- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Immunology Journal
(Discontinued)
ISSN: 1874-2262 ― Volume 10, 2020
Inter-Pathogen Peptide Sharing and the Original Antigenic Sin: Solving a Paradox
Darja Kanduc1, *, Yehuda Shoenfeld2
Abstract
Aims:
To analyse the peptide commonality among viral, bacterial, and protozoan pathogens, and the immunopathologic consequences in the human host.
Methods:
HPV16, HCMV, C. diphtheriae, B. pertussis, C. tetani, T. gondii, and T. cruzi were analysed for common amino acid sequences that are additionally shared with the human host. The pentapeptide, a minimal immune determinant in humoral and cellular immune recognition, was used as a measurement unit of the peptide similarity level. Molecular modeling was applied to compare the amino acid contexts containing common minimal determinants.
Results:
Twenty-nine pentapeptides were found to occur, even hundreds of times, throughout the analyzed pathogen proteomes as well as in the human proteome. Such vast peptide commonalities together with molecular modeling data support the possibility that a pre-existing immune response to a first pathogen can be boosted by a successive exposure to a second different pathogen, i.e., the primary response to a pathogen can be transformed into a secondary response to a previously encountered different pathogen. Two possible consequences emerge. Firstly, no responses might be elicited against the pathogen lastly encountered either by infection or active immunization, but reactions could occur only with the early sensitizing pathogen, which is no more present in the organism. Secondly, the immune response boosted by the pathogen lastly encountered will find a way out by cross-reacting with human proteins.
Conclusion:
This study might explain the “original antigenic sin” phenomenon described seven decades ago [Francis T. Jr. Ann Intern Med 1953;39:203], thus providing explanations for vaccine failures and offering possible clues for designing successful vaccines.
Article Information
Identifiers and Pagination:
Year: 2018Volume: 08
First Page: 16
Last Page: 27
Publisher Id: TOIJ-8-16
DOI: 10.2174/1874226201808010016
Article History:
Received Date: 20/6/2018Revision Received Date: 1/08/2018
Acceptance Date: 4/8/2018
Electronic publication date: 31/08/2018
Collection year: 2018
open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution 4.0 International Public License (CC-BY 4.0), a copy of which is available at: (https://creativecommons.org/licenses/by/4.0/legalcode). This license permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
* Address correspondence to this authors at the Department of Biosciences, Biotechnologies and Biopharmaceutics, University of Bari, Bari 70126, Italy, Tel: +390805231522; E-mail: dkanduc@gmail.com
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 20-6-2018 |
Original Manuscript | Inter-Pathogen Peptide Sharing and the Original Antigenic Sin: Solving a Paradox | |
1. INTRODUCTION
Infections are risk factors for a large spectrum of autoimmune disorders [1George J, Levy Y, Kallenberg CGM, Shoenfeld Y. Infections and Wegener’s granulomatosis-a cause and effect relationship? QJM 1997; 90(5): 367-73.
[http://dx.doi.org/10.1093/qjmed/90.5.367] [PMID: 9205673] -15Grubor NM, Jovanova-Nesic KD, Shoenfeld Y. Liver cystic echinococcosis and human host immune and autoimmune follow-up: A review. World J Hepatol 2017; 9(30): 1176-89.
[http://dx.doi.org/10.4254/wjh.v9.i30.1176] [PMID: 29109850] ]. Likewise, active immunization may also cause collateral autoimmune events [16Segal Y, Shoenfeld Y. Vaccine-induced autoimmunity: The role of molecular mimicry and immune crossreaction. Cell Mol Immunol 2018; 14: 1-9.
[PMID: 29503439] ] and, in addition, result in the production of a waning/weak immunity [17Burdin N, Handy LK, Plotkin SA. What is wrong with pertussis vaccine immunity? The problem of waning effectiveness of pertussis vaccines. Cold Spring Harb Perspect Biol 2017; 9(12): a029454.
[http://dx.doi.org/10.1101/cshperspect.a029454] [PMID: 28289064] -24Xu H, Zhao G, Huang X, et al. CD40-expressing plasmid induces anti-CD40 antibody and enhances immune responses to DNA vaccination. J Gene Med 2010; 12(1): 97-106.
[http://dx.doi.org/10.1002/jgm.1412] [PMID: 19950201] ]. The issue is of crucial importance, especially when considering the necessity of reinforcing the immune defence of the human population from the intensifying assault of old and new microbial threats [25Congdon P. Spatiotemporal frameworks for infectious disease diffusion and epidemiology. Int J Environ Res Public Health 2016; 13(12): E1261.
[http://dx.doi.org/10.3390/ijerph13121261] [PMID: 27999420] -27Lucchese G, Kanduc D. Zika virus and autoimmunity: From microcephaly to Guillain-Barré syndrome, and beyond. Autoimmun Rev 2016; 15(8): 801-8.
[http://dx.doi.org/10.1016/j.autrev.2016.03.020] [PMID: 27019049] ].
In this context, since 2000 [28Natale C, Giannini T, Lucchese A, Kanduc D. Computer-assisted analysis of molecular mimicry between human papillomavirus 16 E7 oncoprotein and human protein sequences. Immunol Cell Biol 2000; 78(6): 580-5.
[http://dx.doi.org/10.1046/j.1440-1711.2000.00949.x] [PMID: 11114967] ], this lab repeatedly documented a high level of peptide sharing between pathogen and human proteins [29Lucchese G, Stufano A, Kanduc D. Proposing low-similarity peptide vaccines against Mycobacterium tuberculosis. J Biomed Biotechnol 2010; 2010: 832341.
[http://dx.doi.org/10.1155/2010/832341] [PMID: 20625421] -37Lucchese G. Understanding neuropsychiatric diseases, analyzing the peptide sharing between infectious agents and the language-associated NMDA 2A protein. Front Psychiatry 2016; 7: 60.
[http://dx.doi.org/10.3389/fpsyt.2016.00060] [PMID: 27148089] ], in this way highlighting the risk for cross-reactivity in the human host following infections or active immunization [38Kanduc D. “Self-nonself” peptides in the design of vaccines. Curr Pharm Des 2009; 15(28): 3283-9.
[http://dx.doi.org/10.2174/138161209789105135] [PMID: 19860677] -41Lucchese G, Kanduc D. Minimal immune determinants connect Zika virus, Human Cytomegalovirus, and Toxoplasma gondii to microcephaly-related human proteins. Am J Reprod Immunol 2017; 77(2): e12608.
[http://dx.doi.org/10.1111/aji.12608] [PMID: 27878897] ].
The present study further analyzes the amino acid (aa) sequences common to potential pathogens evolutionarily different such as viruses, bacteria and protozoans. The specific question faced here is: given the massive peptide overlap that characterizes the protein world [35Kanduc D, Stufano A, Lucchese G, Kusalik A. Massive peptide sharing between viral and human proteomes. Peptides 2008; 29(10): 1755-66.
[http://dx.doi.org/10.1016/j.peptides.2008.05.022] [PMID: 18582510] ], there are peptide commonalities among viruses, bacteria and protozoans that might confound, intensify or weaken the human immune responses that follow infection/active immunization? Searching for answers, we used HPV16 infection/immunization as a research model and the pentapeptide as an operational unit, and define the potential immunologic impact that previous pathogen infections/immunizations might have on the human anti-HPV16 immune responses.
2. METHODS
The following pathogens were analyzed, with Taxonomy ID in parentheses: HPV16, 9 proteins, 2600 aa (333760); HCMV, 168 proteins, 63 460 aa (295027); C. diphtheriae, 2265 proteins, 724 668 aa (257309); C. tetani, 2415 proteins, 809 352 aa (212717); B. pertussis, 3783 proteins, 812 989 aa (520); T. gondii, 8404 proteins, 6 625 207 aa (432359); T. cruzi, 19242 proteins, 9 424 566 aa (353153). H. sapiens proteome consisted of 70941 proteins at the time of this study. Proteomes are described at http://www.uniprot.org/ [42UniProt: the universal protein knowledgebase. Nucleic Acids Res 2017; 45(D1): D158-69.
[http://dx.doi.org/10.1093/nar/gkw1099] [PMID: 27899622] ]. Peptide matching was carried out using PIR Peptide Match program (http://research.bioinformatics.udel.edu/peptidematch/) [43Chen C, Li Z, Huang H, Suzek BE, Wu CH. A fast peptide match service for UniProt knowledgebase. Bioinformatics 2013; 29(21): 2808-9.
[http://dx.doi.org/10.1093/bioinformatics/btt484] [PMID: 23958731] ].
