- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Genetic Characterization of Pepino Mosaic Virus Isolates from Morocco
Amal Souiri1, 2, 3, Mustapha Zemzami3, Hayat Laatiris3, Saaid Amzazi2, Moulay M. Ennaji1, *
Abstract
Introduction:
Throughout the past few years, Pepino Mosaic Virus (PepMV) has rapidly evolved from an emerging virus to endemic pathogen that causes significant losses in tomato crops worldwide. Reliable detection and molecular characterization are very important tools to support disease control. Cross-protection can also be an effective strategy, but the efficacy depends strongly on the genotype. The genetic composition of the PepMV population in Morocco has not yet been determined.
Aims:
The current study aims to genetically characterize twelve PepMV isolates (PepMV-MA), all from different Moroccan tomato production areas, by analyzing nucleotide sequences of a part of the RNA-dependent RNA polymerase (RdRp), Triple Gene Block (TGB) and Coat Protein (CP) genes.
Results:
The sequence analysis of the twelve PepMV-MA isolates shows minor nucleotide differences between them, which implies a homogenous population. The phylogenetic analysis, based on the comparison with the major genotypes, showed that Moroccan PepMV populations share a very high sequence identity, 98%, with the Chilean strain (CH2), while the shared identity with the European strains (EU) is only 85%. Interestingly, Moroccan isolates reveal specific single nucleotide polymorphisms, some of which lead to amino acids changes. These mutations have never been described before, suggesting distinct variants that may enhance aggressiveness and symptomatology.
Conclusion:
Our careful sequence analysis and genotype determination, which placing homogenous Moroccan PepMV strains into CH2 genotype, would be a prerequisite for deploying effective cross-protection strategies for controlling the pathogen in the field.
Article Information
Identifiers and Pagination:
Year: 2019Volume: 13
First Page: 18
Last Page: 28
Publisher Id: TOVJ-13-18
DOI: 10.2174/1874357901913010018
Article History:
Received Date: 24/01/2019Revision Received Date: 20/05/2019
Acceptance Date: 22/05/2019
Electronic publication date: 31/07/2019
Collection year: 2019
open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution 4.0 International Public License (CC-BY 4.0), a copy of which is available at: https://creativecommons.org/licenses/by/4.0/legalcode. This license permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
* Address correspondence to this author at the Laboratory of Virology Microbiology, Quality and Biotechnologies/Ecotoxicology and, Biodiverity - Faculty of Science and Techniques Mohammedia, University Hassan II of Casablanca, Mohammedia 20650, Morocco;
Tel: +212661748862 / +212662013772; E-mail: m.ennaji@yahoo.fr
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 24-01-2019 |
Original Manuscript | Genetic Characterization of Pepino Mosaic Virus Isolates from Morocco | |
1. INTRODUCTION
Pepino Mosaic Virus (PepMV), genus potexvirus, family Alphaflexiviridae [1Adams MJ, Antoniw JF, Bar-Joseph M, et al. The new plant virus family Flexiviridae and assessment of molecular criteria for species demarcation. Arch Virol 2004; 149(5): 1045-60.
[http://dx.doi.org/10.1007/s00705-004-0304-0] [PMID: 15098118] , 2Martelli GP, Adams MJ, Kreuze JF, Dolja VV. Family Flexiviridae: A case study in virion and genome plasticity. Annu Rev Phytopathol 2007; 45: 73-100.
[http://dx.doi.org/10.1146/annurev.phyto.45.062806.094401] [PMID: 17362202] ] was originally identified in Peru on the Pepino plant (melon-pear, Solanum muricatum) in 1974 [3Jones RAC, Koenig R, Lesemann DE. Pepino mosaic virus, a new potexvirus from pepino (Solanum muricatum) Ann Appl Biol 1980; 94: 61-8.
[http://dx.doi.org/10.1111/j.1744-7348.1980.tb03896.x] ]. It later started infecting tomato (Solanum lycopersicum) crops in the Netherlands and the United Kingdom [4Wright D, Mumford R. Pepino mosaic Potexvirus (PepMV): First records in tomato in the United Kingdom. Plant Dis 1999; Notice. No. 89, York: Central Science Laboratory, 5van der Vlugt RAA, Stijger CCMM, Verhoeven JTJ, Lesemann DE, Th J, Lesemann DE. First report of Pepino mosaic virus on tomato. Plant Dis 2000; 84(1): 103.
[http://dx.doi.org/10.1094/PDIS.2000.84.1.103C] [PMID: 30841211] ]. Since then, PepMV has rapidly achieved worldwide distribution, despite the attempts to control its spread. It has been reported in many other European countries, the United States, and Chile [5van der Vlugt RAA, Stijger CCMM, Verhoeven JTJ, Lesemann DE, Th J, Lesemann DE. First report of Pepino mosaic virus on tomato. Plant Dis 2000; 84(1): 103.
[http://dx.doi.org/10.1094/PDIS.2000.84.1.103C] [PMID: 30841211] -14Maroon-Lango CJ, Guaragna MA, Jordan RL, Hammond J, Bandla M, Marquardt SK. Two unique US isolates of Pepino mosaic virus from a limited source of pooled tomato tissue are distinct from a third (European-like) US isolate. Arch Virol 2005; 150(6): 1187-201.
[http://dx.doi.org/10.1007/s00705-005-0495-z] [PMID: 15750864] ]. The virus has also been detected in China [15Zhang Y, Shen ZJ, Zhong J, Lu XL, Cheng G, Li RD. Preliminary characterization of Pepino mosaic virus Shanghai isolate (PepMV-Sh) and its detection with ELISA. Acta Agric Shanghai 2003; 19: 90-2.], the Middle East [16Fakhro A, von Bargen S, Bandte M, Büttner C. Pepino mosaic virus, a first report of a virus infecting tomato in Syria. Phytopathol Mediterr 2010; 49: 99-101.], and South Africa [17Carmichael DJ, Rey MEC, Naidoo S, Cook G, van Heerden SW. First report of Pepino mosaic virus infecting tomato in South Africa. Plant Dis 2011; 95(6): 767.
[http://dx.doi.org/10.1094/PDIS-01-11-0036] [PMID: 30731922] ]. This viral disease has become a limiting factor for tomato production, which has caused important economic losses. In 2009, it was included in the European plant protection organization alert list [18 Pepino mosaic virus. European and Mediterranean Plant Protection Organization http://www.eppo.int/QUARANTINE/Alert_List/viruses/PEPMV0.htm (Panel review date 2014-03)].
PepMV is a single-stranded (ss) RNA genome approxi- matively 6400 nt long, with both a 5’ methylated guanine cap and 3’ polyadenylated tail. It comprises five Open Reading Frames (ORFs), flanked by 5’ and 3’ Untranslated Regions (UTR) [19Aguilar JM, Hernández-Gallardo MD, Cenis JL, Lacasa A, Aranda MA. Complete sequence of the Pepino mosaic virus RNA genome. Arch Virol 2002; 147(10): 2009-15.
[http://dx.doi.org/10.1007/s00705-002-0848-9] [PMID: 12376761] ]. ORF1 encodes the putative RNA dependent RNA polymerase (RdRp) containing methyltransferase, helicase, and polymerase domains. ORFs 2-4 overlap to form the Triple Gene Block (TGB) element, with roles indicated in viral movement, RNA silencing suppression and symptom development [20Hasiów-Jaroszewska B, Borodynko N, Jackowiak P, Figlerowicz M, Pospieszny H. Single mutation converts mild pathotype of the Pepino mosaic virus into necrotic one. Virus Res 2011; 159(1): 57-61.
[http://dx.doi.org/10.1016/j.virusres.2011.04.008] [PMID: 21536084] , 21Morozov SY, Solovyev AG. Triple gene block: Modular design of a multifunctional machine for plant virus movement. J Gen Virol 2003; 84(Pt 6): 1351-66.
[http://dx.doi.org/10.1099/vir.0.18922-0] [PMID: 12771402] ]. ORF5 encodes the Coat Protein (CP), which is involved in movement and encapsidation of the virion [21Morozov SY, Solovyev AG. Triple gene block: Modular design of a multifunctional machine for plant virus movement. J Gen Virol 2003; 84(Pt 6): 1351-66.
[http://dx.doi.org/10.1099/vir.0.18922-0] [PMID: 12771402] ]
The virus causes a wide range of symptoms that reduce the economic value of crops, including typical fruit marbling and leaf or stem necrosis [22Roggero P, Masenga V, Lenzi R, Coghe F, Ena S, Winter S. First report of Pepino mosaic virus in tomato in Italy. Plant Pathol 2001; 50: 798.
[http://dx.doi.org/10.1046/j.1365-3059.2001.00621.x] -24Hanssen IM, Paeleman A, Vandewoestijne E, et al. Pepino mosaic virus isolates and differential symptomatology in tomato. Plant Pathol 2009; 450-60.
[http://dx.doi.org/10.1111/j.1365-3059.2008.02018.x] ]. Many factors, such as genotype [20Hasiów-Jaroszewska B, Borodynko N, Jackowiak P, Figlerowicz M, Pospieszny H. Single mutation converts mild pathotype of the Pepino mosaic virus into necrotic one. Virus Res 2011; 159(1): 57-61.
