- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Genetic Characterization of HIV-1 Strains Among the Injecting Drug Users in Nagaland, India
Roni Sarkar, Reshmi Pal, Baishali Bal, Ranajoy Mullick, Satarupa Sengupta, Kamalesh Sarkar, Sekhar Chakrabarti*
Abstract
Global HIV-1 surveillance has led to the detection of its new recombinant forms. This study was carried out for the first time to elucidate the genetic characterization and evolutionary relationship of HIV-1 strains among injecting drug users of Nagaland, northeastern India. A total of 156 injecting drug users participated in this study voluntarily. Among them 18 were seropositive for HIV-1 (11.5%).
The Heteroduplex Mobility Assay (HMA) of HIV-1 based on p24-p7 region of gag gene and C2-V3 region of env gene revealed 11 samples to be subtype C (gag/env), 1 sample as subtype B (gag/env) and 6 samples to be recombinants between subtype C and B. Also, the sequencing and phylogenetic analysis of gag (p24-p7) and env (C2-V3) genes from eighteen samples of Nagaland IDUs with different global HIV-1 strains showed the presence of Indian, African, Thai and their recombinant forms. However, more recombinant strains based on different genomic regions of HIV-1 were detected using Multiregional Hybridization Assay (MHA) where 8 out of 18 samples were found to be recombinants between subtype C and B. Thus, multiregional hybridization assay along with heteroduplex mobility assay can serve as an efficient tool in the characterization of recombination pattern among the newly emerging HIV-1 recombinants.
Article Information
Identifiers and Pagination:
Year: 2011Volume: 5
First Page: 96
Last Page: 102
Publisher Id: TOVJ-5-96
DOI: 10.2174/1874357901105010096
Article History:
Received Date: 22/2/2011Revision Received Date: 19/4/2011
Acceptance Date: 20/4/2011
Electronic publication date: 15/7/2011
Collection year: 2011
open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http: //creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the National Institute of Cholera and Enteric Diseases, P-33, C.I.T. Road, Scheme-XM, Beliaghata, Calcutta-700010, India; Tel: 91-33-2370 1176; Fax: 91-33-2370 5066; E-mail: drsekharchakrabarti@yahoo.co.in
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 22-2-2011 |
Original Manuscript | Genetic Characterization of HIV-1 Strains Among the Injecting Drug Users in Nagaland, India | |
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) is a double single stranded RNA virus. It belongs to the Family: Retroviridae, Genus: lentivirus and Species: Human immunodeficiency virus 1. India has been witnessing the rapid spread of HIV-1 infection throughout the north-eastern border region since a very long time [1Sarkar S, Mookerjee P, Roy A, et al. Descriptive epidemiology of intravenous heroin users- a new risk group for transmission of HIV in India J Infect 1991; 23: 201-7., 2Weniger BG, Takebe Y, Ou CY, Yamazaki S. The molecular epidemiology of HIV in Asia AIDS 1994; 8: 13-28.]. Nagaland, one of the northeastern states of India, bordering Myanmar and Manipur and close to the “golden triangle”, is one of the major opium and heroin producing areas of south-east Asia. It is the sixth highly prevalent state in the country with a prevalence rate of 1.27 %. Sexual route of transmission has taken predominance in this area with 78.73%, followed by transmission with infected needles and syringes contributing to 12.21 %, as reported by Nagaland states AIDS control society. The high rate of HIV-1 transmission among injecting drug users of Manipur, a neighboring state of Nagaland, through sharing of contaminated needles and syringes was reported earlier [1Sarkar S, Mookerjee P, Roy A, et al. Descriptive epidemiology of intravenous heroin users- a new risk group for transmission of HIV in India J Infect 1991; 23: 201-7., 2Weniger BG, Takebe Y, Ou CY, Yamazaki S. The molecular epidemiology of HIV in Asia AIDS 1994; 8: 13-28.]. Previous studies from Manipur detected the presence of Thai-B as the second major subtype after subtype-C, circulating among IDUs [3Mandal DK, Jana S, Bhattacharya SK, Chakrabarti S. HIV-1 subtypes circulating in eastern and northeastern regions of India AIDS Res Hum Retroviruses 2002; 18: 1219-27.-5Bhanja P, Sengupta S, Singh NY, Sarkar K, Bhattacharya SK, Chakrabarti S. Determination of gag and env subtypes of HIV-1 detected among injecting drug users in Manipur, India: Evidence for intersubtype recombination Virus Res 2005; 114: 149-53.]. Recent studies have revealed the occurrence of different multigenomic variants of HIV-1 through multiregional hybridization assay in Manipur which might pose serious challenge to effective candidate vaccine design against newly evolving HIV-1 viruses in the northeastern states of India [6Sarkar R, Sengupta S, Mullick R, Singh NB, Sarkar K, Chakrabarti S. Implementation of a multiregion hybridization assay to characterize HIV-1 strains detected among Injecting Drug Users in Manipur, India Intervirology 2009; 52: 175-8.]. But the nucleotide sequencing of the whole 9 kb HIV-1 genome is a tedious as well as expensive procedure. However, a breakthrough has been achieved by the combined implementation of heteroduplex mobility assay along with multiregional hybridization assay. This parallel combination has been successful to reveal a detailed picture of emerging recombination patterns in different genomic regions of HIV-1. In order to bring forth the present scenario of HIV-1 strains in north eastern states of India, we have selected Nagaland where the characterization of HIV-1 strains has not been conducted till date. Thus the present study focuses towards the trend of increasing HIV-1 recombinant strains in the northeastern states of India.
