- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Ornithology Journal
(Discontinued)
ISSN: 1874-4532 ― Volume 13, 2020
New Microsatellite Markers for the Common Tern (Sterna hirundo) Developed with 454 Shot-Gun Pyrosequencing
Susann Janowski1, *, Ina Gross1, Hedwig Sauer-Gürth1, Dieter Thomas Tietze1, Markus Grohme2, Marcus Frohme2, Peter Becker3, Michael Wink1
Abstract
Long term studies, focusing on population- and socio-biology research, require the unequivocal identification of individuals. DNA studies with Short Tandem Repeats (STR loci) became a widespread tool in population genetics. We used the next-generation sequencing (NGS) approach with 454 shot-gun pyrosequencing to identify 13 new polymorphic STR loci for the Common Tern, Sterna hirundo. To enlarge the marker set we added two more loci originally developed for Black-legged Kittiwake (Rissa tridactyla) and Red-billed Gull (Chroicocephalus scopulinus) and arranged these 15 loci into three multiplex PCR panels for high throughput genotyping. Loci characterization demonstrated that our marker set is of high quality. A PIC value of about 0.67 and a power of exclusion value of 0.99 were reached. Deviation from Hardy-Weinberg expectations of some loci and low frequencies for null alleles are interpreted as a result of inbreeding and founder effect in the investigated tern colony. We used a test data set of this well-studied breeding colony of Common Tern at Banter Lake, Wilhelmshaven, Germany, to perform a parentage test. Parent-chick relationships, known from the social pedigree of that colony, were compared with genetically calculated ones. In order to test our markers and the used parentage program COLONY, we conducted six competing data sets with varying completeness of included parental genotypes. By including fully sampled parent pairs of known family assignment, results were correct for nest mates, single parents and parent pairs. Our marker set provides a powerful tool to investigate life-time reproductive success and other issues of population and socio-biology for Common Terns, e.g. in the aforementioned colony monitored for decades.
Article Information
Identifiers and Pagination:
Year: 2016Volume: 9
First Page: 50
Last Page: 59
Publisher Id: TOOENIJ-9-50
DOI: 10.2174/1874453201609010050
Article History:
Received Date: 16/05/2016Revision Received Date: 25/10/2016
Acceptance Date: 06/11/2016
Electronic publication date: 19/12/2016
Collection year: 2016
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution-Non-Commercial 4.0 International Public License (CC BY-NC 4.0) (https://creativecommons.org/licenses/by-nc/4.0/legalcode), which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Institute of Pharmacy and Molecular Biotechnology, Heidelberg University, Im Neuenheimer Feld 364, 69120 Heidelberg, Germany; Tel: +49 6221 54-4847; Fax: +49 6221 54-4884; E-mails: susann.janowski@outlook.com, susann.janowski@freenet.de
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 16-05-2016 |
Original Manuscript | New Microsatellite Markers for the Common Tern (Sterna hirundo) Developed with 454 Shot-Gun Pyrosequencing | |
INTRODUCTION
Population and socio-biology research of birds requires long-term studies. Monitoring of populations or breeding colonies over a long period of time depends on the unequivocal identification of individuals. Trapping and re-trapping as well as the utilization of different tagging methods (metal and color rings, pit-tags, wing-tags, radio- and satellite transmitters) are among the most common means. These methods are extremely useful also in evaluating animal movements, especially in migrating species, as well as investigating habitat use in breeding and wintering areas, for example in Montagu’s harriers [1Liminana R, Garcia JT, Gonzalez JM, et al. Philopatry and natal dispersal of Montagu's harriers (Circus pygargus) breeding in Spain: a review of existing data. Eur J Wildl Res 2012; 58: 549-55.
[http://dx.doi.org/10.1007/s10344-011-0602-2] -4Trierweiler C, Mullié WC, Drent RH, et al. A Palaearctic migratory raptor species tracks shifting prey availability within its wintering range in the Sahel. J Anim Ecol 2013; 82(1): 107-20.
[http://dx.doi.org/10.1111/j.1365-2656.2012.02036.x] [PMID: 23137184] ] among many others.
However, some of them are invasive and may have possible influences on the survival of individuals [5Authier M, Peron C, Mante A, Vidal P, Gremillet D. Designing observational biologging studies to assess the causal effect of instrumentation. Methods Ecol Evol 2013; 4: 802-10.
[http://dx.doi.org/10.1111/2041-210X.12075] -7Dann P, Sidhu LA, Jessop R, et al. Effects of flipper bands and injected transponders on the survival of adult Little Penguins Eudyptula minor. Ibis 2014; 156: 73-83.
[http://dx.doi.org/10.1111/ibi.12122] ], which could for example affect interpretation of dispersal, philopatry and survival rate. Furthermore, methodical challenges mayarise from different recovery probabilities.
DNA analyses might be an additional and sometimes more powerful tool to identify individuals. It could improve studies on philopatry and survival and enhances research topics like connectivity and exchange rates between populations as well as genetic diversity within and between (sub)populations [8Vassart M. New tools in conservation genetics. Rev Med Vet (Toulouse) 1995; 146: 171-80.]. They also possess the advantage to provide detailed knowledge of kinship and demographic structure within a particular population or breeding colony, which can be used for life-history analyses. Furthermore, utilizing parentage tests via genetic markers, already uncovered many peculiarities in breeding systems such as extra-pair parentage in many bird species [9Briggs CW, Collopy MW. Extra-pair paternity in Swainson's Hawks. J Field Ornithol 2012; 83: 41-6.
[http://dx.doi.org/10.1111/j.1557-9263.2011.00354.x] -12Schmoll T, Winkel W, Lubjuhn T. Molecular genetic evidence for mixed maternity in broods of the Coal Tit Parus ater. Vogelwarte 2008; 46: 223-7.]. Among genetic applications, microsatellites (short tandem repeats, STR loci) and SNPs (single nucleotide polymorphisms) are presently among the most useful genetic markers for population genetics [13Sunnucks P. Efficient genetic markers for population biology. Trends Ecol Evol (Amst) 2000; 15(5): 199-203.