[http://dx.doi.org/10.1016/j.virusres.2011.04.008] [PMID: 21536084] ] climate [23Spence NJ, Basham J, Mumford RA, Hayman G, Edmondson R, Jones DR. Effect of Pepino mosaic virus on the yield and quality of glasshouse-grown tomatoes in the UK. Plant Pathol 2006; 55: 595-606.
[http://dx.doi.org/10.1111/j.1365-3059.2006.01406.x] ] and cultivar [25Villemson S, Hunt R, Järvekülg L. Pepino mosaic virus – new dangerous pathogen for tomato and potato. Plant Prot Q 2003; 11: 37-40.] may affect the development and intensity of the symptoms.
PepMV is known to infect a relatively broad host range of plants. Most are in the family Solanaceae, but several Solanaceous hosts may not support systemic infection [3Jones RAC, Koenig R, Lesemann DE. Pepino mosaic virus, a new potexvirus from pepino (Solanum muricatum) Ann Appl Biol 1980; 94: 61-8.
[http://dx.doi.org/10.1111/j.1744-7348.1980.tb03896.x] , 26Martin J, Mousserion C. Potato varieties which are sensitive to the tomato strains of Pepino mosaic virus (PepMV). Phytoma Défence Végétaux 2002; 552: 26-8.-28Córdoba MC, Martínez-Priego L, Jordá C. New natural hosts of Pepino mosaic virus in Spain. Plant Dis 2004; 88(8): 906.
[http://dx.doi.org/10.1094/PDIS.2004.88.8.906D] [PMID: 30812528] ].
The virus is transmitted mechanically from plant to plant without the involvement of an obvious vector using feeding behavior. However, bumblebees [29Shipp JL, Buitenhuis R, Stobbs L, Wang K, Kim WS, Ferguson G. Vectoring of pepino mosaic virus by bumble-bees in tomato greenhouses. Ann Appl Biol 2008; 153: 149-55.
[http://dx.doi.org/10.1111/j.1744-7348.2008.00245.x] ], the soil-borne fungus Olpidium virulentus [30Alfaro-Fernández A, Sánchez-Navarro JÁ, Cebrián MC, Cordoba-Selles MC, Pallas V, Jorda C. Simultaneous detection and identification of Pepino mosaic virus (PepMV) isolates by multiplex one-step RT-PCR. Eur J Plant Pathol 2009; 125: 143-58.
[http://dx.doi.org/10.1007/s10658-009-9466-7] ] and whiteflies can operate as vectors only by contact [31Noël P, Hance T, Bragard C. Transmission of the pepino mosaic virus by whitefly. Eur J Plant Pathol 2014; 138: 23-7.
[http://dx.doi.org/10.1007/s10658-013-0313-5] ]. Seed transmission may also play a role in long-distance spread [32Hanssen IM, Mumford R, Blystad DG, et al. Seed transmission of Pepino mosaic virus in tomato. Eur J Plant Pathol 2010; 126: 145-52.
[http://dx.doi.org/10.1007/s10658-009-9528-x] ]. Water was confirmed to be the source of PepMV infection [33Mehle N, Gutiérrez-Aguirre I, Prezelj N, Delić D, Vidic U, Ravnikar M. Survival and transmission of potato virus Y, pepino mosaic virus, and potato spindle tuber viroid in water. Appl Environ Microbiol 2014; 80(4): 1455-62.
[http://dx.doi.org/10.1128/AEM.03349-13] [PMID: 24334672] ].
Five main PepMV genotypes are currently recognized: The Peruvian (LP) which includes the original pepino isolate (SM.74), the European (EU), the American (US1 and US2), Chilean (CH2) and the south Peruvian (PES) [13Ling K-S. Molecular characterization of two Pepino mosaic virus variants from imported tomato seed reveals high levels of sequence identity between Chilean and US isolates. Virus Genes 2007; 34(1): 1-8.
[http://dx.doi.org/10.1007/s11262-006-0003-x] [PMID: 16927118] , 14Maroon-Lango CJ, Guaragna MA, Jordan RL, Hammond J, Bandla M, Marquardt SK. Two unique US isolates of Pepino mosaic virus from a limited source of pooled tomato tissue are distinct from a third (European-like) US isolate. Arch Virol 2005; 150(6): 1187-201.
[http://dx.doi.org/10.1007/s00705-005-0495-z] [PMID: 15750864] , 34Moreno-Pérez MG, Pagán I, Aragón-Caballero L, Cáceres F, Fraile A, García-Arenal F. Ecological and genetic determinants of Pepino Mosaic Virus emergence. J Virol 2014; 88(6): 3359-68.
[http://dx.doi.org/10.1128/JVI.02980-13] [PMID: 24390328] ], with an inter-genotype RNA sequence identity of at least 80%. Hence, PepMV isolates display considerable genomic differences, possibly associated with geographic origins, specific host and high mutation rates of RNA viruses. These differences have been suggested to increase the adaptation ability to new environments [35Domingo E. Viruses at the edge of adaptation. Virology 2000; 270(2): 251-3.
[http://dx.doi.org/10.1006/viro.2000.0320] [PMID: 10792982] ]. However, no correlation has been observed between different PepMV genotypes and the severity of symptom development in infected tomato plants [11Pagán I, Del Carmen Córdoba-Sellés M, Martínez-Priego L, et al. Genetic structure of the population of Pepino Mosaic Virus infecting tomato crops in Spain. Phytopathology 2006; 96(3): 274-9.
[http://dx.doi.org/10.1094/PHYTO-96-0274] [PMID: 18944442] , 24Hanssen IM, Paeleman A, Vandewoestijne E, et al. Pepino mosaic virus isolates and differential symptomatology in tomato. Plant Pathol 2009; 450-60.
[http://dx.doi.org/10.1111/j.1365-3059.2008.02018.x] ]. In contrast, single nucleotide variations have been associated with an aggressive and highly virulent isolate known to cause significant symptoms and damages [36Hasiów-Jaroszewska B, Paeleman A, Ortega-Parra N, et al. Ratio of mutated versus wild-type coat protein sequences in Pepino mosaic virus determines the nature and severity of yellowing symptoms on tomato plants. Mol Plant Pathol 2013; 14(9): 923-33.
[http://dx.doi.org/10.1111/mpp.12059] [PMID: 23855964] ].
Interestingly, a replacement of strains and/or co-existence have characterized the epidemic of PepMV [37Gómez P, Sempere RN, Elena SF, Aranda MA. Mixed infections of Pepino mosaic virus strains modulate the evolutionary dynamics of this emergent virus. J Virol 2009; 83(23): 12378-87.
[http://dx.doi.org/10.1128/JVI.01486-09] [PMID: 19759144] ]. In Europe, EU isolates have been replaced by the CH2 strain. The American US1 genotype has been found on the Canary Islands [38Alfaro-Fernández A, Cebrián MC, Córdoba-Sellés C, Herrera-Vásquez JA, Jordá C. First report of the US1 strain of Pepino mosaic virus in tomato in the Canary Isands, Spain. Plant Dis 2008; 92(11): 1590.
[http://dx.doi.org/10.1094/PDIS-92-11-1590C] [PMID: 30764462] ]. In North America, the EU strain has become predominant [39French CJ, Dubeau C, Bunckle A, et al. Overview of Pepino Mosaic Virus research. Can J Plant Pathol 2008; 30: 373-4.] and has since been replaced by CH2 [40Ling K-S, Li R, Bledsoe M. Pepino mosaic virus genotype shift in North America and development of a loop-mediated isothermal amplification for rapid genotype identification. Virol J 2013; 10: 117.
[http://dx.doi.org/10.1186/1743-422X-10-117] [PMID: 23587202] ]. Thus, mixed infection and recombination between PepMV strains play a role in the dynamic virus evolution [37Gómez P, Sempere RN, Elena SF, Aranda MA. Mixed infections of Pepino mosaic virus strains modulate the evolutionary dynamics of this emergent virus. J Virol 2009; 83(23): 12378-87.
[http://dx.doi.org/10.1128/JVI.01486-09] [PMID: 19759144] , 41Gómez P, Sempere R, Aranda M, Elena S. Phylodynamics of Pepino mosaic virus in spain. Eur J Plant Pathol 2012; 134: 445-9.
[http://dx.doi.org/10.1007/s10658-012-0019-0] ].
Prevention of PepMV disease through strict hygiene measures is currently the main control strategy for the virus in tomato production. Cross-protection can be effective, but only if the conditions are well-defined. The effectiveness depends strongly on the PepMV genotype [32Hanssen IM, Mumford R, Blystad DG, et al. Seed transmission of Pepino mosaic virus in tomato. Eur J Plant Pathol 2010; 126: 145-52.
[http://dx.doi.org/10.1007/s10658-009-9528-x] ].
