- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Systems Biology Journal
(Discontinued)
ISSN: 1876-3928 ― Volume 5, 2014
Ureaplasma Urease Genes have Undergone a Unique Evolutionary Process
Hiromi Nishida*
Abstract
Ureaplasma, a member of mycoplasmas, has a unique ATP synthesis system, which is coupled to the urea hydrolysis. Urease catalyzes the hydrolysis of urea into carbon dioxide and ammonia. Phylogenetic analyses of the urease genes indicated that Ureaplasma urease genes were not gained by recent horizontal transfer and have a unique evolutionary process. Ureaplasma unique ATP synthesis system leaded to breakdown of the glycolysis pathway. Some glycolytic genes are absent and some glycolytic genes are evolving under relaxed selection in Ureaplasma. Probably glycolytic genes can be used as an indicator of ATP synthesis system. Thus, the organisms that have incomplete glycolysis pathway or glycolytic genes evolving under relaxed selection would have an ATP synthesis system independently of the glycolysis.
Article Information
Identifiers and Pagination:
Year: 2009Volume: 2
First Page: 1
Last Page: 7
Publisher Id: TOSYSBJ-2-1
DOI: 10.2174/1876392800902010001
Article History:
Received Date: 1/12/2008Revision Received Date: 16/12/2008
Acceptance Date: 17/12/2008
Electronic publication date: 6/1/2009
Collection year: 2009
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http: //creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Agricultural Bioinformatics Research Unit, Graduate School of Agricultural and Life Sciences, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan; E-mail: hnishida@iu.a.u-tokyo.ac.jp
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 1-12-2008 |
Original Manuscript | Ureaplasma Urease Genes have Undergone a Unique Evolutionary Process | |
Mycoplasmas are widespread in nature as parasites of mammals, reptiles, fishes, arthropods, and plants [1]. During the mycoplasma evolution, gene loss has occurred frequently, resulting in very small genome size [1-3]. The reductive evolution of mycoplasmas is still in progress. The genus Ureaplasma is a member of mycoplasmas, which generates 95% of its ATP using the hydrolysis of urea [4]. Growth of Ureaplasma is dependent on urea [5]. This unique ATP synthesis is not found in the other mycoplasmas. In fact, key enzymes in the glycolytic pathway are absent in Ureaplasma [6]. In addition, some glycolytic genes of Ureaplasma are evolving under relaxed selection [7, 8]. Thus, the glycolysis pathway is collapsing in Ureaplasma.
Urea is hydrolyzed into carbon dioxide and ammonia in many organisms. However, it is unique that the urea hydrolysis is coupled to ATP synthesis in Ureaplasma [9]. This unique system of Ureaplasma leaded to breakdown of the glycolysis pathway. For example, Ureaplasma does not have any genes encoding glucose-6-phosphate isomerae [10]. It suggests that the glycolysis system had been important for ATP synthesis rather than glucose metabolism at least in Ureaplasma (Fig. 1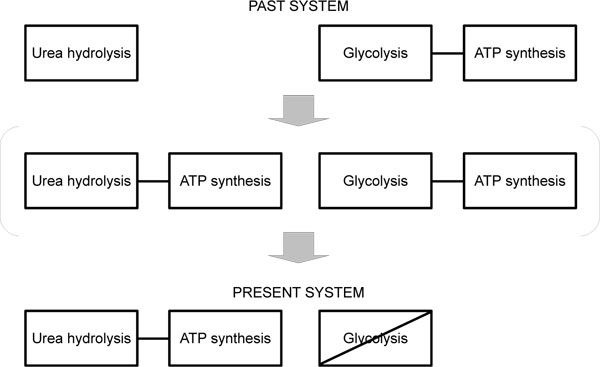 ). Generally the TCA (tricarboxylic acid) cycle is linked to the glycolysis pathway, which has played an important role in ATP synthesis of many organisms and conserved in the course of evolution. On the other hand, the urea hydrolysis is not linked to the glycolysis pathway. Ureaplasma generates ATP through the urea hydrolysis not through the glycolysis. Probably Ureaplasma glycolysis pathway was not able to be broken before the urea hydrolysis was coupled to ATP synthesis. Therefore, after the coupling of the urea hydrolysis and ATP synthesis, dominant ATP synthesis had been changed from through the glycolysis to through the urea hydrolysis during the Ureaplasma evolution (Fig. 1
). Generally the TCA (tricarboxylic acid) cycle is linked to the glycolysis pathway, which has played an important role in ATP synthesis of many organisms and conserved in the course of evolution. On the other hand, the urea hydrolysis is not linked to the glycolysis pathway. Ureaplasma generates ATP through the urea hydrolysis not through the glycolysis. Probably Ureaplasma glycolysis pathway was not able to be broken before the urea hydrolysis was coupled to ATP synthesis. Therefore, after the coupling of the urea hydrolysis and ATP synthesis, dominant ATP synthesis had been changed from through the glycolysis to through the urea hydrolysis during the Ureaplasma evolution (Fig. 1 ).