[http://dx.doi.org/10.1016/S0169-5347(00)01825-5] [PMID: 10782134] -15Herráeza DL, Schäfer H, Mosner J, Fries H-R, Wink M. Comparison of microsatellite and single nucleotide polymorphism markers for the genetic analysis of a Galloway cattle population. Z Naturforsch, C, J Biosci 2005; 60(7-8): 637-43.
[PMID: 16163842] ].
With the development of next-generation sequencing (NGS) approaches, the identification of highly variable markers for non-model species became efficient and cost saving [16Gardner MG, Fitch AJ, Bertozzi T, Lowe AJ. Rise of the machinesrecommendations for ecologists when using next generation sequencing for microsatellite development. Mol Ecol Resour 2011; 11(6): 1093-101.
[http://dx.doi.org/10.1111/j.1755-0998.2011.03037.x] [PMID: 21679314] -18Santana Q, Coetzee M, Steenkamp E, et al. Microsatellite discovery by deep sequencing of enriched genomic libraries. Biotechniques 2009; 46(3): 217-23.
[http://dx.doi.org/10.2144/000113085] [PMID: 19317665] ]. Numerous species-specific marker sets have been developed [19Brennan PL, Hyseni C. Development of microsatellite markers for parentage analysis in the great tinamou (Tinamus major). Mol Ecol Resour 2008; 8(4): 933-5.
[http://dx.doi.org/10.1111/j.1755-0998.2008.02122.x] [PMID: 21585934] -26Topinka JR, May B. Development of polymorphic microsatellite loci in the northern goshawk (Accipiter gentilis) and cross-amplification in other raptor species. Conserv Genet 2004; 5: 861-4.
[http://dx.doi.org/10.1007/s10592-004-1973-7] ] in recent years.
We used NGS to identify new polymorphic STR loci for the Common Tern, Sterna hirundo (Linnaeus, 1758) and developed three multiplex PCR panels for high throughput genotyping. Microsatellites are among the most often used genetic markers to identify individuals for population studies such as paternity testing [9Briggs CW, Collopy MW. Extra-pair paternity in Swainson's Hawks. J Field Ornithol 2012; 83: 41-6.
[http://dx.doi.org/10.1111/j.1557-9263.2011.00354.x] , 27Babb PL, McIntosh AM, Fernandez-Duque E, Di Fiore A, Schurr TG. An optimized microsatellite genotyping strategy for assessing genetic identity and kinship in Azaras owl monkeys (Aotus azarai). Folia Primatol (Basel) 2011; 82(2): 107-17.
[http://dx.doi.org/10.1159/000330564] [PMID: 21912137] -31Seddon N, Amos W, Adcock G, et al. Mating system, philopatry and patterns of kinship in the cooperatively breeding subdesert mesite Monias benschi. Mol Ecol 2005; 14(11): 3573-83.
[http://dx.doi.org/10.1111/j.1365-294X.2005.02675.x] [PMID: 16156824] ] population structure [32Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics 2000; 155(2): 945-59.
[PMID: 10835412] , 33Stumpf KJ, Theimer TC, Mcleod MA, Koronkiewicz TJ. Disparity in population structuring of Southwestern Willow Flycatchers based on geographic distance, movement patterns, and genetic analyses. J Exp Zool A Ecol Genet Physiol 2014; 321: 577-85.] and phylogeography [34Albayrak T, Gonzalez J, Drovetski SV, Wink M. Phylogeography and population structure of Kruper's Nuthatch Sitta krueperi from Turkey based on microsatellites and mitochondrial DNA. J Ornithol 2012; 153: 405-11.
[http://dx.doi.org/10.1007/s10336-011-0756-4] -36Song G, Yu LJ, Gao B, et al. Gene flow maintains genetic diversity and colonization potential in recently range-expanded populations of an Oriental bird, the Light-vented Bulbul (Pycnonotus sinensis, Aves: Pycnonotidae). Divers Distrib 2013; 19: 1248-62.
[http://dx.doi.org/10.1111/ddi.12067] ]. The paramount advantages of codominant multi-allelic markers has been shown [23Martínez-Cruz B, David VA, Godoy JA, et al. Eighteen polymorphic microsatellite markers fort the highly endangered Spanish imperial eagle (Aquila adalberti) and related species. Mol Ecol Notes 2002; 2: 323-6.
[http://dx.doi.org/10.1046/j.1471-8286.2002.00231.x] , 37Gerber S, Mariette S, Streiff R, Bodénès C, Kremer A. Comparison of microsatellites and amplified fragment length polymorphism markers for parentage analysis. Mol Ecol 2000; 9(8): 1037-48.
[http://dx.doi.org/10.1046/j.1365-294x.2000.00961.x] [PMID: 10964223] ].
In order to evaluate the suitability of the marker set for parentage assignments, we characterized the loci regarding common parameters such as number of alleles, observed and expected heterozygosity, deviation from Hardy-Weinberg expectations, polymorphism information content, power of exclusion and null-allele frequencies, as well as parameters concerning identity, sibship and parentage discrimination. To test the ability of parentage assignment in reality we used samples from a well-studied breeding colony of Common Tern. The investigated breeding colony is located on an artificial island on the Banter Lake in Wilhelmshaven, Germany (53°30’ N, 8°06’ E) [38Becker PH, Wendeln H. A new application for transponders in population ecology of the Common Tern. Condor 1997; 99: 534-8.
[http://dx.doi.org/10.2307/1369963] , 39Becker PH, Wendeln H, Gonzalez-Solis J. Population dynamics, recruitment, individual quality and reproductive strategies in Common Terns Sterna hirundo marked with transponders. Ardea 2001; 89: 241-52.]. The widely used parentage program COLONY 2.0 [40Jones OR, Wang J. COLONY: a program for parentage and sibship inference from multilocus genotype data. Mol Ecol Resour 2010; 10(3): 551-5.
[http://dx.doi.org/10.1111/j.1755-0998.2009.02787.x] [PMID: 21565056] ] was used. The test data set comprised samples of a known social pedigree. Since 1992 the social pedigree of the Banter Lake tern colony has been investigated via an ingenious registration system that enables a completely monitored colony. Adults and chicks are marked with subcutaneous transponders that can be recorded automatically at the breeding sites [38Becker PH, Wendeln H. A new application for transponders in population ecology of the Common Tern. Condor 1997; 99: 534-8.
