single Kaplan-Meier survival curve. These methods generally lead to weak predictive power for individual patients. The NIH has defined the statistical guidelines for assessing classification results [3]. Specifically, statistics should be based on false (1 - specificity) and true positive (sensitivity) rates, instead of odds ratios or relative risks. In addition, to avoid over fitting, the developed marker combination in a training sample should be evaluated in a random test sample from the same study [3]. In this study, we constructed a classification system combining several well-known machine-learning algorithms. We evaluated the prediction results according to the new NIH statistics guidelines [3]. This system provides stable and accurate prediction results for individual patients. Using the proposed classification system, the overall accuracy of individualized survival and treatment response predictions is over 95% in a cohort of 269 breast cancer patients.
Substantial efforts have been made to establish the predictive factors for patients with breast cancer during last two decades. The traditional predictive factors are lymph node status, tumor size, histologic type, histologic grade, lymphatic vessel invasion, and hormone receptor status [4]. With the development of molecular biology and cell biology, many new predictive factors have been created, including markers of regulating cell cycle and cell death, Her2/neu, markers of metastasis or metastatic process, lymph node micrometastases, bone marrow micrometastases, and markers of angiogenesis [5]. Although the traditional predictive factors lack the information about the biological diversity of breast cancer and have not reflected the complexity of molecular mechanisms of these diseases, they are still the most valuable criteria for clinicians to decide the relevant therapies [6]. For instance, Adjuvant! (www.adjuvantonline.com) is a prognostic system based on traditional pathological features, including age, ER expression, and grade. It has been independently validated as a reliable aid to clinical decision-making on average breast cancer patients [7]. The newer factors have not yet gained full recognition in the clinic due to their contradictory results and clinical confusion [4,5]. Thus, there is a pressing clinical need to develop a comprehensive profile which includes both traditional and molecular predictive factors to assist clinicians to assess individual breast cancer patient outcomes and to select relevant therapies.
Although lymph node status, tumor size, histologic type, histologic grade, and tumor stage are the most powerful traditional pathologic prognostic and predictive factors for breast cancer patients, the methods for measurements of the traditional pathologic factors have been difficult to standardize [4]. The difference in the measurements could lead to the misclassification of some breast cancer patients. Even under the same standards, the inter-observer variability still occurs frequently among different examiners. Therefore, the traditional factors are inclined to be subjective. To optimize the measurement parameter profile for the individualized survival and treatment response predictions, we explored the efficiency of estrogen receptor (ER), progesterone receptor (PR), and Her2/neu and their association with other objective parameters. The measurement of ER, PR, and Her2/neu is mainly through machine-based analysis, which is objective and amenable to standardization. Our results showed that the sole utilization of these objective factors was unable to achieve highly accurate predictions. In order to provide a comprehensive profile for individualized survival and treatment response predictions in patients with breast cancer, the objective factors were incorporated into the traditional pathologic factors. Our results showed that the combined parameters increased the accuracy of the disease-free survival prediction (p <0.000001) and treatment response prediction (p < 0.14) for individual breast cancer patients.
Recently, it has been reported that predictions based on total protein expression levels of both EGFR and Her2/neu may not be sufficient to reflect the functionality of these proteins in vivo [8,9]. Both EGFR and Her2/neu are members of receptor tyrosine kinases. They affect cell and tissue functions through the induction of signaling transduction pathways from the outside of a cell to its nucleus through protein phosphorylation and deposphorylation. The critical step of these processes is the phosphorylation of tyrosine residues, which reflects the kinase activities for EGFR and Her2/neu. The phosphorylated EGFR and Her2/neu create the binding sites for the downstream signaling proteins. Some of these downstream signaling proteins are enzymes, which are activated upon binding to tyrosine-phosporylated EGFR and Her2/neu, and lead to tumorgenesis. Therefore, overexpression of EGFR and Her2/neu does not necessarily indicate that they are functionally activated. This may explain conflicting results concerning the correlation between overexpression of EGFR and breast cancer prognosis and prediction [8]. The difference between the overexpression and the activation of Her2/neu in vivo may also reveal the mechanisms which explain why some Her2/neu overexpressed breast cancer patients have no response to Her2/neu-targeted trastuzumab therapy. One way to measure the kinase activities of EGFR and Her2/neu is to use anti-phospho-antibodies to detect tyrosine-phosphorylated EGFR and Her2/neu. In this study, we sought to explore the predictive value of activated EGFR and Her2/neu in the survival prediction for individual breast cancer patients.