).
Urease catalyzes the hydrolysis of urea into carbon dioxide and ammonia. Urease has three core subunits, UreA, UreB, and UreC. Generally these three core subunits and the related accessory proteins UreD, UreE, UreF, and UreG are clustered on the genome. The order of the Ureaplasma (belongs to the Firmicutes) urease genes in the cluster is identical to that of the Bacillus sp. TB-90 (belongs to the Firmicutes) urease genes [11], suggesting that an ancestor of the Firmicutes had the urease gene cluster. Some species had lost some urease genes during the evolution. For example, Bacillus subtilis contains urease structural genes but lacks the accessory genes typically required for the maturation of urease [12]. On the other hand, the mycoplasmas except for Ureaplasma completely lack urease genes. I have some questions. Was the Ureaplasma urease genes (gene cluster) transferred horizontally? Or did the mycoplasmas except for Ureaplasma lose urease genes in the course of the mycoplasma evolution? Did the Ureaplasma urease have a unique evolutionary process? In order to answer these questions, I compared the amino acid sequences of urease genes of Ureaplasma with the homologous proteins in this study.
The BLAST program was used to search the GenomeNet website (http://www.genome.jp) for proteins homologous to UreA, UreB, UreC, UreD, UreE, UreF, and UreG of Ureaplasma parvum ATCC 27815 with E-value < 10-23, 10-25, 10-170, 10-35, 10-25, 10-40, and 10-77, respectively. This search led to identification of 30 UreA homologues, 32 UreB homologues, 45 UreC homologues, 17 UreD homologues, 20 UreE homologues, 20 UreF homologues, and 21 UreG homologues. The 31 UreA, 33 UreB, 46 UreC, 18 UreD, 21 UreE, 21 UreF, and 22 UreG proteins were multiple aligned using the CLUSTAL W program on the GenomeNet website. Phylogenetic trees were reconstructed, based on the multiple-alignments with complete deletion of gap sites using the neighbor-joining method of MEGA software [13] with 1000 bootstrap replicates. In addition, phylogenetic