[http://dx.doi.org/10.2307/1369963] ]. Numerous different studies dealing with population ecology have been derived from these data [41Becker PH, Zhang H. Renesting of Common Terns Sterna hirundo in the life history perspective. J Ornithol 2011; 152: 213-25.
[http://dx.doi.org/10.1007/s10336-010-0639-0] -44Wendeln H, Becker PH. Effects of parental quality and effort on the reproduction of Common Turns. J Anim Ecol 1999; 68: 205-14.
[http://dx.doi.org/10.1046/j.1365-2656.1999.00276.x] ]. By investigating this well studied colony, we were able to compare a data set of known kinships (chicks and their social parents) with genetically assigned ones. Furthermore, we wanted to learn about required quality of genotyping data and the reliability of the used parentage program COLONY. This software is able to assign missing parents even without parental genotypes. It defines parents with a symbol (‘#’ for mothers and ‘*’ for fathers) and a consecutive number, which enables sibship determination. For our quality test, we conducted six different parentage tests and compared them with our known pedigree. Data sets for these tests varied in their completeness of available parental genotypes so that COLONY had to construct the pedigree with more or less complete genetic information. Sampling of genetic material is often difficult in wild animals and achieving material from complete populations is often far from being realistic. Hence, assignment results of these competing data sets may give us an idea about the required amount of parental genotypes in parentage assignment tests for future studies. Moreover it could enable researchers to interpret and evaluate likelihood-based parentage studies in general conducted with COLONY. The development of new species-specific STR markers for the Common Tern now also facilitates a long-term population genetic study of this colony, to investigate for example heterozygosity, fitness and inbreeding, among many other topics. New microsatellite markers will help to determine the population genetic structure of the Common Tern such as it has already been done for the endangered Roseate Tern, Sterna dougallii, in the northwestern Atlantic and western Australia [45Szczys P, Hughes CR, Kesseli RV. Novel microsatellite markers used to determine the population genetic structure of the endangered Roseate Tern, Sterna dougallii, in Northwest Atlantic and Western Australia. Conserv Genet 2005; 6: 461-6.
[http://dx.doi.org/10.1007/s10592-004-4975-6] , 46Szczys P, Nisbet IC, Wingate DB. Conservation genetics of the Common Tern (Sterna hirundo) in the North Atlantic region; implications for the critically endangered population at Bermuda. Conserv Genet 2012; 13: 1039-43.
[http://dx.doi.org/10.1007/s10592-012-0351-0] ].
MATERIALS AND METHODOLOGY
Blood Sampling
DNA samples from juvenile and adult Common Terns were collected in a well-monitored colony in the harbour area of Wilhelmshaven (Banter Lake, German North Sea coast). During the breeding season, samples from non-fledged nestlings were obtained from blood of quills from body feathers plucked before fledging or from tissue samples collected from chicks found dead on the ground. Triatomine bugs, Dipetalogaster maxima Uhler, 1894 (Heteroptera, Reduviidae) were used to sample blood from incubating adult birds [47Arnold JM, Oswald SA, Voigt CC, et al. Taking the stress out of blood collection: comparison of field blood-sampling techniques for analysis of baseline corticosterone. J Avian Biol 2008; 39: 588-92.
[http://dx.doi.org/10.1111/j.0908-8857.2008.04265.x] , 48Becker PH, Voigt CC, Arnold JM, Nagel R. A non-invasive technique to bleed incubating birds without trapping: a blood-sucking bug in a hollow egg. J Ornithol 2006; 147: 115-8.
[http://dx.doi.org/10.1007/s10336-005-0027-3] ]. Since most of the breeding pairs were equipped with a transponder, we were able to identify the social parents for each chick. Feathers, blood samples and tissue material from dead birds were stored in an EDTA buffer (10% EDTA, 0.5% NaF, 0.5% thymol, 1%, Tris-HCL, pH = 7.5) at 4 °C until DNA extraction.
DNA Extraction, 454 Pyrosequencing and Primer Development
For DNA extraction we used a standard proteinase K (Merck, Darmstadt) protocol [49Sambrook J, Fritsch EF, Maniatis T. Moleculer clooning: a labaratuary manual. New York: Cold Spring Harbor Laboratory Press 1989.]. 454 shot-gun pyrosequencing (NGS) on a GS Junior sequencer (454 Life Sciences/Roche Applied Science) was used to create a genomic library of the Common Tern, using the GS FLX Titanium Rapid Library Preparation Kit following the manufacturer’s recommendations. Starting material for the sequencing library was 500 ng genomic DNA. Emulsion PCR was carried out with a ratio of two DNA copies per bead. A single sequencing run yielded 56,755 reads with an average read length of 391 (±165) base pairs, totaling ~22.19 Mb of sequence data. Sequence reads were scanned with MSATCOMMANDER 0.8.1 [50Faircloth BC. msatcommander: detection of microsatellite repeat arrays and automated, locus-specific primer design. Mol Ecol Resour 2008; 8(1): 92-4.
[http://dx.doi.org/10.1111/j.1471-8286.2007.01884.x] [PMID: 21585724] ] for repetitive loci. In total, 233 STR motifs (excluding mononucleotide repeats) could be identified. Using Primer3 software [51Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 2000; 132: 365-86.
[PMID: 10547847] ] we found 96 primer pairs of which 80 were unique. Of those, 48 microsatellite loci containing at least six repeats were tested for polymorphisms. Ten Common Tern DNA samples were amplified by PCR under the following conditions: a PCR reaction of 25 µL contained 60 ng of total genomic DNA, 0.4 pmol/µL of each forward and reverse primer, 0.1 mM of dGTP, dCTP and dTTP, as well as 45 µM of dATP, 1.5 x PCR buffer (Bioron), 0.15 units of Top-Taq DNA polymerase (Bioron), 1 µCi [α-33P]-dATP (Amersham Biosciences) and a variable amount of mono distilled water to reach a volume of 25 µL. In a TGradient ThermoCycler (Biometra) we performed the following thermocycling program: initial denaturing for 5 min at 95 °C, followed by 38 cycles of 45 s at 95 °C, 60 s at 50–58 °C, 90 s at 72 °C, followed by a final extension step at 72 °C for 10 min and a cooling step at 15 °C for storage. PCR products were denatured (95 °C for 5 min) for electrophoresis on a vertical high-resolution polyacrylamide gel (Urea 5%) at 65 W for 1.5 h (run length ca. 40 cm). An X-ray film (Hyperfilm-MP; Amersham) was placed on the dried gel for 1–2 days for autoradiography and afterwards was developed with X-ray developer and fixer (Kodak). Six primer pairs showed no amplification products, whereas the remaining 42 amplified well. Twenty-four of them produced several different allele bands and were used for further development steps, including multiplex PCRs. In order to enlarge the marker set, we included six primers developed for Black-legged Kittiwake (Rissa tridactyla) and Red-billed Gull (Chroicocephalus scopulinus) [52Given AD, Mills JA, Baker AJ. Isolation of polymorphic microsatellite loci from the Red-billed Gull (Larus novaehollandiae scopulinus) and amplification in related species. Mol Ecol Notes 2002; 2: 416-8.