MATERIAL AND METHODS
Data Sources
Data for this analysis were obtained from the Biorepository of Clinomics Biosciences Inc, which is a large collection of highly characterized human tissue samples. These samples span a wide range of common diseases, including many forms and stages of cancer, neurological disorders and heart disease. Clinomics has pioneered the development of an emerging new technology known as Tissue Microarray to enable researchers to simultaneously study hundreds of individual tissue samples in parallel, establishing the relative levels of protein expression in those samples and allowing conclusions as to the relevance of these proteins to disease to be made [10,11].
Immunohistochemistry
Immunohistochemistry methods were described previously [12,13]. Briefly, tissue specimens were treated with 3% H2O2 to quench endogenous peroxidase activity, followed by washing with PBS. After washing, the tissue specimens were first incubated with a specific primary antibody and then with a biotinylated secondary antibody. Substrate-Chromogen was applied to the specimens according to manufacturer’s instructions, followed by stained with hematoxylin. The stain was semiquantitatively examined by pathologists (Clinomics BioSciences, Inc.) using the Allred 8-unit system [14]. Staining was scored on a 0 –5 scale, with 0=no staining. Grades of 1 to 5 represent increased intensity of staining with 5 being strong, dark brown staining. For eachtumor, represented by one slide, the tumor epithelialcells proportion score and intensity score were determined. Peritumoral inflammatoryand stromal cells were not included in the evaluation. The proportionscore included the fraction of positively stained tumor cellsand was as follows: 0 = none, 1 = < 1/100th; 2 = 1/100th to1/10th; 3 = 1/10th to 1/3; 4 = 1/3 to 2/3; 5 = > 2/3. Theestimated average staining intensity of the positive tumor cellswas expressed as follows: 0 = none; 1 = weak; 2 = intermediate ;3 = strong [14]. Each protein was measured with six parameters: Cytoplasmic % Intensity defines percent intensity of stain within cytoplasm; Cytoplasmic % Positive defines percent of all cells positive within cytoplasm; Cytoplasmic Total Score defines the product of Cytoplasmic % Intensity and Cytoplasmic % Positive; Nuclear % Intensity defines percent intensity of stain within nucleus; Nuclear % positive defines percent of all cells positive within nucleus; and Nuclear Total Score defines the product of Nuclear % Intensity and nuclear % Positive.
Anti-EGFR antibody, anti-Her2/neu antibody, anti-phos-pho-EGFR antibody (Tyr845), and anti-phosphoHer2/neu (Tyr877) were from Cell Signaling Technology, Inc. (Beverly, MA).
Study Cohort
Information of a breast cancer cohort was extracted from the Cell Signaling Database of Clinomics Biosciences Inc. The study cohort contained 269 breast tumor samples obtained from surgery. The patients had an average age of 63.7 years (ranging from 21 to 95 years). There were 24.9% with Stage 1, 45.0% with Stage 2, 17.8% with Stage 3, and 12.3% with Stage 4 breast cancer. Ninety-one patients (33.8%) were ER negative and 178 (66.2%) were ER positive. Forty-three (16.0%) patients were Her2/neu positive and 226 (84.0%) were Her2/neu negative (assayed by immunohistochemistry). There were 56.9% of patients received lumpectomy and node dissection, and 43.1% of patients received mastectomy and node dissection. Two hundred and two (75.1%) patients accepted CMF chemotherapy. One hundred and fifty-three (56.9%) patients went through localized (breast) radiation therapy. Among 33 patients who developed metastases, 10 patients were responders to the treatments, while 23 were none responders. Treatment response was defined according to RECIS [15]. Responders include complete response (CR) and partial response (PR), and none responders include stable disease (SD) and progressive disease (PR). Among the patients with Stage 0 to 3, 191 patients survived a 5-year disease-free interval, while 44 had recurrence within five years. The remaining patients’ survival information was censored (details in Table 1).
Clinical Outcomes
There were two outcomes included in the current analyses: disease free survival after therapy and treatment response. The experiments were designed to construct prognostic models to predict disease-free survival and treatment response in individual patients with breast cancers. The patient cohort was separated into two groups: one for patients with Stage 0 to 3 (Group I, N = 235), the other for patients with metastatic diseases (Group II, N = 33). All patients in Group II received CMF chemotherapy. For Group I, biomarkers were identified to build prognostic models for 5-year disease-free survival. For Group II, treatment response prediction models were constructed using a separately identified biomarker set.