In Morocco, the current situation of PepMV presence is unknown since no official survey has been conducted until the present. In spite of this fact, there are several interception reports of PepMV on exported Moroccan tomato fruit suggesting the viral presence in the original tomato production areas [18 Pepino mosaic virus. European and Mediterranean Plant Protection Organization http://www.eppo.int/QUARANTINE/Alert_List/viruses/PEPMV0.htm (Panel review date 2014-03)]. PepMV-like symptoms have been observed, between 2009 and 2014, in many tomato fields and green- houses in the Souss, El Jadida, and Skhirat regions of Morocco. The presence of the symptoms was correlated with positive DAS-ELISA results, and confirmed by RT-PCR [42Souiri A, Zemzami M, Laatiris H, Amzazi S, Ennaji MM. First report of Pepino mosaic virus in tomato in Morocco. New Dis Rep 2017; 35: 10.
[http://dx.doi.org/10.5197/j.2044-0588.2017.035.010] ].
The genetic characterization and variations among circulating PepMV strain in Morocco have not been described. Nevertheless, genotype determination is a precondition for deploying an effective cross-protection strategy. This paper gives a careful genetic analysis of PepMV of Moroccan isolates in tomatoes by studying different parts of the genome.
2. METHODS
2.1. Plant Material and Serological Determination of PepMV Presence
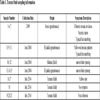
Tomato fruit showing both typical and non-typical symptoms of PepMV infection as well as symptomless tomatoes (n=21) collected between 2009 and 2014 were used in this study. The tomato samples are originated from different production areas in Morocco (Table 1). The experiments were carried out at Laboratory of Sanitary Control, Control Unit of Plants, Domaines Agricoles Maâmora, Sale, Morocco. PepMV was detected by optimized DAS ELISA using monoclonal antibodies produced locally in a previous study (Domaines agricol UCP Maamora, Salé, Morocco). The assay included healthy plant extract as a negative control and PepMV infected plant as a positive control [43Souiri A, Amzazi S, Laatiris H, Ennaji MM, Zemzami M. Serological detection of tomato Pepino mosaic virus in Morocco. J Agr Sci Tech B 2013; 3: 847-52.].
2.2. Viral RNA Extraction
The total RNA was extracted from the infected tomato (90mg of homogenized fruit sample) by using the phe- nol-guanidine isothiocyanate procedure according to the manufacturer instructions (TRIzol® reagent; Invitrogen) [44Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat Protoc 2006; 1(2): 581-5.
[http://dx.doi.org/10.1038/nprot.2006.83] [PMID: 17406285] ]. RNAs obtained were stored at -80°C.
2.3. Primer Selection for PepMV Genome Characterization
Genetic characterization of PepMV isolates was performed by the determination of the nucleotide (nt) sequence for three genomic regions: a part of the RNA-dependent RNA Poly- Merase (RdRp) gene, the Triple Gene Block (TGB), and the Capsid Protein (CP) gene. The genome structure and ampli- fication strategy are illustrated in Fig. (1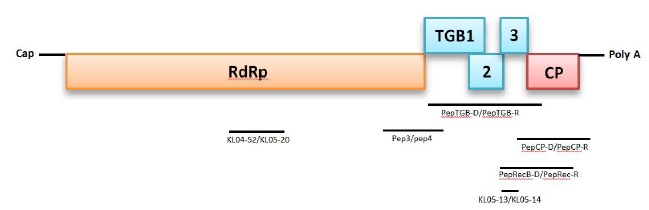 ).
).
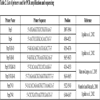
Amplifications of partial, overlapping, or complete gene regions of the RNA genome were performed in one step RT-PCR with the literature primers cited in Table 2.
Most of these primers were used when the EU genotype was circulating [11Pagán I, Del Carmen Córdoba-Sellés M, Martínez-Priego L, et al. Genetic structure of the population of Pepino Mosaic Virus infecting tomato crops in Spain. Phytopathology 2006; 96(3): 274-9.
[http://dx.doi.org/10.1094/PHYTO-96-0274] [PMID: 18944442] ], but they proved to also amplify the CH2 and US isolates. Even when the percentage intergenotype similarity is 80-85%, the targeted regions are conserved [8Hanssen IM, Paeleman A, Wittemans L, et al. Genetic characterization of Pepino Mosaic Virus isolates from Belgian greenhouse tomatoes reveals genetic recombination. Eur J Plant Pathol 2008; 121: 131-46.
[http://dx.doi.org/10.1007/s10658-007-9255-0] , 38Alfaro-Fernández A, Cebrián MC, Córdoba-Sellés C, Herrera-Vásquez JA, Jordá C. First report of the US1 strain of Pepino mosaic virus in tomato in the Canary Isands, Spain. Plant Dis 2008; 92(11): 1590.
[http://dx.doi.org/10.1094/PDIS-92-11-1590C] [PMID: 30764462] ]. For those that varied, we used discriminative primers.
A 624-nt region in the RNA-dependent RNA polymerase (RdRp) gene was amplified using the primers Pep3 and Pep4 [19Aguilar JM, Hernández-Gallardo MD, Cenis JL, Lacasa A, Aranda MA. Complete sequence of the Pepino mosaic virus RNA genome. Arch Virol 2002; 147(10): 2009-15.
[http://dx.doi.org/10.1007/s00705-002-0848-9] [PMID: 12376761] ] used for both CH2+EU strains [11Pagán I, Del Carmen Córdoba-Sellés M, Martínez-Priego L, et al. Genetic structure of the population of Pepino Mosaic Virus infecting tomato crops in Spain. Phytopathology 2006; 96(3): 274-9.
[http://dx.doi.org/10.1094/PHYTO-96-0274] [PMID: 18944442] ]; an 845-nt region, including the complete Capsid Protein (CP) gene, was amplified using primers PepCP-D and PepCP-R [10Mumford RA, Metcalfe EJ. The partial sequencing of the genomic RNA of a UK isolate of Pepino mosaic virus and the comparison of the coat protein sequence with other isolates from Europe and Peru. Arch Virol 2001; 146(12): 2455-60.
[http://dx.doi.org/10.1007/s007050170015] [PMID: 11811692] ], Aguilar et al. [19Aguilar JM, Hernández-Gallardo MD, Cenis JL, Lacasa A, Aranda MA. Complete sequence of the Pepino mosaic virus RNA genome. Arch Virol 2002; 147(10): 2009-15.
[http://dx.doi.org/10.1007/s00705-002-0848-9] [PMID: 12376761] ] reported that a 1,317-nt region encompassing the complete Triple Gene Block (TGB) was amplified using two discriminatory primers PepTGB-D/PepTGB-R, which amplify European genotypes and PepUSTGB-D/PepUSTGB-R that cover American and Chilean genotypes [11Pagán I, Del Carmen Córdoba-Sellés M, Martínez-Priego L, et al. Genetic structure of the population of Pepino Mosaic Virus infecting tomato crops in Spain. Phytopathology 2006; 96(3): 274-9.
[http://dx.doi.org/10.1094/PHYTO-96-0274] [PMID: 18944442] ]. Primers KL05-13/ KL05-14 that target a short region (202-nt) of TGB2-3 of all known PepMV genotypes were used [45Ling K-S, Wintermantel WM, Bledsoe M. Genetic composition of Pepino mosaic virus population in North American greenhouse tomatoes. Plant Dis 2008; 92(12): 1683-8.
[http://dx.doi.org/10.1094/PDIS-92-12-1683] [PMID: 30764290] ]. In addition, a CH2 genotype-specific primer, KL04-52/KL05-20 that amplifies an RdRp helicase domain (642 nt), was utilized to support genotype identification [45Ling K-S, Wintermantel WM, Bledsoe M. Genetic composition of Pepino mosaic virus population in North American greenhouse tomatoes. Plant Dis 2008; 92(12): 1683-8.
[http://dx.doi.org/10.1094/PDIS-92-12-1683] [PMID: 30764290] ]. Finally, to improve sequence data accuracy, an overlapping region between CP and TGB (1028 nt) was amplified using PepRecB-D/PepUSTGB-R [19Aguilar JM, Hernández-Gallardo MD, Cenis JL, Lacasa A, Aranda MA. Complete sequence of the Pepino mosaic virus RNA genome. Arch Virol 2002; 147(10): 2009-15.
[http://dx.doi.org/10.1007/s00705-002-0848-9] [PMID: 12376761] ] (Table 2)
.
2.4. Reverse Transcription Polymerase Chain Reaction (RT-PCR) and Sequencing
RNA extracts from tomato fruit were reverse transcribed using M-MLV reverse transcriptase (Promega) and polymerase chain reaction PCR, amplified using Taq polymerase (Pro- mega).
Briefly, 25µl of the reaction was carried out in a 0.2ml tube containing 1µl of prepared RNA and 5µl of M-MLV buffer 5X, 2µl of MgCl2 (25mM), 1U M-MLV reverse transcriptase, 1000U RNasin, 0.2mM each dNTPs, 0.4 µM each primers and 0.05 U Taq polymerase. The cycling parameter was reverse transcribed for 30 min at 50°C, denaturated for 2 min at 94°C, followed by 40 cycles of denaturation at 94°C for 15s, annealing at 45-52°C depending on the specific primers, for 30 second and extension cycle 68°C for 45s with a final extent cycle of 7 min at 68°C. An aliquot of RT-PCR preparation from each reaction (6µl) was applied onto 1.5% agarose gel for electrophoresis and the RT-PCR product was visualized under UV light.