The immunologic potential of the peptide sharing among infectious pathogens and the human host was investigated using the Immune Epitope Database (IEDB; www.iedb.org) resource [44Vita R, Overton JA, Greenbaum JA, et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res 2015; 43(Database issue): D405-12.
[http://dx.doi.org/10.1093/nar/gku938] [PMID: 25300482] ]. Only epitopes experimentally validated as immunopositive in the human host were considered.
PEP-FOLD3 program (http://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD3/) [45Shen Y, Maupetit J, Derreumaux P, Tufféry P. Improved PEP-FOLD approach for peptide and miniprotein structure prediction J Chem Theor Comput 2014; 10: 4745-58.-47Lamiable A, Thévenet P, Rey J, Vavrusa M, Derreumaux P, Tufféry P. PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res 2016; 44(W1): W449-54.
[http://dx.doi.org/10.1093/nar/gkw329] [PMID: 27131374] ] was used to obtain 3D peptide structures from linear aa sequences.
3. RESULTS AND DISCUSSION
HPV16 was chosen as a research model since papillomavirus infection or active immunization mostly occur in sexually active subjects [48Orlando G, Fasolo M, Mazza F, et al. Risk of cervical HPV infection and prevalence of vaccine-type and other high-risk HPV types among sexually active teens and young women (13-26 years) enrolled in the VALHIDATE study. Hum Vaccin Immunother 2014; 10(4): 986-94.
[http://dx.doi.org/10.4161/hv.27682] [PMID: 24423757] ] or in girls and boys aged 11-12 years [49Committee Opinion No. 704: Human papillomavirus vaccination. Obstet Gynecol 2017; 129: 1155-6.
[http://dx.doi.org/10.1097/AOG.0000000000002111] [PMID: 28538494] ], i.e., in individuals that have an immunologic past of encounters with numerous infectious agents. The pathogens HCMV, C. diphtheriae, C. tetani, B. pertussis, T. gondii, and T. cruzi were chosen to analyze the molecular basis of the impact of previous infections/immunizations since scientific-clinical literature and immunization protocols pose the first encounter with such pathogens early in the human life [50https://www.cdc.gov/vaccines/parents/downloads/parent-ver-sch-0-6yrs.pdf-53Llewellyn MS, Messenger LA, Luquetti AO, et al. Deep sequencing of the Trypanosoma cruzi GP63 surface proteases reveals diversity and diversifying selection among chronic and congenital Chagas disease patients. PLoS Negl Trop Dis 2015; 9(4): e0003458.
[http://dx.doi.org/10.1371/journal.pntd.0003458] [PMID: 25849488] ]. Analyses were conducted as already described in detail [54Lucchese G, Capone G, Kanduc D. Peptide sharing between influenza A H1N1 hemagglutinin and human axon guidance proteins. Schizophr Bull 2014; 40(2): 362-75.
[http://dx.doi.org/10.1093/schbul/sbs197] [PMID: 23378012] , 55Lucchese G. Confronting JC virus and Homo sapiens biological signatures. Front Biosci 2013; 18: 716-24.
[http://dx.doi.org/10.2741/4133] [PMID: 23276955] ] utilizing the pentapeptide as a measurement unit because a grouping of five residues is endowed with immunogenicity and antigenicity, and can act as a minimal immune determinant in humoral and cellular immune recognition [54Lucchese G, Capone G, Kanduc D. Peptide sharing between influenza A H1N1 hemagglutinin and human axon guidance proteins. Schizophr Bull 2014; 40(2): 362-75.
[http://dx.doi.org/10.1093/schbul/sbs197] [PMID: 23378012] -65Li Z, Wang D, Gu Y, et al. Crystal structures of two immune complexes identify determinants for viral infectivity and type-specific neutralization of human papillomavirus. MBio 2017; 8(5): e00787-17.
[http://dx.doi.org/10.1128/mBio.00787-17] [PMID: 28951471] ].
3.1. Occurrences of HPV16 Pentapeptides in HCMV, C. diphtheriae, C. tetani, B. pertussis, T. gondii, T. cruzi, and H. sapiens Proteomes
Primary aa sequences corresponding to the 9 HPV16 proteins were dissected into pentapeptides overlapping by four residues each other (eg, MQVTF, QVTFI, VTFIY, TFIYI, and so forth). Each HPV16 pentapeptide was probed for occurrences within HCMV, C. diphtheriae, C. tetani, B. pertussis, T. gondii, T. cruzi, and H. sapiens proteomes. Results are shown in Table 1.
Remarkably, Table 1 shows that HPV16 shares pentapeptides with all of the 7 analyzed proteomes. Even more relevant, HPV16 is more similar to the human host than to HCMV, by being 94% the pentapeptide identity between HPV16 and the human host in front of the 5% pentapeptide identity with HCMV. In other words, only 152 pentapeptides differentiate HPV16 from the human host, whereas 2278 pentapeptides phenetically divide HPV16 from HCMV.
Moreover, Table 1, column 2, indicates that the shared HPV pentapeptides repeatedly occur in the analyzed proteomes. The distribution and multiple occurrences of HPV16 pentapeptides in the analysed pathogens and in the human host are detailed in Table S1.
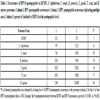
Occurrences of HPV16 pentapeptides in HCMV, C. diphtheriae, C. tetani, B. pertussis, T. gondii, T. cruzi, and H. sapiens proteomes. Column 1: HPV pentapeptide occurrences; Column 2: HPV pentapeptide occurrences (including multiple ones); Column 3: percent of similarity of HPV16 at the pentapeptide level.
3.2. Twenty-nine Pentapeptides are Common to HPV16, HCMV, C. diphtheriae, C. tetani, B. pertussis, T. gondii. T. cruzi, and H. sapiens
The 123 pentapeptides common to HPV16 and HCMV (Tables 1 and S1) were tested for occurrences in C. tetani, C. diphtheriae, B. pertussis, T. gondii. T. cruzi, and the human host. The comparative analysis revealed a common core consisting of 29 pentapeptides, all of which occur at different extent in the 7 analyzed proteomes (Table 2).
As regards the human proteome, the 29 pentapeptides common to the analyzed pathogens are dislocated in 1310 human proteins that are described in Table S2. Ictu oculi, space does not permit a detailed analysis of the human proteins involved in the microbial peptide sharing. An extreme example is the 5-mer GGSGG, which occurs 215 times throughout the human proteome (see Tables 2 and S2).
Immunologically, such an impressive peptide overlap clearly indicates the possibility that the seven analyzed infectious agents might trigger a cross-reactivity network capable of causing multiple and apparently unrelated autoimmune pathologies in the human host, depending on the human proteins involved in the cross-reactions. Biochemically, a noteworthy annotation is that the unexpected, massive, and apparently unexplainable microbial vs human peptide matching may have its evolutionary roots in the critical role played by bacteria and viruses in the origin of the eukaryotic mitochondrion and nucleus, respectively, as detailed in the Endosymbiotic Theory [66Lazcano A, Peretó J. On the origin of mitosing cells: A historical appraisal of Lynn Margulis endosymbiotic theory. J Theor Biol 2017; 434: 80-7.
[http://dx.doi.org/10.1016/j.jtbi.2017.06.036] [PMID: 28684295] ] and the Viral Eukaryogenesis Hypothesis [67Bell PJ. Viral eukaryogenesis: Was the ancestor of the nucleus a complex DNA virus? J Mol Evol 2001; 53(3): 251-6.
[http://dx.doi.org/10.1007/s002390010215] [PMID: 11523012] , 68Forterre P. The origin of viruses and their possible roles in major evolutionary transitions. Virus Res 2006; 117(1): 5-16.
[http://dx.doi.org/10.1016/j.virusres.2006.01.010] [PMID: 16476498] ].
3.3. Immunologic Potential of the Pentapeptide Commonality
In like manner, analysis of the publicly available epitope database IEDB [44Vita R, Overton JA, Greenbaum JA, et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res 2015; 43(Database issue): D405-12.
[http://dx.doi.org/10.1093/nar/gku938] [PMID: 25300482] ] shows an unexpected immunologic potential of the peptide sharing, with 26 out of the 29 pentapeptides being widely, repeatedly, and massively represented in experimentally validated epitopes cataloged as immunopositive in humans (Table S3). Again we observe that a detailed discussion of the possible cross-reactivity scenario is prevented by the high number of epitopes. De facto, even using as an example the least redundant pentapeptide DLLIR (see Table 2), it can be seen that numerous epitope sequences contain DLLIR (Table 3).