Based on the clinical information in the studied cohort, we performed three analyses for both survival prediction and treatment response prediction, respectively (The details are listed in Results section). The clinical-pathological parameters for outcome predictions were chosen using gain Ratio Attribution Evaluation method with software WEKA. The parameters with merit greater than zero in the evaluation were included in the analysis.
Machine Learning Prediction System
Different machine learning algorithms perform differently according to datasets and the given tasks. The evaluation of different pathologic factors based on a single classifier may not lead to an objective conclusion in general. Therefore, we developed a prediction system combining three selected classifiers, a nearest neighbor method (KStar)[16], Random Forests [17], and Neural Networks (MultilayerPerceptron)from the WEKA3.4 software package1 [18]. These algorithms are accurate and stable in clinical outcome predictions on the studied cohort. The prediction accuracy of each classifier is evaluated by 10-fold cross validation, which has the least variance and bias of accuracy estimation among all validation methods including the leave-one-out method [19]. The average prediction accuracy of these classifiers is used as the evaluation criterion for breast cancer outcome predictions. If the prediction accuracy is unbalanced for a certain class label, the F-measure [20] was used as the evaluation criterion instead, which is defined as:
F = 2 × (recall × precision) / (recall + precision)
The average of the F-measure of all class labels was used as the final F-measure for a classifier. The average final F-measure of the three classifiers was used as the evaluation criterion of clinical outcome prediction (see Supplementary Information for details).
Statistical Methods
To assess the significance of the performance of an individual classifier, we computed the probability of the observed prediction accuracy occurring by chance (random prediction using a fair coin flip). The probability of doing at least as well as our prediction models by chance was calculated using Binomial Distribution functions [21] in software package R (http://www.r-project.org/). Statistical significance test was used to assess the significance of different prediction results on the studied cohort.
RESULTS
Identifying Specific High Risk Breast Cancer Patients
In order to identify specific high risk breast cancer patients, we used the clinical information in the studied cohort to predict whether or not a patient would survive a 5-year disease free interval. As we discussed previously, the traditional pathologic factors are inclined to be subjective and ER, PR, Her2/neu, as well as the demographic factors tend to be objective. The clinical information was separated into two categories: one includes the traditional pathologic factors and the other includes ER, PR, Her2/neu, plus the demographic factors. To compare the predictive power of these two different kinds of clinical information in a survival prediction, we conducted following analyses on the studied cohort. (1) Using pathologic factors (histology, stage, pT, pN, and nodes positive) to predict breast cancer survival. (2) Using ER, PR, Her2/neu, age, and smoking to predict breast cancer survival. (3) Using a comprehensive profile (age, histology, stage, pT, pN, nodes positive, ER, PR, Her2/neu, and smoking) to predict breast cancer survival. Our analyses demonstrated that the traditional pathologic factors entailed overall accuracy 94.6% in the survival prediction (Table 2, Analysis 1) and the parameters of ER, PR, Her2/neu, plus patient’s age and smoking status resulted in survival prediction accuracy 82% (Table 2, Analysis 2), indicating that the traditional pathologic parameters are more accurate in the individualized risk assessment (p <0.000007). However, when two kinds of predictive factors were incorporated, the survival prediction accuracy was further increased from 82% to 95.8% (p <0.000001) for individual breast cancer patients (Table 2, Analysis 3). It should also be pointed out that the addition of ER, PR, Her2/neu, plus patient's age and smoking status to the traditional pathologic factors did not significantly (p >0.05) improve the prediction of the traditional factors alone. These results are consistent with the observations in Paik et al. [22]. Our results demonstrated that the identified comprehensive classifier can accurately define a patient’s risk and be used to develop an optimal individualized therapy of breast cancer (see Supplement Information for detailed results).