The amplified PCR products were directly sequenced on both strands using the “Big Dye Terminator v3.1 Cycle Sequencing kit” (Applied Biosystems), according to the manufacturer’s instructions in the Molecular and Functional Genomics Platform (UATRS-CNRST, Rabat, Morocco).
2.5. Sequence and Phylogenetic Analysis
The generated sequences were edited at the ends to elimi- nate the less precise regions. Moreover, overlapped sequences were assembled to cover the longest region, explaining that the final sequence lengths were changed in comparison to the PCR fragments. Sequences were then blasted in the NCBI GenBank using online BLAST 2.0 software server (http://www. ncbi. nih. gov/BLAST/) to confirm the amplified region.
The percentages of nucleotide and deduced amino acid similarities between Moroccan PepMV isolates and reference strains were calculated using EMBOSS Matcher, a pairwise local alignment tool available in (www.ebi.ac.uk). The obtained amino acid (aa) sequences and translations of Open Reading Frames (ORFs) were generated with translation ExPASy tool (http://web.expasy.org/translate/). Multiple sequence alignments of the set of sequences were performed using the BioEdit Software v7.1.9 [46Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 1999; 41: 95-8.] and Clustal W program [47Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994; 22(22): 4673-80.
[http://dx.doi.org/10.1093/nar/22.22.4673] [PMID: 7984417] ]. PepMV nucleotide positions were numbered according to the PepMV reference (Ch2 strain published in GenBank with accession number: DQ000985) [13Ling K-S. Molecular characterization of two Pepino mosaic virus variants from imported tomato seed reveals high levels of sequence identity between Chilean and US isolates. Virus Genes 2007; 34(1): 1-8.
[http://dx.doi.org/10.1007/s11262-006-0003-x] [PMID: 16927118] ].
Complete virus sequences of the representative reference strains of each genotype were selected from the NCBI Gen- Bank Database and used for phylogenetic analysis. Their acce- ssion numbers are: PepMV-Ch2 [DQ000985], PepMV-P19 [HQ650559],PepMV-P22 [HQ650560], PepMV-PK [EF40- 8821], PepMV-Pa [FJ612601], PepMV-SIC1-09 [HQ663891], PepMV-SIC2-09 [HQ663892], PepMV-US2 [AY509927], PepMV-Ch1 [DQ000984], PepMV-US1 [AY509926], PepMV-Sp-13 [AF484251], PepMV-LP-2001 [AJ606361], PepMV-SM.74 [AM109896], PepMV-UK [AF340024], PepMV-Fr [AJ438767], PepMV-Chi2.9 [HG313805], PepMV-Tor9 [HG313806], PepMV- Yur1.5 [HG313807].
Phylogenetic trees were built by MEGA 7 package [48Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 2016; 33(7): 1870-4.
[http://dx.doi.org/10.1093/molbev/msw054] [PMID: 27004904] ] using the Maximum Likelihood method based on the Tamura-Nei model [49Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 1993; 10(3): 512-26.
[PMID: 8336541] ]. They were built with 1,000 bootstrap replicates to test the robustness of the major phylogenetic groups. The data matrix of phylogenetic analyses was deposited into TreeBase under accession URL: http://purl. org / phylo/ treebase/ phylows/study/TB2:S19 550.
3. RESULTS
3.1. PepMV Amplification and Genotyping
The disease symptoms of the collected tomatoes varied among period and location and from typical to atypical. Therefore, viral identification cannot be based on the symptoms. DAS-ELISA and RT-PCR tests showed the same results and detected PepMV in twelve samples from the twenty-one that were collected. It should be noted that all samples detected positive by DAS ELISA showed bands when tested with RT-PCR and those negative in DAS-ELISA were also negative in RT-PCR; false positives and false negatives were absent (Fig. 2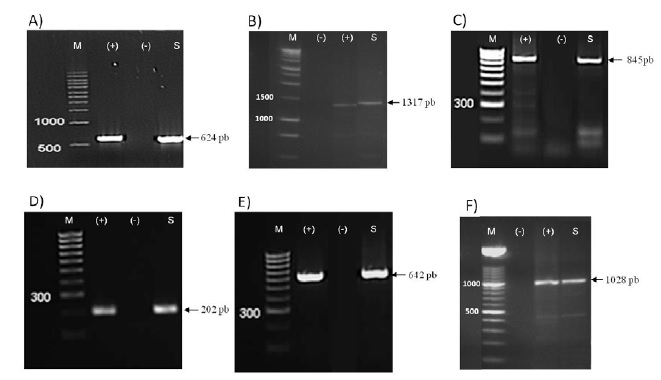 ).
).
This shows the higher sensitivity of the DAS-ELISA assay used for the screening of the PepMV presence [43Souiri A, Amzazi S, Laatiris H, Ennaji MM, Zemzami M. Serological detection of tomato Pepino mosaic virus in Morocco. J Agr Sci Tech B 2013; 3: 847-52.]. Antisera and primers were able to capture all potential PepMV diversity, which is in contrast to the previously reported studies that state the absence of correlation between molecular method and ELISA assays for PepMV detection [30Alfaro-Fernández A, Sánchez-Navarro JÁ, Cebrián MC, Cordoba-Selles MC, Pallas V, Jorda C. Simultaneous detection and identification of Pepino mosaic virus (PepMV) isolates by multiplex one-step RT-PCR. Eur J Plant Pathol 2009; 125: 143-58.
[http://dx.doi.org/10.1007/s10658-009-9466-7] ].
Primers for RT-PCR amplification (Table 2) were selected to cover much of the main genomic regions including RdRP, TGB and CP. For some regions, the amplification was targeted using two or more different primers to produce sufficient high-quality sequence data for each isolate, and to make an accurate assessment of genetic variation and mutation points. Discriminative primers allowed for the determination of the genotype and the exclusion of another in the PCR analysis level. As a result, all the primers were effective in amplifying the samples and produced bands of the expected size, except for PepTGB-D/ PepTGB-R, which specifically target the EU strain, no band has been detected. In addition, the expected RT-PCR product (642 nt) was obtained for all the samples using the CH2 genotype-specific primer (KL04-52/KL05-20). These results indicate that PepMV genetic population is likely to belong to the CH2 genotype rather than EU genotype.
Nucleotide sequences of RdRp, TGB, CP genes, which were determined in this study, completely or partially for the twelve Moroccan PepMV isolates, were deposited into Gen- Bank Database (http://www.ncbi.nlm.nih.gov/GenBank), under the following accession numbers: RdRp [KP761701 to KP761712], RdRp Helicase domain [KT253424 to KT253434], TGB1 [KP761701 to KP761712], TGB2-3 [KU051631 to KU051640] and CP [KT253416 to KT253423].
3.2. Sequence Homology and Phylogenetic Analysis
Sequencing of partial RdRP, partial TGB and complete CP genes of the 12 isolates aims to identify the genotype composition and to assess the genetic diversity of the PepMV population circulating in Morocco. The obtained nucleotide sequences were blasted and compared with the reference sequences retrieved from GenBank belonging to the different genotypes identified around the world. The sequenced fragments covered 48% of the complete virus genome.
All the analysed Moroccan PepMV strains share a nucleotide sequence identity with PepMV-CH2 DQ000985 above 98%. In contrast, they share only 77% similarity with the European strains, while the identity with the US2 strain is 85% and PES 80%. The deduced amino acid sequences of all sequenced PepMV isolates displayed the following identities with the Ch2 isolate: 99.6 -100% for the CP genes, 98.4- 99.2% for the RdRp, 97.0-100%, 97.5-93.3% and 97.6-98.8% for the TGB1, TGB2 and TGB3, respectively.