[http://dx.doi.org/10.1046/j.1471-8286.2002.00261.x] , 53Tirard C, Helfenstein F, Danchin E. Polymorphic microsatellites in the Black-legged Kittiwake Rissa tridactyla. Mol Ecol Notes 2002; 2: 431-3.
[http://dx.doi.org/10.1046/j.1471-8286.2002.00258.x] ].
Primer Labelling, Multiplex PCR and Fragment Length Analysis
Of the 24 newly developed and tested loci 11 failed to amplify properly in multiplex PCRs or showed evidence for null alleles after locus characterization and thus had to be rejected. Furthermore, we tested primers recently isolated from Roseate Terns (Sterna dougallii) that were shown to amplify in Common Terns [45Szczys P, Hughes CR, Kesseli RV. Novel microsatellite markers used to determine the population genetic structure of the endangered Roseate Tern, Sterna dougallii, in Northwest Atlantic and Western Australia. Conserv Genet 2005; 6: 461-6.
[http://dx.doi.org/10.1007/s10592-004-4975-6] , 46Szczys P, Nisbet IC, Wingate DB. Conservation genetics of the Common Tern (Sterna hirundo) in the North Atlantic region; implications for the critically endangered population at Bermuda. Conserv Genet 2012; 13: 1039-43.
[http://dx.doi.org/10.1007/s10592-012-0351-0] ]. Unfortunately these primers did not match to our multiplex PCR panels. Instead, two primers developed for Black-legged Kittiwake and Red-billed Gull [52Given AD, Mills JA, Baker AJ. Isolation of polymorphic microsatellite loci from the Red-billed Gull (Larus novaehollandiae scopulinus) and amplification in related species. Mol Ecol Notes 2002; 2: 416-8.
[http://dx.doi.org/10.1046/j.1471-8286.2002.00261.x] , 53Tirard C, Helfenstein F, Danchin E. Polymorphic microsatellites in the Black-legged Kittiwake Rissa tridactyla. Mol Ecol Notes 2002; 2: 431-3.
[http://dx.doi.org/10.1046/j.1471-8286.2002.00258.x] ] performed well in our test PCRs and were included in the marker set (see result part). With these resulting 15 loci, we established three multiplex PCR panels, each containing five different primer pairs. All forward primers were labeled at the 5’ end with one of three different fluorescent dyes (6-FAM and HEX produced by Eurofins MWG Operon, as well as NED produced by Applied Biosystems). Primers with overlapping allele size ranges were labeled with different dyes, whereas those with non-overlapping ranges were labeled with the same dye.
We used Type-it Microsatellite PCR Kit (Qiagen) for Multiplex PCRs under the following conditions: a reaction volume of 15 µL contained 0.09–0.24 pmol/µL of each forward and reverse primer, 15 ng of total DNA, 7.5 µL of 2x Type-it Multiplex PCR Master Mix and a variable amount of RNase-free water to reach the end-volume of 15 µL. Thermocycling started with an initial denaturing step at 95 °C for 5 min, followed by 29 cycles of 30 s at 95 °C, 90 s at 58 °C for each multiplex set and 72 °C for 30 s, a final elongation step at 60 °C for 30 min and a cooling step at 4 °C.
A 96-well plate of diluted PCR products (2.5 µL PCR product mixed with 7.5 µL sterile filtrated mono distilled water) was analyzed on a Applied Biosystems DNA-Analyzer (ABI 3730) by GATC, Köln, Germany. As internal size standard ET-ROX 500 (Amersham Biosciences) was added to each sample. Peak Scanner Software 2 (Applied Biosystems) was used to analyze result files, which produces decimal numbers for each fragment. We wrote a script in R v3.1.0 [54Team R Core. A language and environment for statistical computing. R Foundation for Statistical Computing 2014, Vienna, Austria, 2014. Available from: http://wwwR-projectorg/ ] to round the decimal numbers for each allele to full allele units in a more objective way. To first define the expected number of alleles per locus, we sorted all occurring unrounded alleles at a specific locus per size and plotted them in a diagram. Allele units were defined when significant gaps between fragment sizes became clear [17Guichoux E, Lagache L, Wagner S, et al. Current trends in microsatellite genotyping. Mol Ecol Resour 2011; 11(4): 591-611.
[http://dx.doi.org/10.1111/j.1755-0998.2011.03014.x] [PMID: 21565126] ]. For each marker, rounding was performed by comparing each allele with the previous one by taking into account the locus-specific repeat-motif length and a pre-defined minimum discriminatory distance. If the distance between two subsequently compared alleles was smaller than the defined distinction distance, both alleles were rounded to the same value. On the contrary, a larger value between two compared alleles resulted in assigning two different alleles. To achieve an accurate rounding result we also manually compared the received rounded alleles with the sizes gained from Peak Scanner Software 2.
STR Characterization and Parentage Assessments
For STR characterization we used 46 adult Common Tern samples. Parentage assignment was conducted with these parents and 85 of their chicks. Samples were collected in the breeding seasons of 2000–2012 and corresponded to 23 different families; they included both parents and their offspring that were produced in the study period. The number of chicks per family varied between a single one and 11 individuals. 56.5% of families produced at least three chicks. All of the analyzed samples were fully genotyped, which means that all alleles of the 15 markers were determined.
For single locus characterization we used CERVUS 3.0 [55Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 2007; 16(5): 1099-106.