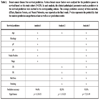
Breast cancer disease free survival prediction. Various breast cancer factors were analyzed for the predictive power in survival based on this study cohort (N=235). In each analysis, the clinical-pathological parameters used as predictors in the survival prediction were marked in the corresponding column. The average prediction accuracy of three methods, (KStar), Random Forests, and Neural Networks, was reported as the final result. P value represents the probability that the random prediction can perform at least as well as our prediction models
Predicting Treatment Response of Each Individual Breast Cancer Patient
In order to assess a breast cancer patient’s treatment response, we used the clinical information in the studied cohort to predict whether or not a patient is a responder to chemotherapy. It enables clinicians to determine the most suitable treatment options for each individual patient. To find out the best predictive parameters for the breast cancer treatment response, we conducted the following analyses on the studied cohort of the 33 patients with metastatic disease: (1) Using traditional pathologic factors (histology, stage, pT, pN, nodes positive, metastasis site, surgery procedure, and chemotherapy) to predict a patient’s response to treatments. (2) Using a patient’s ER, PR, Her2/neu, and demographic information (ER, PR, Her2/neu, age, smoking, surgery procedure, and chemotherapy) to predict a patient’s response to treatments. (3) Using a comprehensive profile (age, histology, stage, pT, pN, nodes positive, metastasis site, ER, PR, Her2/neu, smoking, surgery procedure, and chemotherapy) to predict a patient’s response to treatments. Using the traditional pathologic parameters, the accuracy of treatment response prediction is 94.7% (Table 3, Analysis 1). Using ER, PR, Her2/neu, and the patient’s age and smoking status, the accuracy of treatment response prediction is 87.8% (Table 3, Analysis 2). Again, the traditional pathologic factors entailed higher accuracy in the treatment response prediction (p <0.16). However, when the parameters in Analysis 1 and Analysis 2 were combined, the treatment response prediction accuracy was increased from 87.8% to 95.3% (p <0.14) (Table 3, Analysis 3). Here, the addition of the biomarker panel to the traditional histological features did not significantly improve the prediction accuracy (p >0.05). Our data demonstrate that the comprehensive profile encompassing both the traditional pathologic factors and ER, PR, Heu2/neu plus patient’s age and smoking status was more accurate in predicting the treatment response of breast cancer patients. Our constructed classifier can accurately predict a patient’s response to treatments, which enables the selection of the most suitable combination of therapeutic regimens for each individual breast cancer patient. Our results confirmed previous findings. Kendal previously reported that positive lymph node status is an independent risk factor for breast cancer prognosis [23]. Terry et al. identified the association between prolonged cigarette smoking and increased breast cancer risk [24]
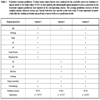
Treatment response prediction. Various breast cancer factors were analyzed for the predictive power in treatment response based on this study cohort (N=33). In each analysis, the clinical-pathological parameters used as predictors in the treatment response prediction were marked in the corresponding column. The average prediction accuracy of three methods, (KStar), Random Forests, and Neural Networks, was reported as the final result. P value represents the probability that the random prediction can perform at least as well as our prediction models
Evaluating the Predictive Power of Total Her2/neu, Total EGFR, Phospho-Her2/neu, and Phospho-EGFR in Beast Cancer Survival
To more accurately reflect the functionalities of these two proteins in vivo. We performed the following analyses on the studied cohort to investigate the relative strength of the predictive power of total Her2/neu, total EGFR, phospho-EGFR and phospho-Her2/neu in breast cancer survival prediction. The six antibody measurements for each phosphorylated protein were used in disease-free survival prediction. As a control, we first used a patient’s age, ER and PR status to predict the patient survival after therapy (Table 4, Analysis 1). In this analysis, all patients were classified as low risk (5-year survival). It indicates that the predictive power of a patient’s age, ER, and PR is weak, because they cannot identify any high risk patients. However, since majority of the patients (82.1%) survived 5 years in the studied cohort, the overall prediction accuracy is 82.1%. In this situation, we cannot use overall prediction accuracy as the evaluation criterion. As a resolution, we used the average F-measure [20] of the 5-year survival and non-5-year survival prediction results as the evaluation criterion. The F-measure is a combined measurement of recall and precision. It thus overcomes the problems caused by unbalanced prediction error rates. We then evaluated the survival prediction model by adding the expression level of total Her2/neu and total EGFR (Table 4, Analysis 2), as well as phospho-Her2/neu and phospho-EGFR (Table 4, Analysis 3), respectively, to the prediction model. Our results showed that the survival prediction accuracy was increased with the addition of total Her2/neu and total EGFR measurements (p <0.15). Furthermore, our analyses showed that the incorporation of phospho-Her2/neu and phospho-EGFR entailed the highest survival prediction accuracy (p <0.05), while the incorporation of total Her2/neu, total EGFR, phospho-Her2/neu and phospho-EGFR resulted in the second highest survival prediction accuracy compared to the control (p <0.09) (Table 4, Analysis 4).