Phylogenetic analyses were performed to confirm the genetic composition of PepMV genotypes circulating in Morocco. Based on the sequence alignment, phylogenetic trees were constructed based on RdRp, TGB, and CP regions, respectively. Representative sequences from the five major genotypes were included (Fig. 3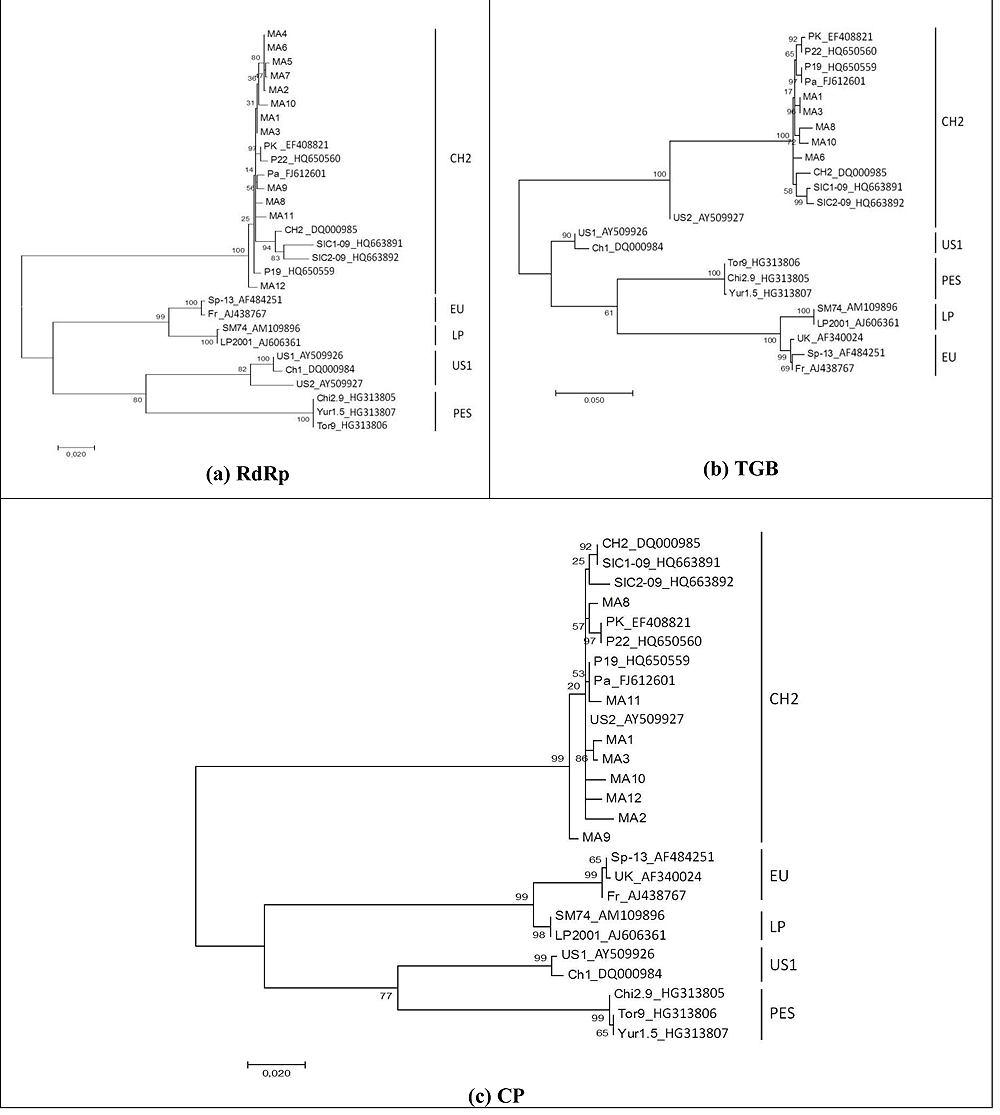 ). Maximum Likelihood trees of these data sets were obtained with 1000 bootstrap repli- cations.
). Maximum Likelihood trees of these data sets were obtained with 1000 bootstrap repli- cations.
The branches leading to genotype groups were sustained by nearly 100% of bootstrap replications in all topologies.
Moroccan PepMV strains were highly related and belonging to a single phylogenetic cluster classified into the CH2 genotype represented by the Chilean reference isolate CH2, PK, Pa, P19, P22, SIC1-09 et SIC2-09. Our data sequences do not cluster with any of the reference US1 genotype, PES genotype, the European reference isolates Fr, SP13, UK or the original Peruvian PepMV strains LP, SM.74 confirming the results of RT-PCR genotyping analysis Fig. (3 ).
).
3.3. PepMV Sequence Variations
The nucleotide variations and predicted amino-acid changes identified in this study are shown in Table 3. The variations of PepMV-MA sequences were mainly found to be concentrated in two regions: the second half of RdRp and CP; TGB1, TGB2 and TGB3 displayed less variation. Collectively, all studied isolates were highly similar between them differing in just a few Single Nucleotide Polymorphisms (SNPs). These nucleotides changes do not affect the phylogenic distribution.
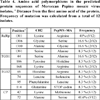
In comparison with the reference genome CH2 (accession number: DQ000985), we found several mutation points in the PepMV-MA sequences. The majority of mutations were substitutions. There was no evidence insertions, or deletions. Most of them were silent, although we found 5 amino acid changes in the RdRp gene, 3 changes in TGB1, TGB2, and CP genes, and only one different amino acid in TGB3. A unique stop codon was identified in MA11 strain, C5452T mutation, which resulted in the change Q→STOP in TGB2 protein (Table 3).
Compared with the EU reference genotypes (Accession Number: AF484251, AF340024 and AJ438767), our results showed that 11 nucleotide variations, specific to European strains and generally absent in CH2 strains, were found in our sequence data set. These common variations are located throughout the genome in the following positions, in RdRp (2527, 4000, 4243), in TGB1-3 (4484, 4502, 5220, 5232, 5310), and in CP (5699, 5827, 6145). Some of these also appear in the PES in the positions 4243, 4484, 5232 and 5699. Moreover, two other single mutations are shared only with PES at the positions 3970 and 5509 (Table 3).
Results of the multiple alignments with the reference genotypes cited in this study are not shown. These specific mutations are highlighted in Table 3. A total of 25 SNPs are observed in a different part of the genome, of which 6 SNPs are distributed in fixed polymorphism sites in the following positions 2338, 4372, 4640, 5310, 5452 and 5785. The other parts of mutations are randomly localized within the genome.
These new nucleotide variations are mostly synonymous, however, 13 amino acid changes related to PepMV-MA sequences are summarized in Table 4. The likelihood of found nucleotide changes due to errors being introduced by reverse transcriptase and sequencing is not excluded and has been reduced in our study. These errors may concern single variations, which are unique and those not repeated within isolates as well as those that are localized out of polymorphism sites. These kinds of nucleotide changes were ignored and not included in the study. The sequencing strategy in both sense and amplifying overlapping fragments increased accuracy of the obtained sequences. Single mutations found in one strand were corrected during the pairwise alignment step, only recurrent mutations in the same position were conserved.

Nucleotide sequence variations, predicted amino-acid changes and specific variations of PepMV Moroccan isolates. Capital letters indicate variants with an amino acid change, Lower-case letters indicate silent mutations, highlighted letters indicate Moroccan specific SNPs, S: silence; the asterisk (*) indicates specific PepMV-MA missense.
4. DISCUSSION
Tomato production is an important crop activity in Morocco and has a significant weight in the general exports [50Aloui A, Kenny L. The cost of compliance with SPS standards for Moroccan exports: A case study. 2005. World Bank Agriculture and Rural Development Discussion Paper. World Bank, Washington DC.] (Greenhouse tomato production is the main system used, which requires hands-on activities including planting, grafting, plant tying, de-leafing, spraying, and fruit picking. These practices may potentially initiate a mechanical transmission of PepMV to healthy plants from a contaminated hand or tool, even with a small number of PepMV-infected plants [32Hanssen IM, Mumford R, Blystad DG, et al. Seed transmission of Pepino mosaic virus in tomato. Eur J Plant Pathol 2010; 126: 145-52.
[http://dx.doi.org/10.1007/s10658-009-9528-x] ]. Outbreaks of the disease were observed in the last few years in many tomato production areas in Morocco and raised concerns of possible seed transmission. Notable progress has been achieved towards understanding the genetic diversity and population structure of PepMV in many countries [10Mumford RA, Metcalfe EJ. The partial sequencing of the genomic RNA of a UK isolate of Pepino mosaic virus and the comparison of the coat protein sequence with other isolates from Europe and Peru. Arch Virol 2001; 146(12): 2455-60.
[http://dx.doi.org/10.1007/s007050170015] [PMID: 11811692] , 11Pagán I, Del Carmen Córdoba-Sellés M, Martínez-Priego L, et al. Genetic structure of the population of Pepino Mosaic Virus infecting tomato crops in Spain. Phytopathology 2006; 96(3): 274-9.
[http://dx.doi.org/10.1094/PHYTO-96-0274] [PMID: 18944442] , 13Ling K-S. Molecular characterization of two Pepino mosaic virus variants from imported tomato seed reveals high levels of sequence identity between Chilean and US isolates. Virus Genes 2007; 34(1): 1-8.
[http://dx.doi.org/10.1007/s11262-006-0003-x] [PMID: 16927118] ]. However, the genetic charac terization of the Moroccan PepMV population remains unknown until now. The present study involved careful analyses of virus isolates circulating in Morocco.
For this purpose, nucleotide sequencing of a part of RdRp, TGB 1-3, and full-lengh CP genes was employed to charac- terize twelve PepMV Moroccan strains (PepMV-MA) (sampled between 2009 and 2014) from tomato fruits exhibiting different symptoms, including isolates from ELISA-positive asymptomatic plants that originated from the main tomato production areas. These genomic regions represent viral genes encoding proteins with distinct functions. In the first step, we amplified the cited genes using a large set of primers that covered 48% of the whole genome. Sequence analysis of isolates sampled from different production areas showed slight differences of about 0.4%, leading to a homogenous popu- lation. This was deduced from the percentage of similarity calculated between Moroccan strains using pairwise align- ments.