trees of UreA, UreB, and UreC proteins were reconstructed, based on the multiple alignments with complete deletion of gap sites using the maximum likelihood method of the PHYLIP program (http://evolution.genetics.washington.edu/ phylip.html) with 100 bootstrap replicates. The JTT model was used as the model of amino acid substitution. Number of times to jumble in the PROML program was 2.
The phylogenetic analyses showed that the KEGG database [14] used in this study did not have closely related protein to UreA and UreB of Ureaplasma (Figs. 2A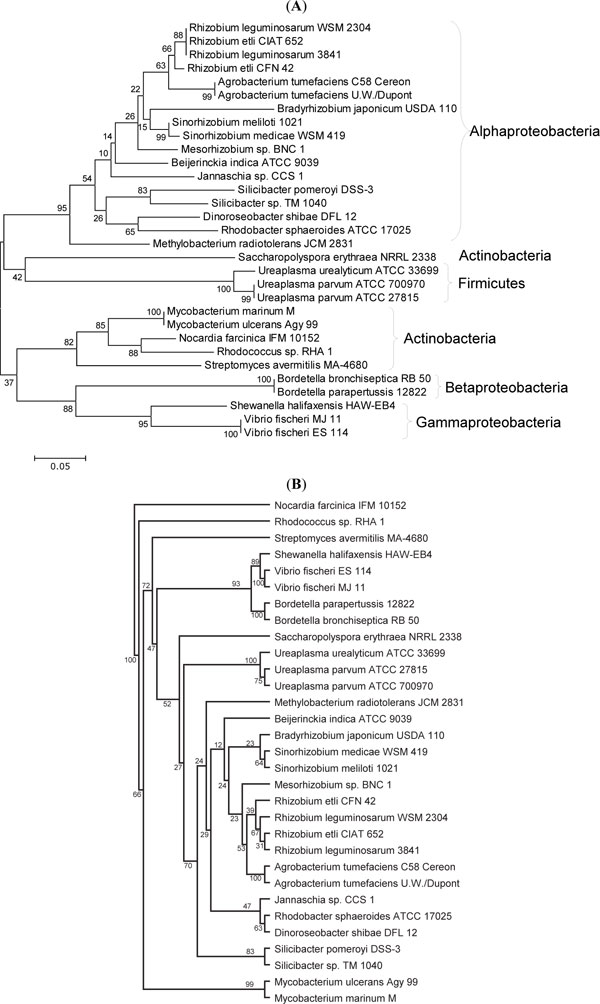 , B
, B and 3A
and 3A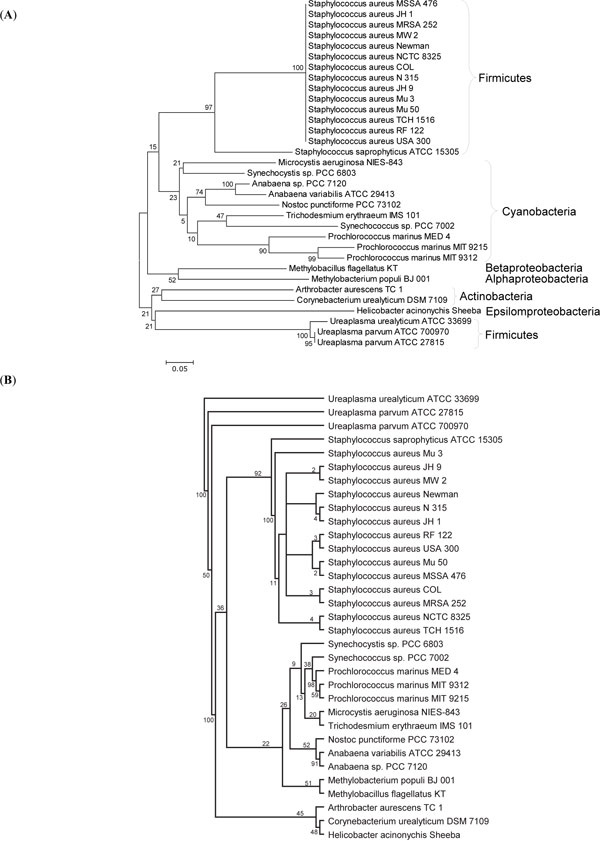 , B
, B ). On the other hand, Ureaplasma UreC is closely related to Streptococcus UreC with 93% bootstrap support in the neighbor-joining tree (Fig. 4A
). On the other hand, Ureaplasma UreC is closely related to Streptococcus UreC with 93% bootstrap support in the neighbor-joining tree (Fig. 4A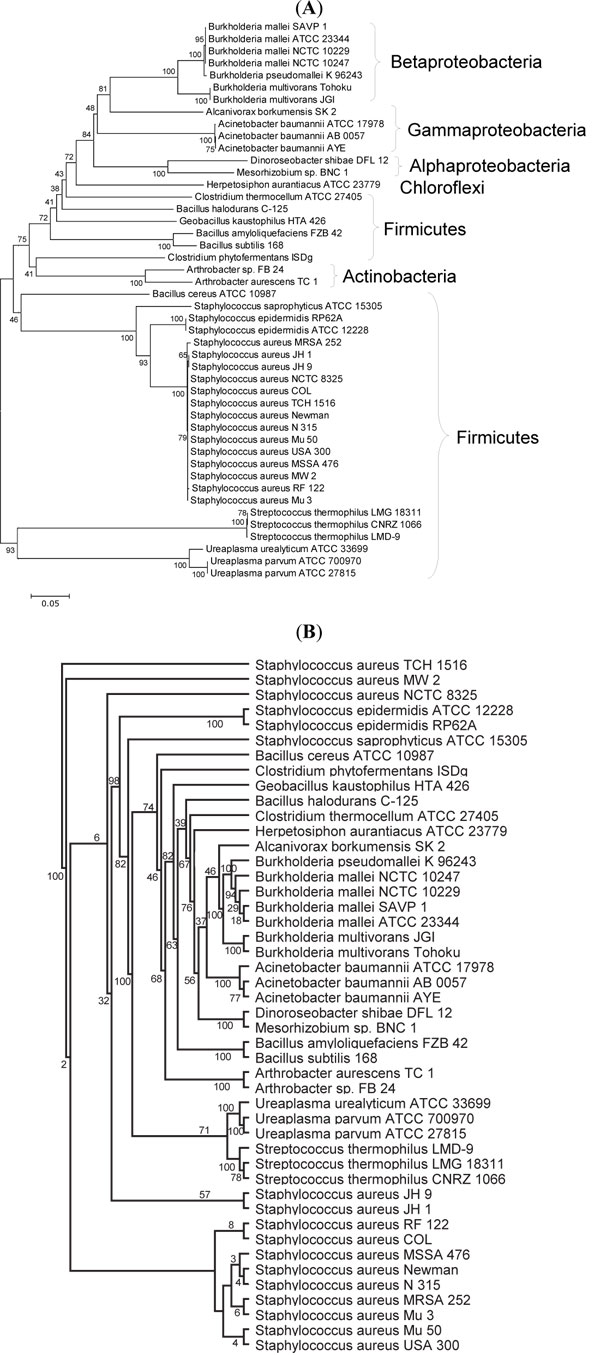 ) and 71% support in the maximum likelihood tree (Fig. 4B
) and 71% support in the maximum likelihood tree (Fig. 4B ). The diverging points of UreA, UreB, and UreC of Ureaplasma are very deep in the neighbor-joining trees (Figs. 2A
). The diverging points of UreA, UreB, and UreC of Ureaplasma are very deep in the neighbor-joining trees (Figs. 2A , 3A
, 3A , 4A
, 4A ), strongly suggesting that Ureaplasma urease genes (gene cluster) were not gained by recent horizontal transfer. It was also supported by the phylogenetic trees of urease accessory proteins (Fig. 5
), strongly suggesting that Ureaplasma urease genes (gene cluster) were not gained by recent horizontal transfer. It was also supported by the phylogenetic trees of urease accessory proteins (Fig. 5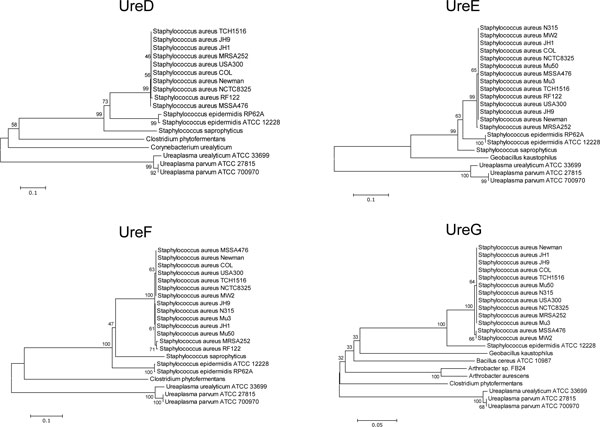 ). Interestingly, Ureaplasma UreB and UreC have Ureaplasma-specific amino acids insertions (Figs. 6
). Interestingly, Ureaplasma UreB and UreC have Ureaplasma-specific amino acids insertions (Figs. 6 , 7