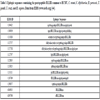
Epitopic sequences containing the pentapeptide DLLIR common to HCMV, C. tetani, C. diphtheriae, B. pertussis, T. gondii, T. cruzi, and H. sapiens. Data from IEDB (www.iedb.org) [44Vita R, Overton JA, Greenbaum JA, et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res 2015; 43(Database issue): D405-12.
[http://dx.doi.org/10.1093/nar/gku938] [PMID: 25300482] ].
3.4. Inter-Pathogen Peptide Sharing and the Human Immune Response Following HPV16 Infection/Immunization: The Questions
In synthesis, Tables 2, S1, S2, and S3 show that most of the 29 pentapeptides occurring among the analyzed pathogens also massively recur in human epitopes and, hence, might be involved in a complex and dense autoimmune cross-reactivity network.
In the context of the HPV16 research model analyzed here, the data reported above suggest that an already primed epitopic peptide network can exist in an individual who undergoes HPV16 infection (or active immunization) during adolescence or in the adult age. Hence, the data raise two main intertwined questions:
− firstly: might immune reactivity against the HPV16 minimal determinants be modified/warped/confounded by previous immune responses against the same determinants present in other pathogen proteins?
− in the second place: following immune reactivity against an HPV determinant, which one(s) among the human proteins containing that same determinant(s) might be hit by immune cross-reactivity ? One, a few or all of them? And why that or those ones?
3.5. The Biochemical Constraint of the Context-Dependent Conformation and the Immunological/Clinical Consequences
Actually, identical pentapeptides in different proteins can have completely different conformations. The same five residues may be part of an α-helix in one protein and part of a β-strand in another protein [69Kabsch W, Sander C. On the use of sequence homologies to predict protein structure: Identical pentapeptides can have completely different conformations. Proc Natl Acad Sci USA 1984; 81(4): 1075-8.
[http://dx.doi.org/10.1073/pnas.81.4.1075] [PMID: 6422466] ]. The same pentapeptide may be embedded in the protein foldings or exposed on the protein surface, and, in general, the pentapeptide conformation in the intact protein is influenced by the 3D aa context that surrounds the pentamer [69Kabsch W, Sander C. On the use of sequence homologies to predict protein structure: Identical pentapeptides can have completely different conformations. Proc Natl Acad Sci USA 1984; 81(4): 1075-8.
[http://dx.doi.org/10.1073/pnas.81.4.1075] [PMID: 6422466] , 70Kabsch W, Sander C. Identical pentapeptides with different backbones. Nature 1985; 317(6034): 207.
[http://dx.doi.org/10.1038/317207a0] [PMID: 4047161] ]. Immunologically, antibodies made against one pentapeptide do not recognize all the possible pentapeptide conformations but react with only one or a few of the pentapeptide conformers depending on the flanking amino acids [71Niman HL, Houghten RA, Walker LE, et al. Generation of protein-reactive antibodies by short peptides is an event of high frequency: Implications for the structural basis of immune recognition. Proc Natl Acad Sci USA 1983; 80(16): 4949-53.
[http://dx.doi.org/10.1073/pnas.80.16.4949] [PMID: 6192445] -73Schulze-Gahmen U, Wilson IA. Monoclonal antibodies against an identical short peptide sequence shared by two unrelated proteins. Pept Res 1989; 2(5): 322-31.
[PMID: 2485209] ].
Keeping on in analysing for simplicity only the HPV pentapeptide DLLIR, we compared the aa context of the pentapeptide DLLIR in pathogen and human proteins. Specifically, we searched for the flanking sequences of the DLLIR pentapeptide, ie, the five NH2- and COOH framing residues, in the pathogen and human proteins (Table 4).
Afterwards, 3D molecular modeling [45Shen Y, Maupetit J, Derreumaux P, Tufféry P. Improved PEP-FOLD approach for peptide and miniprotein structure prediction J Chem Theor Comput 2014; 10: 4745-58.-47Lamiable A, Thévenet P, Rey J, Vavrusa M, Derreumaux P, Tufféry P. PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res 2016; 44(W1): W449-54.
[http://dx.doi.org/10.1093/nar/gkw329] [PMID: 27131374] ] was applied to the resulting 15-mer peptides containing the DLLIR minimal immune determinant (Table 4) in order to single out similar decapentapeptide structures Fig. (1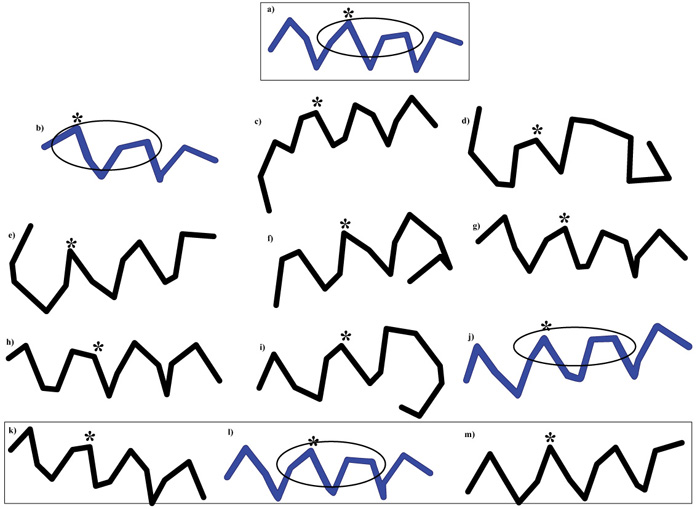 ).
).
Preliminarily, it has to be underscored that the structural modeling data reported in Fig. (1 ) aim at illustrating in a simplified manner the role of peptide conformation in immunological recognition and do not reflect the numerical and structural complexity of the peptide conformers [45Shen Y, Maupetit J, Derreumaux P, Tufféry P. Improved PEP-FOLD approach for peptide and miniprotein structure prediction J Chem Theor Comput 2014; 10: 4745-58.-47Lamiable A, Thévenet P, Rey J, Vavrusa M, Derreumaux P, Tufféry P. PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res 2016; 44(W1): W449-54.
) aim at illustrating in a simplified manner the role of peptide conformation in immunological recognition and do not reflect the numerical and structural complexity of the peptide conformers [45Shen Y, Maupetit J, Derreumaux P, Tufféry P. Improved PEP-FOLD approach for peptide and miniprotein structure prediction J Chem Theor Comput 2014; 10: 4745-58.-47Lamiable A, Thévenet P, Rey J, Vavrusa M, Derreumaux P, Tufféry P. PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res 2016; 44(W1): W449-54.
[http://dx.doi.org/10.1093/nar/gkw329] [PMID: 27131374] , 69Kabsch W, Sander C. On the use of sequence homologies to predict protein structure: Identical pentapeptides can have completely different conformations. Proc Natl Acad Sci USA 1984; 81(4): 1075-8.
[http://dx.doi.org/10.1073/pnas.81.4.1075] [PMID: 6422466] -73Schulze-Gahmen U, Wilson IA. Monoclonal antibodies against an identical short peptide sequence shared by two unrelated proteins. Pept Res 1989; 2(5): 322-31.
[PMID: 2485209] ]. Given this necessary caveat, we observe that Fig. (1 ) shows that the structural conformation of the common DLLIR peptide varies in decapentapeptides listed in Table 4, with a few conformers being more similar than others to the HPV16 decapentapeptide nkplcDLLIRcincq (see in Fig. 1
) shows that the structural conformation of the common DLLIR peptide varies in decapentapeptides listed in Table 4, with a few conformers being more similar than others to the HPV16 decapentapeptide nkplcDLLIRcincq (see in Fig. 1 , decapeptide structures in blue: b) HCMV mDLLIRlgfll; j) T. cruzi isafyDLLIRqltvs; and l) H. sapiens faylrDLLIRthmqn, with the epitopic DLLIR site encircled in the oval).
, decapeptide structures in blue: b) HCMV mDLLIRlgfll; j) T. cruzi isafyDLLIRqltvs; and l) H. sapiens faylrDLLIRthmqn, with the epitopic DLLIR site encircled in the oval).
Immunologically, Fig. (1 ) implies that, following HPV16 infection or active immunization, an anti-HPV16 DLLIR response could represent a booster for memory cells already primed to react rapidly and powerfully against DLLIR present in previously encountered structures, i.e., against DLLIR in b) and j) decapentapeptides described in Fig. (1
) implies that, following HPV16 infection or active immunization, an anti-HPV16 DLLIR response could represent a booster for memory cells already primed to react rapidly and powerfully against DLLIR present in previously encountered structures, i.e., against DLLIR in b) and j) decapentapeptides described in Fig. (1 ). Otherwise said, the primary response against HPV16 DLLIR might result into a secondary response against conformers of infectious agents encountered early in the life. Hence, a first consequence is that the anti-HPV16 infection/vaccination will miss the goal of defending the body from HPV16.