[http://dx.doi.org/10.1111/j.1365-294X.2007.03089.x] [PMID: 17305863] ] to estimate number of alleles (Na), observed and expected heterozygosity values (Hobs and Hexp) as well as polymorphism information content (PIC). Moreover, the program was used to estimate PIC, non-exclusion probabilities for parents, individual identity and sib identity probabilities for the whole marker set. Genepop V4 [56Rousset F. genepop007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour 2008; 8(1): 103-6.
[http://dx.doi.org/10.1111/j.1471-8286.2007.01931.x] [PMID: 21585727] ] was used to calculate exact values for Hardy-Weinberg expectations (HWE) with a Bonferroni correction for multiple comparisons (probability test: dememorization steps: 100000, batches: 500, iterations per batch: 10000) and to estimate null allele frequencies. PowerStats V12 (Promega Corporation 2000) was used to determine power of exclusion values (PE) for each locus and across all loci. COLONY 2.0 [40Jones OR, Wang J. COLONY: a program for parentage and sibship inference from multilocus genotype data. Mol Ecol Resour 2010; 10(3): 551-5.
[http://dx.doi.org/10.1111/j.1755-0998.2009.02787.x] [PMID: 21565056] ] was chosen for parentage assignment. We did not add information about full-sib relationships for computation, which is facultative in the program’s pre-setting. A polygamous mating system was assumed for both sexes in the calculation, since samples from half-sibs of different years were present in the data set. To test our markers and COLONY software for assignment quality we prepared six contrasting data sets. While genotypes for nestlings remained complete in all of the six calculations, entire used parental genotypes varied: Calculation one (23M.23F) contained all available parental genotypes, the second one (11M.23F) contained 100% of paternal genotypes but only 11 randomly selected ones for mothers. The third one (11F.23M) contained 100% of maternal genotypes but only 11 randomly selected ones for fathers. Calculation four (0M.23F) contained only paternal genotypes while data set 0F.23M only maternal ones and data set 0M.0F no parental genotypes at all. Assignment results concerning correct allocation of nest mates and parent pairs were compared with information from the social pedigree. A family was defined according to chicks that were sampled in the same nest and their social parents that were identified through transponder readings. Additionally to COLONY software we confirmed the parentage assignments by eye and checked for sex-linked loci. This was a necessary controlling step regarding the reliability of the new marker set.
RESULTS
By means of next-generation sequencing 13 new polymorphic STR loci could be isolated for the Common Tern. We added two markers (locus K32, isolated from Black-legged Kittiwake [53Tirard C, Helfenstein F, Danchin E. Polymorphic microsatellites in the Black-legged Kittiwake Rissa tridactyla. Mol Ecol Notes 2002; 2: 431-3.
[http://dx.doi.org/10.1046/j.1471-8286.2002.00258.x] ] and locus RBG18, isolated from Red-billed Gull [52Given AD, Mills JA, Baker AJ. Isolation of polymorphic microsatellite loci from the Red-billed Gull (Larus novaehollandiae scopulinus) and amplification in related species. Mol Ecol Notes 2002; 2: 416-8.
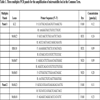
[http://dx.doi.org/10.1046/j.1471-8286.2002.00261.x] ]) and established three different multiplex panels, containing together 15 loci (Table 1).
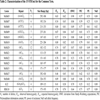
In Table 2 we characterized each locus. Mean number of alleles per locus is 8 and a calculation of PIC across all loci revealed a value of 0.67. Only three loci (MsSh10, MsSh37 and K32) showed PIC values below 0.50. Significant deviation from Hardy-Weinberg expectations was found for MsSh18 and MsSh23. Furthermore, null alleles may exist in locus MsSh07, MsSh20, MsSh21, MsSh23 and MsSh31, when setting the acceptance boundary to 0.05. None of the loci appeared to be sex-linked. Sex-linked loci would result in accumulated homozygote alleles in one sex, while the other one mainly would appear heterozygote.
Parentage Analysis
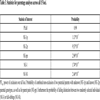
Different statistics for parentage analyses and identity tests are represented in Table 3. A power of exclusion (PE) value over all loci of about 0.99 was reached. The inclusion of parental genotypes in kinship calculation leads to a much higher probability of excluding an unrelated candidate parent from parentage of an arbitrary offspring than without including parental genotypes. We assume that individual identities (NE-I) and full-sibling relationships (NE-SI) are highly reliable.
We analyzed parentage of 85 juvenile Common Terns and corresponding 46 parents (23 females and 23 males). In order to test the program as well as the suitability of our marker set, six assignment runs were conducted. Results are shown in Table 4, where each run differs by the inclusion of involved parental genotypes. For the case of providing all maternal and paternal genotypes (23M, 23F), COLONY 2.0 identified 23 different parent-chick families with their expected sibship and parent-pair combinations. By omitting 12 maternal genotypes (11M, 23F) maternal and parent pair assignment became incorrect for 1.2% of all chicks, while nest mates and fathers were correctly assigned to 100%. The erroneous assignment corresponds to one nest with only one chick. For the case of omitting 12 paternal genotypes from calculation (11F, 23M), only maternal genotypes could be correctly assigned for 100% of the cases. In two nests with two chicks the assignments failed. When supplying only genotypes of a single parental sex (0M, 23F and 0F, 23M), assignment was accurate for the supplied one but incorrect for the other sex and parent-pair combinations in 3.5% of all chicks. In total, 91.3% of nest mates were assorted correctly in both cases. In the first calculation, assignment was incorrect for one nest with one chick, one nest with two chicks and one nest with three chicks. For the second calculation mistakes were found in two nests with two chicks. Finally, assignment results became more prone to errors when no parental genotypes were provided at all. Additionally to the use of COLONY, we confirmed the parentage assignments by eye. Mismatches among genotypes of expected parents were not present.
DISCUSSION AND CONCLUSION
Using 454 pyrosequencing we identified 13 new loci that provide a useful tool for genetic analyses and parentage tests of Common Terns. Only three loci showed PIC values less than 0.5, which implies a moderate polymorphic-information content for them. Nevertheless, the whole marker set is highly informative, due to an overall high PIC value and high discrimination probabilities for parents and sibs. Genepop software revealed deviation from Hardy-Weinberg expectations for two loci and a slight to moderate possibility of existing null alleles for five loci. We interpret these findings as a result of the small initial colony size. The colony originated from a small founder population so that inbreeding is possible [57Ludwig S, Becker PH. Immigration prevents inbreeding in a growing colony of a long-lived and philopatric seabird. Ibis 2012; 154: 74-84.