, 7 ). Thus, Ureaplasma urease gene cluster has a unique evolutionary process and Ureaplasma has a unique urea hydrolysis system coupling to ATP synthesis. Maybe Ureaplasma ancestor had used urease in order to live in the urea-rich environment. During the evolution, the urea hydrolysis was coupled to ATP synthesis system. After the event, glycolysis pathway has not been essential for ATP synthesis in Ureaplasma. Some glycolytic genes are absent and some are evolving under relaxed selection in Ureaplasma. Probaly the establishment of the unique ATP synthesis system triggered those changes. If so, glycolytic genes can be used as an indicator of ATP synthesis system. Thus, the organisms that have incomplete glycolysis pathway or glycolytic genes evolving under relaxed selection would have a unique ATP synthesis system independently of the glycolysis.
). Thus, Ureaplasma urease gene cluster has a unique evolutionary process and Ureaplasma has a unique urea hydrolysis system coupling to ATP synthesis. Maybe Ureaplasma ancestor had used urease in order to live in the urea-rich environment. During the evolution, the urea hydrolysis was coupled to ATP synthesis system. After the event, glycolysis pathway has not been essential for ATP synthesis in Ureaplasma. Some glycolytic genes are absent and some are evolving under relaxed selection in Ureaplasma. Probaly the establishment of the unique ATP synthesis system triggered those changes. If so, glycolytic genes can be used as an indicator of ATP synthesis system. Thus, the organisms that have incomplete glycolysis pathway or glycolytic genes evolving under relaxed selection would have a unique ATP synthesis system independently of the glycolysis.
 |
Fig. (6) Ureaplasma UreB specific amino acids insertion region. Ureaplasma has a specific 9-amino acids insertion (positions 65-73) in the multiple alignment of 33 UreB proteins. |
 |
Fig. (7) Ureaplasma UreC specific amino acids insertion region. Ureaplasma has a specific 25-amino acids insertion (positions 554-578) in the multiple alignment of 33 UreC proteins. |