). Otherwise said, the primary response against HPV16 DLLIR might result into a secondary response against conformers of infectious agents encountered early in the life. Hence, a first consequence is that the anti-HPV16 infection/vaccination will miss the goal of defending the body from HPV16.
 |
Fig. (1) Context-dependent conformations of decapentapeptides containing DLLIR. Decapentapeptide sequences with the common 5-mer DLLIR in capital letters: A) HPV16: nkplcDLLIRcincq; B) HCMV: mDLLIRlgfll; C) C. diphtheriae: rrglgDLLIRmglhh; D) C. tetani: ismdkDLLIRedetl; E) B. pertussis: gdrhtDLLIRfaeac; F) B. pertussis: valglDLLIRrdsgg; G) T. gondii: aerlyDLLIReifvr; H) T. gondii: dasslDLLIRenlkr; I) T. gondii: dearnDLLIRstdqg; J) T. cruzi: isafyDLLIRqltvs; K) H. sapiens: fpllrDLLIRshlqd; L) H. sapiens: faylrDLLIRthmqn; m) H. sapiens: qegfwDLLIRlrgqg. Asterisk indicates the position of the first Leu in the shared pentapeptide DLLIR. The HCMV mDLLIRlgfll lacks a 5-aa sequence at the NH2 terminus (see Table 4). For simplicity, only 3 out of the 11 T. gondii decapeptapeptides containing DLLIR (see Table 4) are reported. Epitope structures similar to that of the HPV decapentapeptide are given in blue, with the oval encircling the DLLIR sequence. PEP-FOLD3 [45Shen Y, Maupetit J, Derreumaux P, Tufféry P. Improved PEP-FOLD approach for peptide and miniprotein structure prediction J Chem Theor Comput 2014; 10: 4745-58.-47Lamiable A, Thévenet P, Rey J, Vavrusa M, Derreumaux P, Tufféry P. PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res 2016; 44(W1): W449-54. [http://dx.doi.org/10.1093/nar/gkw329] [PMID: 27131374] ] was used to obtain the 3D peptide structures. |
Then, a second consequence is that the anamnestic, high affinity, high avidity immune responses triggered by HPV16 infection/active immunization against immune determinants already primed in the immunological memory of the subject will remain without a target. Indeed, no reaction can occur with the early sensitizing infectious agents that evoked the primary response and are no more present in the organism (that is, HCMV and T. cruzi in the examples discussed above).
Therefore, a third consequence is that the boostered anamnestic, immediate, high-avidity, high-affinity immune responses will take the road towards available crossreactive targets. In the case in point, towards the human similar conformer, i.e., the human Septin-9 pentapeptide in the faylrDLLIRthmqn sequence (see Fig. 1 ) with potential heavy autoimmune consequences on the human host. Actually, altered Septin-9 leads to focal neuropathy characterized by recurrent episodes of brachial plexus neuropathy, with muscle weakness and atrophy preceded by severe pain in the affected arm [74Sudo K, Ito H, Iwamoto I, Morishita R, Asano T, Nagata K. SEPT9 sequence alternations causing hereditary neuralgic amyotrophy are associated with altered interactions with SEPT4/SEPT11 and resistance to Rho/Rhotekin-signaling. Hum Mutat 2007; 28(10): 1005-13.
) with potential heavy autoimmune consequences on the human host. Actually, altered Septin-9 leads to focal neuropathy characterized by recurrent episodes of brachial plexus neuropathy, with muscle weakness and atrophy preceded by severe pain in the affected arm [74Sudo K, Ito H, Iwamoto I, Morishita R, Asano T, Nagata K. SEPT9 sequence alternations causing hereditary neuralgic amyotrophy are associated with altered interactions with SEPT4/SEPT11 and resistance to Rho/Rhotekin-signaling. Hum Mutat 2007; 28(10): 1005-13.
[http://dx.doi.org/10.1002/humu.20554] [PMID: 17546647] , 75Hannibal MC, Ruzzo EK, Miller LR, et al. SEPT9 gene sequencing analysis reveals recurrent mutations in hereditary neuralgic amyotrophy. Neurology 2009; 72(20): 1755-9.
[http://dx.doi.org/10.1212/WNL.0b013e3181a609e3] [PMID: 19451530] ].
CONCLUSION
In the 40’, Francis and Colleagues [76Francis T, Salk JE, Quilligan JJ. Experience with vaccination against influenza in the spring of 1947: A preliminary report. Am J Public Health Nations Health 1947; 37(8): 1013-6.
[http://dx.doi.org/10.2105/AJPH.37.8.1013] [PMID: 18016577] , 77Davenport FM, Hennessy AV, Francis T Jr. Epidemiologic and immunologic significance of age distribution of antibody to antigenic variants of influenza virus. J Exp Med 1953; 98(6): 641-56.
[http://dx.doi.org/10.1084/jem.98.6.641] [PMID: 13109114] ] observed that, when influenza A vaccines were given to children and young and older adults, the antibody response did not correspond to the vaccinating influenza antigen. Rather, it seemed to depend on the age of the recipients, each group responding with antibodies that reacted best with the virus subtype they experienced first in their life. That is, humans vaccinated against an influenza strain produced antibodies of higher titer against a different influenza strain that was their first childhood experience of influenza, even if that strain happened to be absent from the vaccine. The phenomenon was named ‘original antigenic sin’ [76Francis T, Salk JE, Quilligan JJ. Experience with vaccination against influenza in the spring of 1947: A preliminary report. Am J Public Health Nations Health 1947; 37(8): 1013-6.
[http://dx.doi.org/10.2105/AJPH.37.8.1013] [PMID: 18016577] -78Francis T. On the doctrine of original antigenic sin. Proc Am Philos Soc 1960; 104: 572-8.] and is also known as the Hoskins effect [79Hoskins TW, Davies JR, Smith AJ, Miller CL, Allchin A. Assessment of inactivated influenza-A vaccine after three outbreaks of influenza A at christ’s hospital. Lancet 1979; 1(8106): 33-5.
[http://dx.doi.org/10.1016/S0140-6736(79)90468-9] [PMID: 83475] ]. As clearly described by Fazekas de St. Groth and Webster [80 Fazekas de St. Groth S, Webster RG. The antibody response. In: Eds.: GEW Wolstenholme, J. Knight, Cellular Biology of Myxovirus Infections. CIBA Foundation Symposium. Little, Brown and Company, Boston, pp. 246-271, 1964.], ‘Immunological memory, then, seems to cover families of antigens rather than only the particular member involved in the primary response. Such a mechanism makes for rapid anamnestic response, at the expense of specificity’.
The original antigenic sin was interpreted as the cause of the absence of effect of revaccinations [76Francis T, Salk JE, Quilligan JJ. Experience with vaccination against influenza in the spring of 1947: A preliminary report. Am J Public Health Nations Health 1947; 37(8): 1013-6.
[http://dx.doi.org/10.2105/AJPH.37.8.1013] [PMID: 18016577] -78Francis T. On the doctrine of original antigenic sin. Proc Am Philos Soc 1960; 104: 572-8.]. However, after seventy years, the molecular basis and the mechanism underlying the antigenic sin phenomenon remained unknown [81Monto AS, Malosh RE, Petrie JG, Martin ET. The doctrine of original antigenic sin: Separating good from evil. J Infect Dis 2017; 215(12): 1782-8.
[http://dx.doi.org/10.1093/infdis/jix173] [PMID: 28398521] ].
More recently, Lucchese and Kanduc [40Lucchese G, Kanduc D. The Guillain–Barrè peptide signatures: From Zika virus to Campylobacter, and beyond. Virus Adaptation and Treatment 2017; 9: 1-11.
[http://dx.doi.org/10.2147/VAAT.S124535] , 41Lucchese G, Kanduc D. Minimal immune determinants connect Zika virus, Human Cytomegalovirus, and Toxoplasma gondii to microcephaly-related human proteins. Am J Reprod Immunol 2017; 77(2): e12608.