[http://dx.doi.org/10.1111/j.1474-919X.2011.01199.x] ]. Nevertheless, the present marker set is of very good quality, since identification of individuals, siblings and parent-pairs is given. This is evidenced by our comparison of expected (social) and assigned (genetic) kinships, when varying the amount of included parental genotypes. The comparison of our six test data sets shows that, by providing genetic information of all possible parents, kinship can be resolved to a very high degree and resamples the expected pedigree. Without any parental genotypes, assignment result for nest mates, single parents as well as for parent pairs is less reliable, compared to the known pedigree. This finding is not surprising, since relatedness is normally much higher in small colonies (such as the investigated one) than in large populations. A moderate sampling success of all possible parents where at least one parent of a pair is available or one sex could be sampled completely is the most realistic scenario for many population studies. Under this condition, we achieved in our test data sets a value of 96.5–100% correctness for single and parent pairs. In that way, we conclude that our new marker set is able to discriminate adequate between closely related individuals under realistic sampling conditions. Therefore, we recommend the following points to enable correct assignment results in wild populations and without known pedigree information: Development of species specific, high quality markers, utilization of fully genotyped samples without missing alleles, supplemented as much as possible by corresponding parental genotypes. Furthermore, families having at least three chicks and at least one parental genotype very likely produce unequivocal identifications. Adhering to such a strict data selection the COLONY software should be able to assign the correct missing parent in most cases. Hence, correct full- and half-sibship determination will become possible. Since in most biological studies, availability of parental genotypes is the most crucial point and samples of parent pairs are often incomplete, COLONY software is a powerful program to identify the missing parent.
The three multiplex STR panels will provide a cost and time saving approach to genotype large numbers of individuals of the Common Tern. They will supplement and expand the research on the social pedigree of the breeding colony on the Banter Lake. Our aim is to genotype thousands of samples in a long-term population study. Together with the automated transponder based identification system, it will enable us to receive a more detailed insight into breeding system, life-time reproductive success, kinship and family structures within this colony.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
ACKNOWLEDGEMENTS
We are grateful to the many volunteers from Institute of Avian Research “Vogelwarte Helgoland”, Wilhelmshaven, Germany, for their aid and support during field work. We thank Götz Wagenknecht for DNA extraction. Sequencing was financed within the project “GenoSeq” funded by the Ministry of Science, Research and Culture of the Federal State of Brandenburg (Germany) in the EFRE program “Science and technology transfer for innovation” (FKZ 80143246). The studies on Common Tern population ecology were funded by the DFG (BE 916/5 to 9). G. Schaub (Ruhr University Bochum, Germany) provided the bugs for blood sampling of adult birds. We also thank all apprentices in the lab of M. Wink for helping with primer development and genotyping.
REFERENCES
| [1] | Liminana R, Garcia JT, Gonzalez JM, et al. Philopatry and natal dispersal of Montagu's harriers (Circus pygargus) breeding in Spain: a review of existing data. Eur J Wildl Res 2012; 58: 549-55. [http://dx.doi.org/10.1007/s10344-011-0602-2] |
| [2] | Liminana R, Soutullo A, Urios V. Autumn migration of Montagu's harriers Circus pygargus tracked by satellite telemetry. J Ornithol 2007; 148: 517-23. [http://dx.doi.org/10.1007/s10336-007-0182-9] |
| [3] | Trierweiler C, Klaassen RH, Drent RH, et al. Migratory connectivity and population-specific migration routes in a long-distance migratory bird. Proc R Soc Biol Sci Ser B. 281. [http://dx.doi.org/10.1098/rspb.2013.2897] |
| [4] | Trierweiler C, Mullié WC, Drent RH, et al. A Palaearctic migratory raptor species tracks shifting prey availability within its wintering range in the Sahel. J Anim Ecol 2013; 82(1): 107-20. [http://dx.doi.org/10.1111/j.1365-2656.2012.02036.x] [PMID: 23137184] |
| [5] | Authier M, Peron C, Mante A, Vidal P, Gremillet D. Designing observational biologging studies to assess the causal effect of instrumentation. Methods Ecol Evol 2013; 4: 802-10. [http://dx.doi.org/10.1111/2041-210X.12075] |
| [6] | Trefry SA, Diamond AW, Jesson LK. Wing marker woes: a case study and meta-analysis of the impacts of wing and patagial tags. J Ornithol 2013; 154: 1-11. [http://dx.doi.org/10.1007/s10336-012-0862-y] |
| [7] | Dann P, Sidhu LA, Jessop R, et al. Effects of flipper bands and injected transponders on the survival of adult Little Penguins Eudyptula minor. Ibis 2014; 156: 73-83. [http://dx.doi.org/10.1111/ibi.12122] |
| [8] | Vassart M. New tools in conservation genetics. Rev Med Vet (Toulouse) 1995; 146: 171-80. |
| [9] | Briggs CW, Collopy MW. Extra-pair paternity in Swainson's Hawks. J Field Ornithol 2012; 83: 41-6. [http://dx.doi.org/10.1111/j.1557-9263.2011.00354.x] |