Using the sequence data of the targeted segments, phylogenetic relationships among PepMV isolates provided in this study were evaluated along with representative isolates of all major PepMV genotypes identified around the world. These analyses revealed that the PepMV strains infecting Moroccan tomatoes belong to the CH2 genotype. Topologies of trees show that strains of the same genotypes are clearly clustered together, except for some differences showing that US2 strain falls into the same cluster as CH2 in CP tree. Nucleotide sequence comparisons were suggested previously that US2 is a recombinant of US1 and CH2 [8Hanssen IM, Paeleman A, Wittemans L, et al. Genetic characterization of Pepino Mosaic Virus isolates from Belgian greenhouse tomatoes reveals genetic recombination. Eur J Plant Pathol 2008; 121: 131-46.
[http://dx.doi.org/10.1007/s10658-007-9255-0] ]. The difference between the topologies of the 3 trees, is explained by the conservation of each region, the most conserved is CP, then TGB and the less conserved is RdRp. Despite these genome differences, the PepMV strains are grouped following 5 clades corresponding to each genotype.
The same result was obtained with the discriminative genotyping RT-PCR assays using specific primers for CH2 [13Ling K-S. Molecular characterization of two Pepino mosaic virus variants from imported tomato seed reveals high levels of sequence identity between Chilean and US isolates. Virus Genes 2007; 34(1): 1-8.
[http://dx.doi.org/10.1007/s11262-006-0003-x] [PMID: 16927118] ] and for EU [19Aguilar JM, Hernández-Gallardo MD, Cenis JL, Lacasa A, Aranda MA. Complete sequence of the Pepino mosaic virus RNA genome. Arch Virol 2002; 147(10): 2009-15.
[http://dx.doi.org/10.1007/s00705-002-0848-9] [PMID: 12376761] ] genotypes. The combination of these two primers, targeting a different part of the genome, was useful not only in amplification but also in the determination of CH2 genotype presence and the exclusion of EU genotype from our strains at PCR analysis level.
The genotypic characterization of the twelve PepMV isolates in Morocco is in accordance with the current situation of CH2 genotype prevalence and geographical distribution. Until 2008, the EU genotype was considered the most common genotype in European tomato production greenhouses [19Aguilar JM, Hernández-Gallardo MD, Cenis JL, Lacasa A, Aranda MA. Complete sequence of the Pepino mosaic virus RNA genome. Arch Virol 2002; 147(10): 2009-15.
[http://dx.doi.org/10.1007/s00705-002-0848-9] [PMID: 12376761] ], [13Ling K-S. Molecular characterization of two Pepino mosaic virus variants from imported tomato seed reveals high levels of sequence identity between Chilean and US isolates. Virus Genes 2007; 34(1): 1-8.
[http://dx.doi.org/10.1007/s11262-006-0003-x] [PMID: 16927118] ]. Later, CH2 was reported at high levels of prevalence in many European countries [8Hanssen IM, Paeleman A, Wittemans L, et al. Genetic characterization of Pepino Mosaic Virus isolates from Belgian greenhouse tomatoes reveals genetic recombination. Eur J Plant Pathol 2008; 121: 131-46.
[http://dx.doi.org/10.1007/s10658-007-9255-0] ]. Considering the geographical closeness, the prevalence of CH2 genotype in Morocco can be considered similar to the case history of PepMV in European countries. The hypothesis of a possible earlier existence of EU strains can be argued by the presence of a few specific EU genotype mutations noticed in the studied sequence data.
Questions regarding how CH2 genotype was introduced and why it is so widespread still arise. Previous studies showed that this PepMV genotype was found in tomato seeds [45Ling K-S, Wintermantel WM, Bledsoe M. Genetic composition of Pepino mosaic virus population in North American greenhouse tomatoes. Plant Dis 2008; 92(12): 1683-8.
[http://dx.doi.org/10.1094/PDIS-92-12-1683] [PMID: 30764290] ] , and many young plants were already infected at an early stage. Their delivery for cultivation in greenhouses was considered as a potential reason for the large-scale occurrence in CH2. This information suggested that CH2 has a biological advantage over and spreads faster than the EU genotype [8Hanssen IM, Paeleman A, Wittemans L, et al. Genetic characterization of Pepino Mosaic Virus isolates from Belgian greenhouse tomatoes reveals genetic recombination. Eur J Plant Pathol 2008; 121: 131-46.
[http://dx.doi.org/10.1007/s10658-007-9255-0] ].
Identification of the CH2 genotype in Moroccan tomato plants indicated the likelihood that transmission of PepMV-CH2 from contaminated seeds to plants has occurred. There- fore, it is important to plant PepMV-free certified commercial tomato seeds.
This study has characterized PepMV-MA isolates as a homogenous population due to the minor differences between sequences. Mutations are mostly silent; missense point mutations, leading to amino acid changes, were also observed. Compared with the reference CH2 genotype, a total of 40 SNPs were found in the 3100 nt sequenced PepMV genome, resulting in 16 differences at the predicted amino acid level. The SNPs were not concentrated in a specific region of the genome. In spite of this, the capsid protein was more conserved.
A remarkable finding made in this study was the presence among these variations of new mutations specific to our isolates, which have never been described before within the representative strains of the major genotypes. These SNPs were responsible for 13 amino acid changes (Table 4), of which one mutation led to a non-functional TGB2 protein, a movement protein. This stop-codon mutation in TGB2 can alter virus cell-to-cell movement [51Ju HJ, Brown JE, Ye CM, Verchot-Lubicz J. Mutations in the central domain of potato virus X TGBp2 eliminate granular vesicles and virus cell-to-cell trafficking. J Virol 2007; 81(4): 1899-911.
[http://dx.doi.org/10.1128/JVI.02009-06] [PMID: 17151124] ].
Interestingly, MA 2, 4, 5, 6, 7 appear to have multiple conserved variants in comparison to the other Morocco strains (Table 3). This correlates to pathogen location (souss greenhouses) and date of collection (2009) as shown in Table 1. A spread of closely related strains to neighboring green- houses is also possible.
Mutational analysis conducted by Hanssen et al. (2009) revealed a limited number of RNA sequence mutations and few nonsynonymous substitutions, reflecting the strong purifying selection. Most of the mutations that take place have no clear biological relevance [24Hanssen IM, Paeleman A, Vandewoestijne E, et al. Pepino mosaic virus isolates and differential symptomatology in tomato. Plant Pathol 2009; 450-60.
[http://dx.doi.org/10.1111/j.1365-3059.2008.02018.x] ].
Previous studies showed that the presence of single nucleotide substitutions in the CH2 strain have a big influence on the symptomology in tomato plants (necrotic and yellowing) [36Hasiów-Jaroszewska B, Paeleman A, Ortega-Parra N, et al. Ratio of mutated versus wild-type coat protein sequences in Pepino mosaic virus determines the nature and severity of yellowing symptoms on tomato plants. Mol Plant Pathol 2013; 14(9): 923-33.
[http://dx.doi.org/10.1111/mpp.12059] [PMID: 23855964] , 52Hasiów-Jaroszewska B, Minicka J, Borodynko N, Pospieszny H. The genetic determinants of symptoms induction by Pepino Mosaic virus in tomato. ISHS Acta Hortic 2015; 1069: 39-43.
[http://dx.doi.org/10.17660/ActaHortic.2015.1069.4] ]. A single amino acid change in the TGB3 protein (K67E) is involved in necrosis induction by PepMV-P19 isolate. This necrotic isolate of CH2 was found in Poland [20Hasiów-Jaroszewska B, Borodynko N, Jackowiak P, Figlerowicz M, Pospieszny H. Single mutation converts mild pathotype of the Pepino mosaic virus into necrotic one. Virus Res 2011; 159(1): 57-61.
[http://dx.doi.org/10.1016/j.virusres.2011.04.008] [PMID: 21536084] , 53Minicka J, Rymelska N, Elena SF, Czerwoniec A, Hasiów-Jaroszewska B. Molecular evolution of Pepino mosaic virus during long-term passaging in different hosts and its impact on virus virulence. Ann Appl Biol 2015; 166: 389-401.
[http://dx.doi.org/10.1111/aab.12179] ]. Two separate mutations in the Coat Protein (CP) gene (E155K) and D166G in (CP) of other isolates identified in several countries have been identified as determinants of yellowing symptoms [36Hasiów-Jaroszewska B, Paeleman A, Ortega-Parra N, et al. Ratio of mutated versus wild-type coat protein sequences in Pepino mosaic virus determines the nature and severity of yellowing symptoms on tomato plants. Mol Plant Pathol 2013; 14(9): 923-33.
[http://dx.doi.org/10.1111/mpp.12059] [PMID: 23855964] ].
In the studied Moroccan PepMV population, none of these points of mutation were found. This does not exclude the possible contribution of these mutations in symptom diversity from the fact that symptoms of sampled tomato fruits varied from the typical fruit marbling to asymptomatic. A better understanding of the relationship between viral diversity and symptoms was not the focus of this study and is difficult to access with a limited number of isolates.
The emergence of new strains and the appearance of new genetic types by mutation and recombination events represent a high potential risk and should be considered by developing efficient control strategies. Cross-protection represents a viable and attractive option for PepMV control.