[http://dx.doi.org/10.1111/aji.12608] [PMID: 27878897] ] suggested that the massive sharing of minimal epitopic determinants among pathogens – namely Zika virus, Epstein-Barr virus, Cytomegalovirus, Influenza virus, Campylobacter jejuni, and Mycoplasma pneumoniae, among others – and the consequent potential cross-reactivity might represent the molecular basis and mechanism by which different infections over time can irrevocably imprint the host immunological memory, thus leading to subsequent anamnestic, misled, immune responses. Said data [40Lucchese G, Kanduc D. The Guillain–Barrè peptide signatures: From Zika virus to Campylobacter, and beyond. Virus Adaptation and Treatment 2017; 9: 1-11.
[http://dx.doi.org/10.2147/VAAT.S124535] , 41Lucchese G, Kanduc D. Minimal immune determinants connect Zika virus, Human Cytomegalovirus, and Toxoplasma gondii to microcephaly-related human proteins. Am J Reprod Immunol 2017; 77(2): e12608.
[http://dx.doi.org/10.1111/aji.12608] [PMID: 27878897] ] and the present ones add biochemical evidence in supporting the view that ‘a cell, which has once produced antibody, is not only preempted to performing the same task again but does this on the slightest provocation by accepting what we may regard as not quite appropriate stimuli’ [80 Fazekas de St. Groth S, Webster RG. The antibody response. In: Eds.: GEW Wolstenholme, J. Knight, Cellular Biology of Myxovirus Infections. CIBA Foundation Symposium. Little, Brown and Company, Boston, pp. 246-271, 1964.].
In essence, our studies suggest that inter-pathogen peptide commonality and the consequent peptide cross-reactivity underlie the “original antigenic sin”, in this way explaining not only the failures of vaccination protocols [19Steinman RM, Pope M. Exploiting dendritic cells to improve vaccine efficacy. J Clin Invest 2002; 109(12): 1519-26.
[http://dx.doi.org/10.1172/JCI0215962] [PMID: 12070296] -26van der Meer JW. The infectious disease challenges of our time. Front Public Health 2013; 1: 7.
[http://dx.doi.org/10.3389/fpubh.2013.00007] [PMID: 24350179] , 82Wiedermann U, Garner-Spitzer E, Wagner A. Primary vaccine failure to routine vaccines: Why and what to do? Hum Vaccin Immunother 2016; 12(1): 239-43.
[http://dx.doi.org/10.1080/21645515.2015.1093263] [PMID: 26836329] -100Ramsay M, Brown K. The public health implications of secondary measles vaccine failure. J Prim Health Care 2013; 5(2): 92.
[PMID: 23748388] ] but also the burden of vaccine-associated adverse events [1George J, Levy Y, Kallenberg CGM, Shoenfeld Y. Infections and Wegener’s granulomatosis-a cause and effect relationship? QJM 1997; 90(5): 367-73.
[http://dx.doi.org/10.1093/qjmed/90.5.367] [PMID: 9205673] -17Burdin N, Handy LK, Plotkin SA. What is wrong with pertussis vaccine immunity? The problem of waning effectiveness of pertussis vaccines. Cold Spring Harb Perspect Biol 2017; 9(12): a029454.
[http://dx.doi.org/10.1101/cshperspect.a029454] [PMID: 28289064] , 101https://vaers.hhs.gov/data/datasets.html]. Indeed, last and perhaps most important is the observation that the massive microbial vs human peptide overlap reiterates the concept that only vaccines based on peptide sequences uniquely owned by the infectious agent and absent in the human proteome may lead to safe, effective, and specific immunotherapies [102Lucchese G, Stufano A, Kanduc D. Proteome-guided search for influenza A B-cell epitopes. FEMS Immunol Med Microbiol 2009; 57(1): 88-92.
[http://dx.doi.org/10.1111/j.1574-695X.2009.00582.x] [PMID: 19659580] -108Kanduc D. Epitopic peptides with low similarity to the host proteome: Towards biological therapies without side effects. Expert Opin Biol Ther 2009; 9(1): 45-53.
[http://dx.doi.org/10.1517/14712590802614041] [PMID: 19063692] ].
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies that are base of this research.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICTS OF INTEREST
DK declares no conflicts. YS appears as a medical consultant in vaccine compensation court, USA.
ACKNOWLEDGEMENTS
Declared none.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publishers Website along with the published article.
REFERENCES
| [1] | George J, Levy Y, Kallenberg CGM, Shoenfeld Y. Infections and Wegener’s granulomatosis-a cause and effect relationship? QJM 1997; 90(5): 367-73. [http://dx.doi.org/10.1093/qjmed/90.5.367] [PMID: 9205673] |
| [2] | Asherson RA, Shoenfeld Y. The role of infection in the pathogenesis of catastrophic antiphospholipid syndrome-molecular mimicry? J Rheumatol 2000; 27(1): 12-4. [PMID: 10648011] |
| [3] | Zandman-Goddard G, Shoenfeld Y. SLE and infections. Clin Rev Allergy Immunol 2003; 25(1): 29-40. [http://dx.doi.org/10.1385/CRIAI:25:1:29] [PMID: 12794259] |
| [4] | Zandman-Goddard G, Shoenfeld Y. Infections and SLE. Autoimmunity 2005; 38(7): 473-85. [http://dx.doi.org/10.1080/08916930500285352] [PMID: 16373252] |
| [5] | Pordeus V, Szyper-Kravitz M, Levy RA, Vaz NM, Shoenfeld Y. Infections and autoimmunity: A panorama. Clin Rev Allergy Immunol 2008; 34(3): 283-99. [http://dx.doi.org/10.1007/s12016-007-8048-8] [PMID: 18231878] |
| [6] | Doria A, Sarzi-Puttini P, Shoenfeld Y. Infections, rheumatism and autoimmunity: The conflicting relationship between humans and their environment. Autoimmun Rev 2008; 8(1): 1-4. [http://dx.doi.org/10.1016/j.autrev.2008.07.014] [PMID: 18707029] |
| [7] | Tozzoli R, Barzilai O, Ram M, et al. Infections and autoimmune thyroid diseases: Parallel detection of antibodies against pathogens with proteomic technology. Autoimmun Rev 2008; 8(2): 112-5. [http://dx.doi.org/10.1016/j.autrev.2008.07.013] [PMID: 18700170] |
| [8] | Belizna CC, Hamidou MA, Levesque H, Guillevin L, Shoenfeld Y. Infection and vasculitis. Rheumatology (Oxford) 2009; 48(5): 475-82. [http://dx.doi.org/10.1093/rheumatology/kep026] [PMID: 19258377] |
| [9] | Kivity S, Agmon-Levin N, Blank M, Shoenfeld Y. Infections and autoimmunity--friends or foes? Trends Immunol 2009; 30(8): 409-14. [http://dx.doi.org/10.1016/j.it.2009.05.005] [PMID: 19643667] |
| [10] | Arnson Y, Amital H, Guiducci S, et al. The role of infections in the immunopathogenesis of systemic sclerosis--evidence from serological studies. Ann N Y Acad Sci 2009; 1173: 627-32. [http://dx.doi.org/10.1111/j.1749-6632.2009.04808.x] [PMID: 19758208] |
| [11] | Lidar M, Langevitz P, Shoenfeld Y. The role of infection in inflammatory bowel disease: Initiation, exacerbation and protection. Isr Med Assoc J 2009; 11(9): 558-63. [PMID: 19960852] |
| [12] | Grossman C, Dovrish Z, Shoenfeld Y, Amital H. Do infections facilitate the emergence of systemic sclerosis? Autoimmun Rev 2011; 10(5): 244-7. [http://dx.doi.org/10.1016/j.autrev.2010.09.010] [PMID: 20863912] |
| [13] | Kivity S, Arango MT, Ehrenfeld M, et al. Infection and autoimmunity in Sjogren’s syndrome: A clinical study and comprehensive review. J Autoimmun 2014; 51: 17-22. [http://dx.doi.org/10.1016/j.jaut.2014.02.008] [PMID: 24637076] |