| [10] | Fridolfsson A-K, Gyllensten UB, Jakobsson S. Microsatellite markers for paternity testing in the Willow Warbler Phylloscopus trochilus: high frequency of extra-pair young in an island population. Hereditas 1997; 126: 127-32. [http://dx.doi.org/10.1111/j.1601-5223.1997.00127.x] |
| [11] | Martínez JJ, de Aranzamendi MC, Masello JF, Bucher EH. Genetic evidence of extra-pair paternity and intraspecific brood parasitism in the monk parakeet. Front Zool 2013; 10(1): 68. [http://dx.doi.org/10.1186/1742-9994-10-68] [PMID: 24209709] |
| [12] | Schmoll T, Winkel W, Lubjuhn T. Molecular genetic evidence for mixed maternity in broods of the Coal Tit Parus ater. Vogelwarte 2008; 46: 223-7. |
| [13] | Sunnucks P. Efficient genetic markers for population biology. Trends Ecol Evol (Amst) 2000; 15(5): 199-203. [http://dx.doi.org/10.1016/S0169-5347(00)01825-5] [PMID: 10782134] |
| [14] | Jarne P, Lagoda PJ. Microsatellites, from molecules to populations and back. Trends Ecol Evol (Amst) 1996; 11(10): 424-9. [http://dx.doi.org/10.1016/0169-5347(96)10049-5] [PMID: 21237902] |
| [15] | Herráeza DL, Schäfer H, Mosner J, Fries H-R, Wink M. Comparison of microsatellite and single nucleotide polymorphism markers for the genetic analysis of a Galloway cattle population. Z Naturforsch, C, J Biosci 2005; 60(7-8): 637-43. [PMID: 16163842] |
| [16] | Gardner MG, Fitch AJ, Bertozzi T, Lowe AJ. Rise of the machinesrecommendations for ecologists when using next generation sequencing for microsatellite development. Mol Ecol Resour 2011; 11(6): 1093-101. [http://dx.doi.org/10.1111/j.1755-0998.2011.03037.x] [PMID: 21679314] |
| [17] | Guichoux E, Lagache L, Wagner S, et al. Current trends in microsatellite genotyping. Mol Ecol Resour 2011; 11(4): 591-611. [http://dx.doi.org/10.1111/j.1755-0998.2011.03014.x] [PMID: 21565126] |
| [18] | Santana Q, Coetzee M, Steenkamp E, et al. Microsatellite discovery by deep sequencing of enriched genomic libraries. Biotechniques 2009; 46(3): 217-23. [http://dx.doi.org/10.2144/000113085] [PMID: 19317665] |
| [19] | Brennan PL, Hyseni C. Development of microsatellite markers for parentage analysis in the great tinamou (Tinamus major). Mol Ecol Resour 2008; 8(4): 933-5. [http://dx.doi.org/10.1111/j.1755-0998.2008.02122.x] [PMID: 21585934] |
| [20] | Busch JD, Katzner TE, Bragin E, Keims P. Tetranucleotide microsatellites for Aquila and Haliaeetus eagles. Mol Ecol Notes 2005; 5: 29-41. [http://dx.doi.org/10.1111/j.1471-8286.2004.00823.x] |
| [21] | Ferreira JM, Burnham-Curtis M, Schunck F, et al. Development of novel polymorphic microsatellite markers for four bird species exploited by the illegal wildlife trade in Brazil. Conserv Genet Resour 2015; 7: 435-6. [http://dx.doi.org/10.1007/s12686-014-0389-z] |
| [22] | King AG, Cooke GM, Johnson RN, Major RE. Development and multiplex genotyping of eighteen novel microsatellite markers for a threatened saltmarsh bird Epthianura albifrons (Meliphagidae). Conserv Genet Resour 2012; 4: 893-6. [http://dx.doi.org/10.1007/s12686-012-9667-9] |
| [23] | Martínez-Cruz B, David VA, Godoy JA, et al. Eighteen polymorphic microsatellite markers fort the highly endangered Spanish imperial eagle (Aquila adalberti) and related species. Mol Ecol Notes 2002; 2: 323-6. [http://dx.doi.org/10.1046/j.1471-8286.2002.00231.x] |
| [24] | Steeves TE, Hale ML, Gemmell NJ. Development of polymorphic microsatellite markers for the New Zealand black stilt (Himantopus novaezelandiae) and cross-amplification in the pied stilt (Himantopus himantopus leucocephalus). Mol Ecol Resour 2008; 8(5): 1105-7. [http://dx.doi.org/10.1111/j.1755-0998.2008.02170.x] [PMID: 21585985] |
| [25] | Tingay RE, Dawson D, Pandhal J, et al. Isolation of 22 new Haliaeetus microsatellite loci and their characterization in the critically endangered Madagascar fish-eagle (Haliaeetus vociferoides) and three other Haliaeetus eagle species. Mol Ecol Notes 2007; 7: 711-5. [http://dx.doi.org/10.1111/j.1471-8286.2007.01690.x] |
| [26] | Topinka JR, May B. Development of polymorphic microsatellite loci in the northern goshawk (Accipiter gentilis) and cross-amplification in other raptor species. Conserv Genet 2004; 5: 861-4. [http://dx.doi.org/10.1007/s10592-004-1973-7] |
| [27] | Babb PL, McIntosh AM, Fernandez-Duque E, Di Fiore A, Schurr TG. An optimized microsatellite genotyping strategy for assessing genetic identity and kinship in Azaras owl monkeys (Aotus azarai). Folia Primatol (Basel) 2011; 82(2): 107-17. [http://dx.doi.org/10.1159/000330564] [PMID: 21912137] |
| [28] | Durrant KL, Hughes JM. Differing rates of extra-group paternity between two populations of the Australian magpie (Gymnorhina tibicen). Behav Ecol Sociobiol 2005; 57: 536-45. [http://dx.doi.org/10.1007/s00265-004-0883-5] |
| [29] | Overeem KR, Gabriel PO, Zirpoli JA, Black JM. Steller sex: infidelity and sexual selection in a social Corvid (Cyanocitta stelleri). PLoS One 2014; 9(8): e105257. [http://dx.doi.org/10.1371/journal.pone.0105257] [PMID: 25148039] |
| [30] | Sakaoka K, Suzuki I, Kasugai N, Fukumoto Y. Paternity testing using microsatellite DNA markers in captive Adélie penguins (Pygoscelis adeliae). Zoo Biol 2014; 33(5): 463-70. [PMID: 25157452] |
| [31] | Seddon N, Amos W, Adcock G, et al. Mating system, philopatry and patterns of kinship in the cooperatively breeding subdesert mesite Monias benschi. Mol Ecol 2005; 14(11): 3573-83. [http://dx.doi.org/10.1111/j.1365-294X.2005.02675.x] [PMID: 16156824] |
| [32] | Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics 2000; 155(2): 945-59. [PMID: 10835412] |