The predictive power of EGFR, Her2/neu, phospho-EGFR, phospho-Her2/neu in breast cancer survival assessment on this study cohort (N=235). Six parameters for antibody measurements were included in the analysis of phospho-EGFR, phospho-Her2/neu, and EGFR. P value represents the statistical significance of the improvement over the control analysis
Association of Phospho-Heu2/neu and Phospho-EGFR in Survival Prediction of Individual Breast Cancer Patients
Both Her2/neu and EGFR belong to the EGFR family. There is growing evidence showing that the interaction among EGFR family members contributes to oncogenic transformation in human tumors. Studies found that human tumors are prone to co-express multi-EGFR family members, which are correlated to the more aggressive phenotypes, enhanced transforming activities, and a poor clinical outcome [25]. Therefore, we compared the predictive power of the combination of phospho-Her2/neu and phospho-EGFR vs. either phospho-Her2/neu or phospho-EGFR. The six antibody measurements for each phosphorylated protein were used in disease-free survival prediction. The protein expression patterns of phospho-EGFR and phospho-Her2/neu in clinical samples are shown in Fig. (1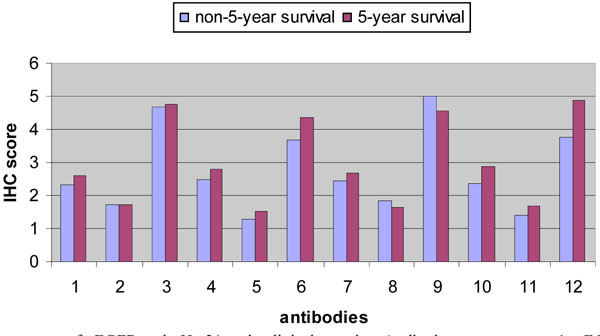 ). Among the measured antibody scores, phospho-EGFR Cytoplasmic % Positive, phospho-EGFR Nuclear % Positive, phospho-EGFR Nuclear % Intensity, phospho-Her2/neu Cytoplasmic % Positive, phospho-Her2/neu Cytoplasmic % Intensity, phospho-Her2/neu Nuclear % Positive, and phospho-Her2/neu Nuclear % Intensity were differentially expressed among non-5-year survival and 5-year survival (p <0.05). Furthermore, we found that age-ER-PR-phospho-Her2/neu-phospho-EGFR was more accurate in predicting the survival of breast cancer patients than either age-ER-PR-phospho-Her2/neu or age-ER-PR-phospho-EGFR (Table 5). Our results imply that some interaction may exist between phospholated Her2/neu and phospholated EGFR in breast cancer. These observations are consistent with the clinical results in Her2/neu-targented trastuzumab treatment for breast cancer patients. It was found that some breast cancer patients with Her2/neu overexpression had no response to the treatment. In vitro experiments showed that the response to trastuzumab treatment was dependent not only on Her2/neu expression, but on the expression of other EGFR family member as well [26]. Our results are consistent with the recent publication showing that there is a simultaneous phosphorylation of Her2/neu and EGFR in Her2/neu overexpressed metastatic breast cancer [27]. Taken together, our results demonstrated that the combination of phospho-Her2/neu and phospho-EGFR is more accurate in survival prediction of individual breast cancer patients than either phospho-Her2/neu (p <0.10) or phospho-EGFR (p <0.21).
). Among the measured antibody scores, phospho-EGFR Cytoplasmic % Positive, phospho-EGFR Nuclear % Positive, phospho-EGFR Nuclear % Intensity, phospho-Her2/neu Cytoplasmic % Positive, phospho-Her2/neu Cytoplasmic % Intensity, phospho-Her2/neu Nuclear % Positive, and phospho-Her2/neu Nuclear % Intensity were differentially expressed among non-5-year survival and 5-year survival (p <0.05). Furthermore, we found that age-ER-PR-phospho-Her2/neu-phospho-EGFR was more accurate in predicting the survival of breast cancer patients than either age-ER-PR-phospho-Her2/neu or age-ER-PR-phospho-EGFR (Table 5). Our results imply that some interaction may exist between phospholated Her2/neu and phospholated EGFR in breast cancer. These observations are consistent with the clinical results in Her2/neu-targented trastuzumab treatment for breast cancer patients. It was found that some breast cancer patients with Her2/neu overexpression had no response to the treatment. In vitro experiments showed that the response to trastuzumab treatment was dependent not only on Her2/neu expression, but on the expression of other EGFR family member as well [26]. Our results are consistent with the recent publication showing that there is a simultaneous phosphorylation of Her2/neu and EGFR in Her2/neu overexpressed metastatic breast cancer [27]. Taken together, our results demonstrated that the combination of phospho-Her2/neu and phospho-EGFR is more accurate in survival prediction of individual breast cancer patients than either phospho-Her2/neu (p <0.10) or phospho-EGFR (p <0.21).