| [14] | Rinaldi M, Perricone C, Ortega-Hernandez OD, Perricone R, Shoenfeld Y. Immune thrombocytopaenic purpura: An autoimmune cross-link between infections and vaccines. Lupus 2014; 23(6): 554-67. [http://dx.doi.org/10.1177/0961203313499959] [PMID: 24763539] |
| [15] | Grubor NM, Jovanova-Nesic KD, Shoenfeld Y. Liver cystic echinococcosis and human host immune and autoimmune follow-up: A review. World J Hepatol 2017; 9(30): 1176-89. [http://dx.doi.org/10.4254/wjh.v9.i30.1176] [PMID: 29109850] |
| [16] | Segal Y, Shoenfeld Y. Vaccine-induced autoimmunity: The role of molecular mimicry and immune crossreaction. Cell Mol Immunol 2018; 14: 1-9. [PMID: 29503439] |
| [17] | Burdin N, Handy LK, Plotkin SA. What is wrong with pertussis vaccine immunity? The problem of waning effectiveness of pertussis vaccines. Cold Spring Harb Perspect Biol 2017; 9(12): a029454. [http://dx.doi.org/10.1101/cshperspect.a029454] [PMID: 28289064] |
| [18] | Koopman JS, Henry CJ, Park JH, Eisenberg MC, Ionides EL, Eisenberg JN. Dynamics affecting the risk of silent circulation when oral polio vaccination is stopped. Epidemics 2017; 20: 21-36. [http://dx.doi.org/10.1016/j.epidem.2017.02.013] [PMID: 28283373] |
| [19] | Steinman RM, Pope M. Exploiting dendritic cells to improve vaccine efficacy. J Clin Invest 2002; 109(12): 1519-26. [http://dx.doi.org/10.1172/JCI0215962] [PMID: 12070296] |
| [20] | Grossmann C, Tenbusch M, Nchinda G, et al. Enhancement of the priming efficacy of DNA vaccines encoding dendritic cell-targeted antigens by synergistic toll-like receptor ligands. BMC Immunol 2009; 10: 43. [http://dx.doi.org/10.1186/1471-2172-10-43] [PMID: 19650904] |
| [21] | Nara PL, Tobin GJ, Chaudhuri AR, et al. How can vaccines against influenza and other viral diseases be made more effective? PLoS Biol 2010; 8(12): e1000571. [http://dx.doi.org/10.1371/journal.pbio.1000571] [PMID: 21203586] |
| [22] | Didierlaurent AM, Collignon C, Bourguignon P, et al. Enhancement of adaptive immunity by the human vaccine adjuvant AS01 depends on activated dendritic cells. J Immunol 2014; 193(4): 1920-30. [http://dx.doi.org/10.4049/jimmunol.1400948] [PMID: 25024381] |
| [23] | Coban C, Kobiyama K, Aoshi T, et al. Novel strategies to improve DNA vaccine immunogenicity. Curr Gene Ther 2011; 11(6): 479-84. [http://dx.doi.org/10.2174/156652311798192815] [PMID: 22023477] |
| [24] | Xu H, Zhao G, Huang X, et al. CD40-expressing plasmid induces anti-CD40 antibody and enhances immune responses to DNA vaccination. J Gene Med 2010; 12(1): 97-106. [http://dx.doi.org/10.1002/jgm.1412] [PMID: 19950201] |
| [25] | Congdon P. Spatiotemporal frameworks for infectious disease diffusion and epidemiology. Int J Environ Res Public Health 2016; 13(12): E1261. [http://dx.doi.org/10.3390/ijerph13121261] [PMID: 27999420] |
| [26] | van der Meer JW. The infectious disease challenges of our time. Front Public Health 2013; 1: 7. [http://dx.doi.org/10.3389/fpubh.2013.00007] [PMID: 24350179] |
| [27] | Lucchese G, Kanduc D. Zika virus and autoimmunity: From microcephaly to Guillain-Barré syndrome, and beyond. Autoimmun Rev 2016; 15(8): 801-8. [http://dx.doi.org/10.1016/j.autrev.2016.03.020] [PMID: 27019049] |
| [28] | Natale C, Giannini T, Lucchese A, Kanduc D. Computer-assisted analysis of molecular mimicry between human papillomavirus 16 E7 oncoprotein and human protein sequences. Immunol Cell Biol 2000; 78(6): 580-5. [http://dx.doi.org/10.1046/j.1440-1711.2000.00949.x] [PMID: 11114967] |
| [29] | Lucchese G, Stufano A, Kanduc D. Proposing low-similarity peptide vaccines against Mycobacterium tuberculosis. J Biomed Biotechnol 2010; 2010: 832341. [http://dx.doi.org/10.1155/2010/832341] [PMID: 20625421] |
| [30] | Lucchese G, Stufano A, Kanduc D. Proteome-guided search for influenza A B-cell epitopes. FEMS Immunol Med Microbiol 2009; 57(1): 88-92. [http://dx.doi.org/10.1111/j.1574-695X.2009.00582.x] [PMID: 19659580] |
| [31] | Kanduc D, Tessitore L, Lucchese G, Kusalik A, Farber E, Marincola FM. Sequence uniqueness and sequence variability as modulating factors of human anti-HCV humoral immune response. Cancer Immunol Immunother 2008; 57(8): 1215-23. [http://dx.doi.org/10.1007/s00262-008-0456-y] [PMID: 18256830] |
| [32] | Lucchese A, Serpico R, Crincoli V, Shoenfeld Y, Kanduc D. Sequence uniqueness as a molecular signature of HIV-1-derived B-cell epitopes. Int J Immunopathol Pharmacol 2009; 22(3): 639-46. [http://dx.doi.org/10.1177/039463200902200309] [PMID: 19822080] |
| [33] | Kanduc D, Serpico R, Lucchese A, Shoenfeld Y. Correlating low-similarity peptide sequences and HIV B-cell epitopes. Autoimmun Rev 2008; 7(4): 291-6. [http://dx.doi.org/10.1016/j.autrev.2007.11.001] [PMID: 18295732] |
| [34] | Kanduc D. Immunogenicity in peptide-immunotherapy: From self/nonself to similar/dissimilar sequences. Adv Exp Med Biol 2008; 640: 198-207. [http://dx.doi.org/10.1007/978-0-387-09789-3_15] [PMID: 19065793] |
| [35] | Kanduc D, Stufano A, Lucchese G, Kusalik A. Massive peptide sharing between viral and human proteomes. Peptides 2008; 29(10): 1755-66. [http://dx.doi.org/10.1016/j.peptides.2008.05.022] [PMID: 18582510] |
| [36] | Polito A, Polimeno R, Kanduc D. Peptide sharing between parvovirus B19 and DNA methylating/histone modifying enzymes: A potential link to childhood acute lymphoblastic leukemia. Int J Pediatr Child Health 2017; 5: 29-39. |
| [37] | Lucchese G. Understanding neuropsychiatric diseases, analyzing the peptide sharing between infectious agents and the language-associated NMDA 2A protein. Front Psychiatry 2016; 7: 60. [http://dx.doi.org/10.3389/fpsyt.2016.00060] [PMID: 27148089] |
| [38] | Kanduc D. “Self-nonself” peptides in the design of vaccines. Curr Pharm Des 2009; 15(28): 3283-9. [http://dx.doi.org/10.2174/138161209789105135] [PMID: 19860677] |
| [39] | Kanduc D. Peptide cross-reactivity: The original sin of vaccines. Front Biosci (Schol Ed) 2012; 4: 1393-401. [http://dx.doi.org/10.2741/s341] [PMID: 22652881] |
| [40] | Lucchese G, Kanduc D. The Guillain–Barrè peptide signatures: From Zika virus to Campylobacter, and beyond. Virus Adaptation and Treatment 2017; 9: 1-11. [http://dx.doi.org/10.2147/VAAT.S124535] |
| [41] | Lucchese G, Kanduc D. Minimal immune determinants connect Zika virus, Human Cytomegalovirus, and Toxoplasma gondii to microcephaly-related human proteins. Am J Reprod Immunol 2017; 77(2): e12608. [http://dx.doi.org/10.1111/aji.12608] [PMID: 27878897] |
| [42] | UniProt: the universal protein knowledgebase. Nucleic Acids Res 2017; 45(D1): D158-69. [http://dx.doi.org/10.1093/nar/gkw1099] [PMID: 27899622] |
| [43] | Chen C, Li Z, Huang H, Suzek BE, Wu CH. A fast peptide match service for UniProt knowledgebase. Bioinformatics 2013; 29(21): 2808-9. [http://dx.doi.org/10.1093/bioinformatics/btt484] [PMID: 23958731] |
| [44] | Vita R, Overton JA, Greenbaum JA, et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res 2015; 43(Database issue): D405-12. [http://dx.doi.org/10.1093/nar/gku938] [PMID: 25300482] |