| [33] | Stumpf KJ, Theimer TC, Mcleod MA, Koronkiewicz TJ. Disparity in population structuring of Southwestern Willow Flycatchers based on geographic distance, movement patterns, and genetic analyses. J Exp Zool A Ecol Genet Physiol 2014; 321: 577-85. |
| [34] | Albayrak T, Gonzalez J, Drovetski SV, Wink M. Phylogeography and population structure of Kruper's Nuthatch Sitta krueperi from Turkey based on microsatellites and mitochondrial DNA. J Ornithol 2012; 153: 405-11. [http://dx.doi.org/10.1007/s10336-011-0756-4] |
| [35] | Barrientos R, Kvist L, Barbosa A, et al. Refugia, colonization and diversification of an arid-adapted bird: coincident patterns between genetic data and ecological niche modelling. Mol Ecol 2014; 23(2): 390-407. [http://dx.doi.org/10.1111/mec.12588] [PMID: 24215522] |
| [36] | Song G, Yu LJ, Gao B, et al. Gene flow maintains genetic diversity and colonization potential in recently range-expanded populations of an Oriental bird, the Light-vented Bulbul (Pycnonotus sinensis, Aves: Pycnonotidae). Divers Distrib 2013; 19: 1248-62. [http://dx.doi.org/10.1111/ddi.12067] |
| [37] | Gerber S, Mariette S, Streiff R, Bodénès C, Kremer A. Comparison of microsatellites and amplified fragment length polymorphism markers for parentage analysis. Mol Ecol 2000; 9(8): 1037-48. [http://dx.doi.org/10.1046/j.1365-294x.2000.00961.x] [PMID: 10964223] |
| [38] | Becker PH, Wendeln H. A new application for transponders in population ecology of the Common Tern. Condor 1997; 99: 534-8. [http://dx.doi.org/10.2307/1369963] |
| [39] | Becker PH, Wendeln H, Gonzalez-Solis J. Population dynamics, recruitment, individual quality and reproductive strategies in Common Terns Sterna hirundo marked with transponders. Ardea 2001; 89: 241-52. |
| [40] | Jones OR, Wang J. COLONY: a program for parentage and sibship inference from multilocus genotype data. Mol Ecol Resour 2010; 10(3): 551-5. [http://dx.doi.org/10.1111/j.1755-0998.2009.02787.x] [PMID: 21565056] |
| [41] | Becker PH, Zhang H. Renesting of Common Terns Sterna hirundo in the life history perspective. J Ornithol 2011; 152: 213-25. [http://dx.doi.org/10.1007/s10336-010-0639-0] |
| [42] | Bouwhuis S, Vedder O, Becker PH. Sex-specific pathways of parental age effects on offspring lifetime reproductive success in a long-lived seabird. Evolution 2015; 69(7): 1760-71. [http://dx.doi.org/10.1111/evo.12692] [PMID: 26095174] |
| [43] | Gonzalez-Solis J, Wendeln H, Becker PH. Within and between season nest-site and mate fidelity in Common Terns (Sterna hirundo). J Ornithol 1999; 140: 491-8. [http://dx.doi.org/10.1007/BF01650993] |
| [44] | Wendeln H, Becker PH. Effects of parental quality and effort on the reproduction of Common Turns. J Anim Ecol 1999; 68: 205-14. [http://dx.doi.org/10.1046/j.1365-2656.1999.00276.x] |
| [45] | Szczys P, Hughes CR, Kesseli RV. Novel microsatellite markers used to determine the population genetic structure of the endangered Roseate Tern, Sterna dougallii, in Northwest Atlantic and Western Australia. Conserv Genet 2005; 6: 461-6. [http://dx.doi.org/10.1007/s10592-004-4975-6] |
| [46] | Szczys P, Nisbet IC, Wingate DB. Conservation genetics of the Common Tern (Sterna hirundo) in the North Atlantic region; implications for the critically endangered population at Bermuda. Conserv Genet 2012; 13: 1039-43. [http://dx.doi.org/10.1007/s10592-012-0351-0] |
| [47] | Arnold JM, Oswald SA, Voigt CC, et al. Taking the stress out of blood collection: comparison of field blood-sampling techniques for analysis of baseline corticosterone. J Avian Biol 2008; 39: 588-92. [http://dx.doi.org/10.1111/j.0908-8857.2008.04265.x] |
| [48] | Becker PH, Voigt CC, Arnold JM, Nagel R. A non-invasive technique to bleed incubating birds without trapping: a blood-sucking bug in a hollow egg. J Ornithol 2006; 147: 115-8. [http://dx.doi.org/10.1007/s10336-005-0027-3] |
| [49] | Sambrook J, Fritsch EF, Maniatis T. Moleculer clooning: a labaratuary manual. New York: Cold Spring Harbor Laboratory Press 1989. |
| [50] | Faircloth BC. msatcommander: detection of microsatellite repeat arrays and automated, locus-specific primer design. Mol Ecol Resour 2008; 8(1): 92-4. [http://dx.doi.org/10.1111/j.1471-8286.2007.01884.x] [PMID: 21585724] |
| [51] | Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 2000; 132: 365-86. [PMID: 10547847] |
| [52] | Given AD, Mills JA, Baker AJ. Isolation of polymorphic microsatellite loci from the Red-billed Gull (Larus novaehollandiae scopulinus) and amplification in related species. Mol Ecol Notes 2002; 2: 416-8. [http://dx.doi.org/10.1046/j.1471-8286.2002.00261.x] |
| [53] | Tirard C, Helfenstein F, Danchin E. Polymorphic microsatellites in the Black-legged Kittiwake Rissa tridactyla. Mol Ecol Notes 2002; 2: 431-3. [http://dx.doi.org/10.1046/j.1471-8286.2002.00258.x] |
| [54] | Team R Core. A language and environment for statistical computing. R Foundation for Statistical Computing 2014, Vienna, Austria, 2014. Available from: http://wwwR-projectorg/ |
| [55] | Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 2007; 16(5): 1099-106. [http://dx.doi.org/10.1111/j.1365-294X.2007.03089.x] [PMID: 17305863] |
| [56] | Rousset F. genepop007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour 2008; 8(1): 103-6. [http://dx.doi.org/10.1111/j.1471-8286.2007.01931.x] [PMID: 21585727] |
| [57] | Ludwig S, Becker PH. Immigration prevents inbreeding in a growing colony of a long-lived and philopatric seabird. Ibis 2012; 154: 74-84. [http://dx.doi.org/10.1111/j.1474-919X.2011.01199.x] |