DISCUSSION
Breast cancer is a complex and heterogeneous disease, encompassing a wide range of pathologic entities and molecular profiles [28]. It is crucial for the physicians to accurately define a patient’s risk of developing metastatic and recurrent diseases at diagnosis, which will determine the clinical course for the given patient, i.e. which patient should receive expensive and toxic adjuvant therapy, and which patient should avoid over-treatment. Furthermore, it is critical for the physicians to determine which combination of treatment is most suitable for each individual patient. However, it still remains challenging to make accurate predictive assessment of a patient’s risk or response to certain treatment regimens. This paper presents an accurate and robust model system for individualized survival and treatment response predictions for patients with breast cancer. Our results demonstrated that our model system can achieve overall accuracy above 95% in individualized breast cancer outcome predictions.
Previous studies of constructing predictive assessment models of breast cancer outcome were based on different criteria of patient selection and different data analysis methods. As different data analysis methods may generate different results leading to discrepant conclusions, it is difficult to perform generic comparisons of various predictive factors and ensure the repeatable accuracy of constructed outcome prediction models, making it difficult for the comprehensive evaluation and comparison of various predictive factors [28]. To develop a standardized and comprehensive profile for accurate clinical outcome predictions, we evaluated various factors in prediction assessments on the same patient cohort using a model system which combines three well known machine learning algorithms, Kstar, random forests, and neural networks. These three methods are all based on sound computational algorithms and have been widely deployed in various applications. They all provided accurate and stable results in this study. In order to provide an objective evaluation of various predictive factors in general, we used the average performance of these three methods as the evaluation criterion of the predictive assessments on the same patient cohort. It thus provides consistent, generic, and applicable evaluation with regard to the selection of patients and methods of data analysis. In addition, it ensures the accuracy and stability of the performance of the proposed prediction system despite the variability in data.
In this study, we also sought to apply new molecular knowledge and techniques in the evaluation of breast cancer predictive factors. Both EGFR and Her2/neu belong to the epidermal growth factor receptor (EGFR) superfamily. The phosphorylation in tyrosines is an essential step for the activation of EGFR and Her2/neu proteins, and is a strict requirement for these proteins to transform cells. Overexpression of EGFR and Her2/neu does not necessary indicate they are functionally activated. The binding of downstream proteins, such as Src, PI3K, Ras, Shc, and PLC-γ, to the tyrosine-phosphorylated EGFR and Her2/neu, induces a cascade of signal transduction events, ultimately leading to cell transformation [29]. Therefore, the expression levels of phosphorylation of EGFR and Her2/neu may more accurately reflect the status of these enzymes in the tumorgenesis of individual breast cancer patients. Indeed, studies of the correlation of EGFR overexpression with breast cancer prognosis and prediction have yielded conflicting results [8]. Some reports showed that the overexpression of EGFR was correlative to the adverse prognosis of breast cancer while other reports showed EGFR overexpression was not a good prognosis factor [8]. Study of the profiles of Her2/neu overexpression in a large group of patients found that patients with overexpression of unphosphoryled Her2/neu have a similar prognosis as those patients without the overexpression of Her2/neu [30]. These results suggest that the overexpression of phosphorylated EGFR and Her2/neu may be more relevant in predicting the clinical outcomes of individual breast cancer patients than the overexpression of total EGFR and Her2/neu. Our results showed that age-ER-PR-phospho-Her2/neu-phospho-EGFR is more accurate in predicting the survival of individual breast cancer patients than age-ER-PR-Her2/neu-EGFR. Our results are consistent with the recently published observation, showing that both phosphorylated Her2/neu and phosphorylated EGFR are more efficient in predicting the clinical outcome of patients undergoing trastuzumab treatment [31].
In conclusion, we found that the incorporation of ER, PR, and Her2/neu, as well as other objective parameters, into the traditional pathologic factors is highly accurate to perform individualized survival and treatment response predictions for breast cancer patients. We also demonstrated that the measurements of active EGFR and Her2/neu via anti-phospho-antibodies are more predictive in the survival prediction than the measurements of overexpression of total EGFR and Her2/neu proteins. Nevertheless, it should be noted that the protein expression values were measured using immunohistochemistry assays, which are based on assessments of pathologists and are semi-quantitative. With the advances in protein array technology, the methodology presented in this study could be used to analyze the data generated using new technologies. Furthermore, it should be noted that the results presented in this paper need to be validated using independent datasets.
DISCLAIMER
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the National Institute for Occupational Safety and Health