| [45] | Shen Y, Maupetit J, Derreumaux P, Tufféry P. Improved PEP-FOLD approach for peptide and miniprotein structure prediction J Chem Theor Comput 2014; 10: 4745-58. |
| [46] | Lamiable A, Thevenet P, Tufféry P. A critical assessment of hidden Markov model sub-optimal sampling strategies applied to the generation of peptide 3D models. J Comput Chem 2016; 37(21): 2006-16. [http://dx.doi.org/10.1002/jcc.24422] [PMID: 27317417] |
| [47] | Lamiable A, Thévenet P, Rey J, Vavrusa M, Derreumaux P, Tufféry P. PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res 2016; 44(W1): W449-54. [http://dx.doi.org/10.1093/nar/gkw329] [PMID: 27131374] |
| [48] | Orlando G, Fasolo M, Mazza F, et al. Risk of cervical HPV infection and prevalence of vaccine-type and other high-risk HPV types among sexually active teens and young women (13-26 years) enrolled in the VALHIDATE study. Hum Vaccin Immunother 2014; 10(4): 986-94. [http://dx.doi.org/10.4161/hv.27682] [PMID: 24423757] |
| [49] | Committee Opinion No. 704: Human papillomavirus vaccination. Obstet Gynecol 2017; 129: 1155-6. [http://dx.doi.org/10.1097/AOG.0000000000002111] [PMID: 28538494] |
| [50] | https://www.cdc.gov/vaccines/parents/downloads/parent-ver-sch-0-6yrs.pdf |
| [51] | Zhang Q, Gao Y, Peng Y, et al. Epidemiological survey of human cytomegalovirus antibody levels in children from Southeastern China. Virol J 2014; 11: 123. [http://dx.doi.org/10.1186/1743-422X-11-123] [PMID: 24996226] |
| [52] | Dubey JP, Lago EG, Gennari SM, Su C, Jones JL. Toxoplasmosis in humans and animals in Brazil: High prevalence, high burden of disease, and epidemiology. Parasitology 2012; 139(11): 1375-424. [http://dx.doi.org/10.1017/S0031182012000765] [PMID: 22776427] |
| [53] | Llewellyn MS, Messenger LA, Luquetti AO, et al. Deep sequencing of the Trypanosoma cruzi GP63 surface proteases reveals diversity and diversifying selection among chronic and congenital Chagas disease patients. PLoS Negl Trop Dis 2015; 9(4): e0003458. [http://dx.doi.org/10.1371/journal.pntd.0003458] [PMID: 25849488] |
| [54] | Lucchese G, Capone G, Kanduc D. Peptide sharing between influenza A H1N1 hemagglutinin and human axon guidance proteins. Schizophr Bull 2014; 40(2): 362-75. [http://dx.doi.org/10.1093/schbul/sbs197] [PMID: 23378012] |
| [55] | Lucchese G. Confronting JC virus and Homo sapiens biological signatures. Front Biosci 2013; 18: 716-24. [http://dx.doi.org/10.2741/4133] [PMID: 23276955] |
| [56] | Frank A. Immunology and Evolution of Infectious Disease 2002. |
| [57] | Lucchese G, Kanduc D. Single amino acid repeats connect viruses to neurodegeneration. Curr Drug Discov Technol 2014; 11(3): 214-9. [http://dx.doi.org/10.2174/1570163811666140212112300] [PMID: 24521198] |
| [58] | Zeng W, Pagnon J, Jackson DC. The C-terminal pentapeptide of LHRH is a dominant B cell epitope with antigenic and biological function. Mol Immunol 2007; 44(15): 3724-31. [http://dx.doi.org/10.1016/j.molimm.2007.04.004] [PMID: 17512595] |
| [59] | Kanduc D. Pentapeptides as minimal functional units in cell biology and immunology. Curr Protein Pept Sci 2013; 14(2): 111-20. [http://dx.doi.org/10.2174/1389203711314020003] [PMID: 23305312] |
| [60] | Kanduc D. Homology, similarity, and identity in peptide epitope immunodefinition. J Pept Sci 2012; 18(8): 487-94. [http://dx.doi.org/10.1002/psc.2419] [PMID: 22696298] |
| [61] | Raychaudhuri S, Sandor C, Stahl EA, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet 2012; 44(3): 291-6. [http://dx.doi.org/10.1038/ng.1076] [PMID: 22286218] |
| [62] | Xiao N, Cao J, Zhou H, Ding SQ, Kong LY, Li JN. Identification of three novel B-cell epitopes of VMH protein from vibrio mimicus by screening a phage display peptide library. Vet Immunol Immunopathol 2016; 182: 22-8. [http://dx.doi.org/10.1016/j.vetimm.2016.09.005] [PMID: 27863546] |
| [63] | El-Turk F, Newby FN, De Genst E, et al. Structural effects of two camelid nanobodies directed to distinct C-terminal epitopes on α-Synuclein. Biochemistry 2016; 55(22): 3116-22. [http://dx.doi.org/10.1021/acs.biochem.6b00149] [PMID: 27096466] |
| [64] | Cui Z, Zhao MH, Jia XY, et al. Antibodies to α5 chain of collagen IV are pathogenic in Goodpasture’s disease. J Autoimmun 2016; 70: 1-11. [http://dx.doi.org/10.1016/j.jaut.2016.04.001] [PMID: 27117167] |
| [65] | Li Z, Wang D, Gu Y, et al. Crystal structures of two immune complexes identify determinants for viral infectivity and type-specific neutralization of human papillomavirus. MBio 2017; 8(5): e00787-17. [http://dx.doi.org/10.1128/mBio.00787-17] [PMID: 28951471] |
| [66] | Lazcano A, Peretó J. On the origin of mitosing cells: A historical appraisal of Lynn Margulis endosymbiotic theory. J Theor Biol 2017; 434: 80-7. [http://dx.doi.org/10.1016/j.jtbi.2017.06.036] [PMID: 28684295] |
| [67] | Bell PJ. Viral eukaryogenesis: Was the ancestor of the nucleus a complex DNA virus? J Mol Evol 2001; 53(3): 251-6. [http://dx.doi.org/10.1007/s002390010215] [PMID: 11523012] |
| [68] | Forterre P. The origin of viruses and their possible roles in major evolutionary transitions. Virus Res 2006; 117(1): 5-16. [http://dx.doi.org/10.1016/j.virusres.2006.01.010] [PMID: 16476498] |
| [69] | Kabsch W, Sander C. On the use of sequence homologies to predict protein structure: Identical pentapeptides can have completely different conformations. Proc Natl Acad Sci USA 1984; 81(4): 1075-8. [http://dx.doi.org/10.1073/pnas.81.4.1075] [PMID: 6422466] |
| [70] | Kabsch W, Sander C. Identical pentapeptides with different backbones. Nature 1985; 317(6034): 207. [http://dx.doi.org/10.1038/317207a0] [PMID: 4047161] |
| [71] | Niman HL, Houghten RA, Walker LE, et al. Generation of protein-reactive antibodies by short peptides is an event of high frequency: Implications for the structural basis of immune recognition. Proc Natl Acad Sci USA 1983; 80(16): 4949-53. [http://dx.doi.org/10.1073/pnas.80.16.4949] [PMID: 6192445] |
| [72] | Wilson IA, Haft DH, Getzoff ED, Tainer JA, Lerner RA, Brenner S. Identical short peptide sequences in unrelated proteins can have different conformations: A testing ground for theories of immune recognition. Proc Natl Acad Sci USA 1985; 82(16): 5255-9. [http://dx.doi.org/10.1073/pnas.82.16.5255] [PMID: 2410917] |
| [73] | Schulze-Gahmen U, Wilson IA. Monoclonal antibodies against an identical short peptide sequence shared by two unrelated proteins. Pept Res 1989; 2(5): 322-31. [PMID: 2485209] |
| [74] | Sudo K, Ito H, Iwamoto I, Morishita R, Asano T, Nagata K. SEPT9 sequence alternations causing hereditary neuralgic amyotrophy are associated with altered interactions with SEPT4/SEPT11 and resistance to Rho/Rhotekin-signaling. Hum Mutat 2007; 28(10): 1005-13. [http://dx.doi.org/10.1002/humu.20554] [PMID: 17546647] |
| [75] | Hannibal MC, Ruzzo EK, Miller LR, et al. SEPT9 gene sequencing analysis reveals recurrent mutations in hereditary neuralgic amyotrophy. Neurology 2009; 72(20): 1755-9. [http://dx.doi.org/10.1212/WNL.0b013e3181a609e3] [PMID: 19451530] |