Recently in 2015, the National Food Safety Office (ONSSA) authorized the sale of the PMV®-01 [54Hoogenven JP. Besluit van de Staatssecretaris van Economische Zaken van 7 oktober 2013, nr 13168344, Wet ewasbeschermingsmiddelen en biociden ter bestrijding van Pepinomozaïekvirus in de productieteelt van tomaat (Tijdelijke vrijstelling voor het gewasbeschermingsmiddel PMV®-01 ter bescherming van de onbelichte teelt van tomaat, toe te passen in de productieteelt). 2013. Staatscourant, 29627] in Morocco; a vaccine that protects against aggressive PepMV infections using a cross-protection method. This vaccine includes a mild CH2 strain (isolate 1906). During the tomato 2015-2016 seasons, a mass vaccination campaign of plant was launched to prevent production and quality losses. Satisfactory results were validated for these tests; this campaign was extended for 2016-17 [55 Agriculture du maghreb. PMV®-01 the sustainable and efficient solution for a tomato crop without Pepino, Agriculture du maghreb, 2016, 96: 24-25].
CONCLUSION
The findings of this work contribute to an understanding of the genetic composition of Pepino mosaic virus in Morocco. The highly efficient mechanical transmission of this disease has made the implementation of cultural practices unsuccessful in preventing its spread. Inoculating tomato plants with a stable mild strain of PepMV at an early stage may help reduce losses. The characterization of the Moroccan population into the CH2 genotype, with a relatively low genetic variability, may lead to comfortable use of the recent proposal vaccination strategy using a mild strain of CH2 PepMV. In addition, the efforts should be concentrated on the phytosanitary control for both exportation and importation for both tomato fruit and seeds.
LIST OF ABBREVIATIONS
| PepMV | = Pepino Mosaic Virus |
| RdRp | = RNA-dependent RNA Polymerase |
| TGB | = Triple Gene Block |
| CP | = Coat Protein |
| DAS ELISA | = Double Antibody Sandwich Enzyme Linked Immuno Sorbent Assay |
| RT-PCR | = Reverse Transcription Polymerase Chain Reaction |
| SNP | = Single Nucleotide Polymorphisms. |
AUTHOR'S CONTRIBUTIONS
AS designed the protocol of the study, carried out PCR, sequence analyses and phylogeny and drafted the manuscript. MZ supported the study and provided the viral isolates. HL provided technical assistance in laboratory. SA participate in the design of the study and gave the opinion in the manuscript. MME conceived of the study, participated in the design, reviewed the final manuscript and assumes coordination. All authors have read and approved the submitted manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The datasets supporting the conclusions of this article are included within the article.
FUNDING
This work was supported by Domaines Agricoles – UCP (Plant Control Unit) Maâmora, Sale Morocco, University Hassan II of Casablanca and Ministry of Higher Education of Morocco.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
We are grateful to the virology laboratory staff of the University Hassan II of Casablanca, the staff of UCP (Plant Control Unit) (Domaines Agricoles - Maâmora Sale, Morocco) and the staff of the UATRS-CNRST, Rabat, Morocco.
REFERENCES
| [1] | Adams MJ, Antoniw JF, Bar-Joseph M, et al. The new plant virus family Flexiviridae and assessment of molecular criteria for species demarcation. Arch Virol 2004; 149(5): 1045-60. [http://dx.doi.org/10.1007/s00705-004-0304-0] [PMID: 15098118] |
| [2] | Martelli GP, Adams MJ, Kreuze JF, Dolja VV. Family Flexiviridae: A case study in virion and genome plasticity. Annu Rev Phytopathol 2007; 45: 73-100. [http://dx.doi.org/10.1146/annurev.phyto.45.062806.094401] [PMID: 17362202] |
| [3] | Jones RAC, Koenig R, Lesemann DE. Pepino mosaic virus, a new potexvirus from pepino (Solanum muricatum) Ann Appl Biol 1980; 94: 61-8. [http://dx.doi.org/10.1111/j.1744-7348.1980.tb03896.x] |
| [4] | Wright D, Mumford R. Pepino mosaic Potexvirus (PepMV): First records in tomato in the United Kingdom. Plant Dis 1999; Notice. No. 89, York: Central Science Laboratory |
| [5] | van der Vlugt RAA, Stijger CCMM, Verhoeven JTJ, Lesemann DE, Th J, Lesemann DE. First report of Pepino mosaic virus on tomato. Plant Dis 2000; 84(1): 103. [http://dx.doi.org/10.1094/PDIS.2000.84.1.103C] [PMID: 30841211] |
| [6] | Cotillon AC, Girard M, Ducouret S. Complete nucleotide sequence of the genomic RNA of a French isolate of Pepino mosaic virus (PepMV). Brief report. Arch Virol 2002; 147(11): 2231-8. [http://dx.doi.org/10.1007/s00705-002-0873-8] [PMID: 12417957] |
| [7] | French CJ, Bouthillier M, Bernardy M, et al. First Report of Pepino mosaic virus in Canada and the United States. Plant Dis 2001; 85(10): 1121. [http://dx.doi.org/10.1094/PDIS.2001.85.10.1121B] [PMID: 30823296] |
| [8] | Hanssen IM, Paeleman A, Wittemans L, et al. Genetic characterization of Pepino Mosaic Virus isolates from Belgian greenhouse tomatoes reveals genetic recombination. Eur J Plant Pathol 2008; 121: 131-46. [http://dx.doi.org/10.1007/s10658-007-9255-0] |
| [9] | Hasiów B, Borodynko N, Pospieszny H. Complete genomic RNA sequence of the Polish Pepino mosaic virus isolate belonging to the US2 strain. Virus Genes 2008; 36(1): 209-14. [http://dx.doi.org/10.1007/s11262-007-0171-3] [PMID: 17934803] |
| [10] | Mumford RA, Metcalfe EJ. The partial sequencing of the genomic RNA of a UK isolate of Pepino mosaic virus and the comparison of the coat protein sequence with other isolates from Europe and Peru. Arch Virol 2001; 146(12): 2455-60. [http://dx.doi.org/10.1007/s007050170015] [PMID: 11811692] |
| [11] | Pagán I, Del Carmen Córdoba-Sellés M, Martínez-Priego L, et al. Genetic structure of the population of Pepino Mosaic Virus infecting tomato crops in Spain. Phytopathology 2006; 96(3): 274-9. [http://dx.doi.org/10.1094/PHYTO-96-0274] [PMID: 18944442] |
| [12] | Pospieszny H, Borodynko N, Palczewska M. First record of Pepino mosaic virus in Poland. J Plant Dis Prot 2003; 100: 97. |
| [13] | Ling K-S. Molecular characterization of two Pepino mosaic virus variants from imported tomato seed reveals high levels of sequence identity between Chilean and US isolates. Virus Genes 2007; 34(1): 1-8. [http://dx.doi.org/10.1007/s11262-006-0003-x] [PMID: 16927118] |
| [14] | Maroon-Lango CJ, Guaragna MA, Jordan RL, Hammond J, Bandla M, Marquardt SK. Two unique US isolates of Pepino mosaic virus from a limited source of pooled tomato tissue are distinct from a third (European-like) US isolate. Arch Virol 2005; 150(6): 1187-201. [http://dx.doi.org/10.1007/s00705-005-0495-z] [PMID: 15750864] |
| [15] | Zhang Y, Shen ZJ, Zhong J, Lu XL, Cheng G, Li RD. Preliminary characterization of Pepino mosaic virus Shanghai isolate (PepMV-Sh) and its detection with ELISA. Acta Agric Shanghai 2003; 19: 90-2. |
| [16] | Fakhro A, von Bargen S, Bandte M, Büttner C. Pepino mosaic virus, a first report of a virus infecting tomato in Syria. Phytopathol Mediterr 2010; 49: 99-101. |
| [17] | Carmichael DJ, Rey MEC, Naidoo S, Cook G, van Heerden SW. First report of Pepino mosaic virus infecting tomato in South Africa. Plant Dis 2011; 95(6): 767. [http://dx.doi.org/10.1094/PDIS-01-11-0036] [PMID: 30731922] |
| [18] | Pepino mosaic virus. European and Mediterranean Plant Protection Organization http://www.eppo.int/QUARANTINE/Alert_List/viruses/PEPMV0.htm (Panel review date 2014-03) |
| [19] | Aguilar JM, Hernández-Gallardo MD, Cenis JL, Lacasa A, Aranda MA. Complete sequence of the Pepino mosaic virus RNA genome. Arch Virol 2002; 147(10): 2009-15. [http://dx.doi.org/10.1007/s00705-002-0848-9] [PMID: 12376761] |
| [20] | Hasiów-Jaroszewska B, Borodynko N, Jackowiak P, Figlerowicz M, Pospieszny H. Single mutation converts mild pathotype of the Pepino mosaic virus into necrotic one. Virus Res 2011; 159(1): 57-61. [http://dx.doi.org/10.1016/j.virusres.2011.04.008] [PMID: 21536084] |
| [21] | Morozov SY, Solovyev AG. Triple gene block: Modular design of a multifunctional machine for plant virus movement. J Gen Virol 2003; 84(Pt 6): 1351-66. [http://dx.doi.org/10.1099/vir.0.18922-0] [PMID: 12771402] |
| [22] | Roggero P, Masenga V, Lenzi R, Coghe F, Ena S, Winter S. First report of Pepino mosaic virus in tomato in Italy. Plant Pathol 2001; 50: 798. [http://dx.doi.org/10.1046/j.1365-3059.2001.00621.x] |
| [23] | Spence NJ, Basham J, Mumford RA, Hayman G, Edmondson R, Jones DR. Effect of Pepino mosaic virus on the yield and quality of glasshouse-grown tomatoes in the UK. Plant Pathol 2006; 55: 595-606. [http://dx.doi.org/10.1111/j.1365-3059.2006.01406.x] |
| [24] | Hanssen IM, Paeleman A, Vandewoestijne E, et al. Pepino mosaic virus isolates and differential symptomatology in tomato. Plant Pathol 2009; 450-60. [http://dx.doi.org/10.1111/j.1365-3059.2008.02018.x] |
| [25] | Villemson S, Hunt R, Järvekülg L. Pepino mosaic virus – new dangerous pathogen for tomato and potato. Plant Prot Q 2003; 11: 37-40. |
| [26] | Martin J, Mousserion C. Potato varieties which are sensitive to the tomato strains of Pepino mosaic virus (PepMV). Phytoma Défence Végétaux 2002; 552: 26-8. |
| [27] | Salomone A, Roggero P. Host range, seed transmission and detection by ELISA and lateral flow of an Italian isolate of Pepino mosaic virus. J Plant Pathol 2002; 84: 65-8. |
| [28] | Córdoba MC, Martínez-Priego L, Jordá C. New natural hosts of Pepino mosaic virus in Spain. Plant Dis 2004; 88(8): 906. [http://dx.doi.org/10.1094/PDIS.2004.88.8.906D] [PMID: 30812528] |
| [29] | Shipp JL, Buitenhuis R, Stobbs L, Wang K, Kim WS, Ferguson G. Vectoring of pepino mosaic virus by bumble-bees in tomato greenhouses. Ann Appl Biol 2008; 153: 149-55. [http://dx.doi.org/10.1111/j.1744-7348.2008.00245.x] |
| [30] | Alfaro-Fernández A, Sánchez-Navarro JÁ, Cebrián MC, Cordoba-Selles MC, Pallas V, Jorda C. Simultaneous detection and identification of Pepino mosaic virus (PepMV) isolates by multiplex one-step RT-PCR. Eur J Plant Pathol 2009; 125: 143-58. [http://dx.doi.org/10.1007/s10658-009-9466-7] |
| [31] | Noël P, Hance T, Bragard C. Transmission of the pepino mosaic virus by whitefly. Eur J Plant Pathol 2014; 138: 23-7. [http://dx.doi.org/10.1007/s10658-013-0313-5] |
| [32] | Hanssen IM, Mumford R, Blystad DG, et al. Seed transmission of Pepino mosaic virus in tomato. Eur J Plant Pathol 2010; 126: 145-52. [http://dx.doi.org/10.1007/s10658-009-9528-x] |
| [33] | Mehle N, Gutiérrez-Aguirre I, Prezelj N, Delić D, Vidic U, Ravnikar M. Survival and transmission of potato virus Y, pepino mosaic virus, and potato spindle tuber viroid in water. Appl Environ Microbiol 2014; 80(4): 1455-62. [http://dx.doi.org/10.1128/AEM.03349-13] [PMID: 24334672] |
| [34] | Moreno-Pérez MG, Pagán I, Aragón-Caballero L, Cáceres F, Fraile A, García-Arenal F. Ecological and genetic determinants of Pepino Mosaic Virus emergence. J Virol 2014; 88(6): 3359-68. [http://dx.doi.org/10.1128/JVI.02980-13] [PMID: 24390328] |
| [35] | Domingo E. Viruses at the edge of adaptation. Virology 2000; 270(2): 251-3. [http://dx.doi.org/10.1006/viro.2000.0320] [PMID: 10792982] |
| [36] | Hasiów-Jaroszewska B, Paeleman A, Ortega-Parra N, et al. Ratio of mutated versus wild-type coat protein sequences in Pepino mosaic virus determines the nature and severity of yellowing symptoms on tomato plants. Mol Plant Pathol 2013; 14(9): 923-33. [http://dx.doi.org/10.1111/mpp.12059] [PMID: 23855964] |
| [37] | Gómez P, Sempere RN, Elena SF, Aranda MA. Mixed infections of Pepino mosaic virus strains modulate the evolutionary dynamics of this emergent virus. J Virol 2009; 83(23): 12378-87. [http://dx.doi.org/10.1128/JVI.01486-09] [PMID: 19759144] |
| [38] | Alfaro-Fernández A, Cebrián MC, Córdoba-Sellés C, Herrera-Vásquez JA, Jordá C. First report of the US1 strain of Pepino mosaic virus in tomato in the Canary Isands, Spain. Plant Dis 2008; 92(11): 1590. [http://dx.doi.org/10.1094/PDIS-92-11-1590C] [PMID: 30764462] |
| [39] | French CJ, Dubeau C, Bunckle A, et al. Overview of Pepino Mosaic Virus research. Can J Plant Pathol 2008; 30: 373-4. |
| [40] | Ling K-S, Li R, Bledsoe M. Pepino mosaic virus genotype shift in North America and development of a loop-mediated isothermal amplification for rapid genotype identification. Virol J 2013; 10: 117. [http://dx.doi.org/10.1186/1743-422X-10-117] [PMID: 23587202] |
| [41] | Gómez P, Sempere R, Aranda M, Elena S. Phylodynamics of Pepino mosaic virus in spain. Eur J Plant Pathol 2012; 134: 445-9. [http://dx.doi.org/10.1007/s10658-012-0019-0] |
| [42] | Souiri A, Zemzami M, Laatiris H, Amzazi S, Ennaji MM. First report of Pepino mosaic virus in tomato in Morocco. New Dis Rep 2017; 35: 10. [http://dx.doi.org/10.5197/j.2044-0588.2017.035.010] |
| [43] | Souiri A, Amzazi S, Laatiris H, Ennaji MM, Zemzami M. Serological detection of tomato Pepino mosaic virus in Morocco. J Agr Sci Tech B 2013; 3: 847-52. |
| [44] | Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat Protoc 2006; 1(2): 581-5. [http://dx.doi.org/10.1038/nprot.2006.83] [PMID: 17406285] |
| [45] | Ling K-S, Wintermantel WM, Bledsoe M. Genetic composition of Pepino mosaic virus population in North American greenhouse tomatoes. Plant Dis 2008; 92(12): 1683-8. [http://dx.doi.org/10.1094/PDIS-92-12-1683] [PMID: 30764290] |
| [46] | Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 1999; 41: 95-8. |
| [47] | Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994; 22(22): 4673-80. [http://dx.doi.org/10.1093/nar/22.22.4673] [PMID: 7984417] |
| [48] | Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 2016; 33(7): 1870-4. [http://dx.doi.org/10.1093/molbev/msw054] [PMID: 27004904] |
| [49] | Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 1993; 10(3): 512-26. [PMID: 8336541] |
| [50] | Aloui A, Kenny L. The cost of compliance with SPS standards for Moroccan exports: A case study. 2005. World Bank Agriculture and Rural Development Discussion Paper. World Bank, Washington DC. |
| [51] | Ju HJ, Brown JE, Ye CM, Verchot-Lubicz J. Mutations in the central domain of potato virus X TGBp2 eliminate granular vesicles and virus cell-to-cell trafficking. J Virol 2007; 81(4): 1899-911. [http://dx.doi.org/10.1128/JVI.02009-06] [PMID: 17151124] |
| [52] | Hasiów-Jaroszewska B, Minicka J, Borodynko N, Pospieszny H. The genetic determinants of symptoms induction by Pepino Mosaic virus in tomato. ISHS Acta Hortic 2015; 1069: 39-43. [http://dx.doi.org/10.17660/ActaHortic.2015.1069.4] |
| [53] | Minicka J, Rymelska N, Elena SF, Czerwoniec A, Hasiów-Jaroszewska B. Molecular evolution of Pepino mosaic virus during long-term passaging in different hosts and its impact on virus virulence. Ann Appl Biol 2015; 166: 389-401. [http://dx.doi.org/10.1111/aab.12179] |
| [54] | Hoogenven JP. Besluit van de Staatssecretaris van Economische Zaken van 7 oktober 2013, nr 13168344, Wet ewasbeschermingsmiddelen en biociden ter bestrijding van Pepinomozaïekvirus in de productieteelt van tomaat (Tijdelijke vrijstelling voor het gewasbeschermingsmiddel PMV®-01 ter bescherming van de onbelichte teelt van tomaat, toe te passen in de productieteelt). 2013. Staatscourant, 29627 |
| [55] | Agriculture du maghreb. PMV®-01 the sustainable and efficient solution for a tomato crop without Pepino, Agriculture du maghreb, 2016, 96: 24-25 |