| [76] | Francis T, Salk JE, Quilligan JJ. Experience with vaccination against influenza in the spring of 1947: A preliminary report. Am J Public Health Nations Health 1947; 37(8): 1013-6. [http://dx.doi.org/10.2105/AJPH.37.8.1013] [PMID: 18016577] |
| [77] | Davenport FM, Hennessy AV, Francis T Jr. Epidemiologic and immunologic significance of age distribution of antibody to antigenic variants of influenza virus. J Exp Med 1953; 98(6): 641-56. [http://dx.doi.org/10.1084/jem.98.6.641] [PMID: 13109114] |
| [78] | Francis T. On the doctrine of original antigenic sin. Proc Am Philos Soc 1960; 104: 572-8. |
| [79] | Hoskins TW, Davies JR, Smith AJ, Miller CL, Allchin A. Assessment of inactivated influenza-A vaccine after three outbreaks of influenza A at christ’s hospital. Lancet 1979; 1(8106): 33-5. [http://dx.doi.org/10.1016/S0140-6736(79)90468-9] [PMID: 83475] |
| [80] | Fazekas de St. Groth S, Webster RG. The antibody response. In: Eds.: GEW Wolstenholme, J. Knight, Cellular Biology of Myxovirus Infections. CIBA Foundation Symposium. Little, Brown and Company, Boston, pp. 246-271, 1964. |
| [81] | Monto AS, Malosh RE, Petrie JG, Martin ET. The doctrine of original antigenic sin: Separating good from evil. J Infect Dis 2017; 215(12): 1782-8. [http://dx.doi.org/10.1093/infdis/jix173] [PMID: 28398521] |
| [82] | Wiedermann U, Garner-Spitzer E, Wagner A. Primary vaccine failure to routine vaccines: Why and what to do? Hum Vaccin Immunother 2016; 12(1): 239-43. [http://dx.doi.org/10.1080/21645515.2015.1093263] [PMID: 26836329] |
| [83] | Kubba AK, Taylor P, Graneek B, Strobel S. Non-responders to hepatitis B vaccination: A review. Commun Dis Public Health 2003; 6(2): 106-12. [PMID: 12889288] |
| [84] | Weinberger B, Keller M, Fischer KH, et al. Decreased antibody titers and booster responses in tick-borne encephalitis vaccinees aged 50-90 years. Vaccine 2010; 28(20): 3511-5. [http://dx.doi.org/10.1016/j.vaccine.2010.03.024] [PMID: 20332047] |
| [85] | McDermott AB, Cohen SB, Zuckerman JN, Madrigal JA. Hepatitis B third-generation vaccines: Improved response and conventional vaccine non-response--evidence for genetic basis in humans. J Viral Hepat 1998; 5(Suppl. 2): 9-11. [http://dx.doi.org/10.1046/j.1365-2893.1998.0050s2009.x] [PMID: 9857354] |
| [86] | Garner-Spitzer E, Wagner A, Paulke-Korinek M, et al. Tick-borne encephalitis (TBE) and hepatitis B nonresponders feature different immunologic mechanisms in response to TBE and influenza vaccination with involvement of regulatory T and B cells and IL-10. J Immunol 2013; 191(5): 2426-36. [http://dx.doi.org/10.4049/jimmunol.1300293] [PMID: 23872054] |
| [87] | Pawelec G, Larbi A. Immunity and ageing in man: Annual review 2006/2007. Exp Gerontol 2008; 43(1): 34-8. [PMID: 17977683] |
| [88] | Siegrist CA, Aspinall R. B-cell responses to vaccination at the extremes of age. Nat Rev Immunol 2009; 9(3): 185-94. [http://dx.doi.org/10.1038/nri2508] [PMID: 19240757] |
| [89] | Boraschi D, Italiani P. Immunosenescence and vaccine failure in the elderly: Strategies for improving response. Immunol Lett 2014; 162(1 Pt B): 346-53. [http://dx.doi.org/10.1016/j.imlet.2014.06.006] [PMID: 24960535] |
| [90] | Grubeck-Loebenstein B. Fading immune protection in old age: Vaccination in the elderly. J Comp Pathol 2010; 142(Suppl. 1): S116-9. [http://dx.doi.org/10.1016/j.jcpa.2009.10.002] [PMID: 19959180] |
| [91] | Boyd SD, Jackson KJL. Predicting vaccine responsiveness. Cell Host Microbe 2015; 17(3): 301-7. [http://dx.doi.org/10.1016/j.chom.2015.02.015] [PMID: 25766292] |
| [92] | Bond KA, Franklin LJ, Sutton B, Firestone SM. Q-Vax Q fever vaccine failures, Victoria, Australia 1994-2013. Vaccine 2017; 35(51): 7084-7. [http://dx.doi.org/10.1016/j.vaccine.2017.10.088] [PMID: 29132996] |
| [93] | Breakwell L, Moturi E, Helgenberger L, et al. Measles outbreak associated with vaccine failure in adults-Federated States of Micronesia, February-August 2014. MMWR Morb Mortal Wkly Rep 2015; 64(38): 1088-92. [http://dx.doi.org/10.15585/mmwr.mm6438a7] [PMID: 26421903] |
| [94] | Matar R, Hong E, Levy C, et al. Vaccine failure after meningococcal C conjugate vaccine may be linked to decline of bactericidal titers and absence of herd immunity. Pediatr Infect Dis J 2015; 34(10): 1142-3. [http://dx.doi.org/10.1097/INF.0000000000000833] [PMID: 26367811] |
| [95] | Naylor C, Lu M, Haque R, et al. Environmental enteropathy, oral vaccine failure and growth faltering in infants in Bangladesh. EBioMedicine 2015; 2(11): 1759-66. [http://dx.doi.org/10.1016/j.ebiom.2015.09.036] [PMID: 26870801] |
| [96] | Cherry JD. Epidemic pertussis and acellular pertussis vaccine failure in the 21st century. Pediatrics 2015; 135(6): 1130-2. [http://dx.doi.org/10.1542/peds.2014-4118] [PMID: 25941310] |
| [97] | Moinho R, Brett A, Ferreira G, Lemos S. Pneumococcal vaccine failure: Can it be a primary immunodeficiency? BMJ Case Rep 2014; 2014: bcr2014204714. [http://dx.doi.org/10.1136/bcr-2014-204714] [PMID: 24925540] |
| [98] | McMichael A, Picker LJ, Moore JP, Burton DR. Another HIV vaccine failure: Where to next? Nat Med 2013; 19(12): 1576-7. [http://dx.doi.org/10.1038/nm.3413] [PMID: 24309655] |
| [99] | Mahalingam S, Herring BL, Halstead SB. Call to action for dengue vaccine failure. Emerg Infect Dis 2013; 19(8): 1335-7. [http://dx.doi.org/10.3201/eid1908.121864] [PMID: 23876389] |
| [100] | Ramsay M, Brown K. The public health implications of secondary measles vaccine failure. J Prim Health Care 2013; 5(2): 92. [PMID: 23748388] |
| [101] | https://vaers.hhs.gov/data/datasets.html |
| [102] | Lucchese G, Stufano A, Kanduc D. Proteome-guided search for influenza A B-cell epitopes. FEMS Immunol Med Microbiol 2009; 57(1): 88-92. [http://dx.doi.org/10.1111/j.1574-695X.2009.00582.x] [PMID: 19659580] |
| [103] | Kanduc D. Immunogenicity, immunopathogenicity, and immunotolerance in one graph. Anticancer Agents Med Chem 2015; 15(10): 1264-8. [http://dx.doi.org/10.2174/1871520615666150716105543] [PMID: 26179265] |
| [104] | Kanduc D, Shoenfeld Y. From HBV to HPV: Designing vaccines for extensive and intensive vaccination campaigns worldwide. Autoimmun Rev 2016; 15(11): 1054-61. [http://dx.doi.org/10.1016/j.autrev.2016.07.030] [PMID: 27490205] |
| [105] | Kanduc D. Peptimmunology: Immunogenic peptides and sequence redundancy. Curr Drug Discov Technol 2005; 2(4): 239-44. [http://dx.doi.org/10.2174/157016305775202946] [PMID: 16475920] |
| [106] | Kanduc D. The self/nonself issue: A confrontation between proteomes. Self Nonself 2010; 1(3): 255-8. [http://dx.doi.org/10.4161/self.1.3.11897] [PMID: 21487482] |
| [107] | Kanduc D. HCV: Written in our DNA. Self Nonself 2011; 2(2): 108-13. [http://dx.doi.org/10.4161/self.2.2.15795] [PMID: 22299062] |
| [108] | Kanduc D. Epitopic peptides with low similarity to the host proteome: Towards biological therapies without side effects. Expert Opin Biol Ther 2009; 9(1): 45-53. [http://dx.doi.org/10.1517/14712590802614041] [PMID: 19063692] |