MATERIALS AND METHODOLOGY
Study Design
It was a community based cross-sectional study. Male IDUs of ages 31- 40 years were contacted through secondary informant method followed by snowballing. All IDUs were explained about the purpose of this study and requested to participate voluntarily. Experienced social workers interviewed them to study their demography, risk behavior and risk perception using a field tested questionnaire. Interview was followed by collection of about 5 ml blood in vacationer containing EDTA. HIV was tested by enzyme linked immunosorbent assay (Immunogenetics, Belgium) followed by tridot assay (Standard Diagnostics, Bioline, Korea). Peripheral blood mononuclear cells were separated from the whole blood by Ficoll-Hypaque gradient centrifugation [7Boyam A. Separation of leucocytes from blood and bone marrow Scand J Clin Lab Invest 1968; 21: 77-81.]. DNA was extracted by using the QIAamp DNA Blood Mini Kit 250 (QIAGEN, Germany, Hilden) according to the manufacturer’s protocol.
Heteroduplex Mobility Assay
The HIV-1 strains were subjected to gag and env Heteroduplex Mobility Assay as described earlier [5Bhanja P, Sengupta S, Singh NY, Sarkar K, Bhattacharya SK, Chakrabarti S. Determination of gag and env subtypes of HIV-1 detected among injecting drug users in Manipur, India: Evidence for intersubtype recombination Virus Res 2005; 114: 149-53., 8Heyndrix L, Janssens W, Zekeng L, et al. Simplified strategy for detection of recombinant HIV-1 group M isolates by gag/ env heteroduplex mobility assay J Virol 2000; 74: 363-70.]. Briefly, the amplicons (460bp, 500bp) for gag (p24-p7) and env (C2-V3) were amplified through nested PCR using standard primers (NIH AIDS Research and Reference Reagent Program, NIH, USA). For gag HMA, 4.5μl of the amplicon of the unknown sample was mixed with 4.5μl of the reference amplicon in presence of 1μl of 10x annealing buffer (1M NaCl, 100Mm Tris-HCl [pH 7.8], 20Mm EDTA). For env HMA, 5μl of the amplicon was mixed with 5μl of the reference amplicon in the presence of 1.1μl of 10x annealing buffer. It was then denatured at 94°C for 2 minutes followed by renaturation by snap freezing in ice to form Heteroduplex molecules. The mixture were then loaded on a 5% Polyacrylamide Gel (for gag-HMA, 5% Polyacrylamide / 20% Urea; 5% polyacrylamide for env) in 1X TBE buffer and electrophoresed at 250 V for 2 hours 30 minutes. Based on the relative mobility of the heteroduplex molecules towards homoduplexes, subtypes were assigned. Heteroduplex molecules formed between the unknown sample and the most closely related subtype exhibited the fastest mobility.
Multiregional Hybridization Assay
The MHAbce v.2 study was a Taqman assay based on real time PCR method performed using probes designed from eight different genomic regions (p17, pro, rt, int, tat, gp120, gp41 and gpnef) of HIV-1 as described earlier [6Sarkar R, Sengupta S, Mullick R, Singh NB, Sarkar K, Chakrabarti S. Implementation of a multiregion hybridization assay to characterize HIV-1 strains detected among Injecting Drug Users in Manipur, India Intervirology 2009; 52: 175-8., 9Kijak GH, Tovanabutra S, Sanders-Buell E, et al. Distinguishing molecular forms of HIV-1 in Asia with a high-throughput, fluorescent genotyping assay, MHAbce v.2 Virology 2007; 358: 178-91.]. Briefly, the first round PCR was conducted at ABI9600 Thermal cycler (Applied Biosystems) with outer primers designed for specific genomic regions, 10x PCR buffer, 25mM MgCl2, 10mM dNTPs and 1U AmpliTaq Gold DNA polymerase (Applied Biosystems) in a final volume of 50µl. The second round PCR was carried out in 96 well ABI PRISM 7900HT sequence detection system (Applied Biosystems) using inner primers and probes designed for specific genomic regions in a final volume of 25µl. The probes were labeled with either 6-carboxy fluorescein (FAM) or 6-carboxytetramethylrhodamine (TET) at the 5’ end and Black Hole Quencher (BHQ) at the 3’ end. The fluorescence intensity was measured by SDS v.2 software (Applied Biosystem, USA). The PCR product generated was checked by conducting dissociation curve analysis through SybrGreen PCR Master Mix (Applied Biosystems) which distinguishes PCR product from primer dimmers of lower thermal stability.
DNA Sequencing, Phylogenetic and Simplot Analysis
Amplicons of the gag & env gene segments were purified by a QIA quick PCR purification kit (QIAGEN, Germany, and Hilden) and were subjected to cycle sequencing reactions using fluorescent dye-labeled dideoxy nucleotides in an ABI PRISM 3100 automated sequencer following the manufacturer’s protocol. The sequences were edited manually using BIOEDIT sequence alignment editor program (version 5.0.6; Department of Microbiology, North Carolina State University) [http://www.mbio.ncsu.edu/Bioedit/BioDoc.pdf]. The edited sequences were Blast searched and further aligned with the reference sequences from different geographic regions available in the HIV database (http://www.hiv.lanl.gov/content/index) for phylogenetic analysis using the Molecular evolutionary genetics analysis software version 4 (MEGA 4) [10Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis software version 4 Mol Biol Evol 2007; 24: 1596-9.].
Gene Bank Accession Numbers
The Gen- Bank accession numbers for the nucleotide sequences of gag (p24-p7) and env (C2-V3) reported in this paper are EU541498, EU526648- EU526656, EU526658- EU526666, EU526668- EU526669, HM-130667, HQ897949- HQ897962.
RESULTS
HMA Analysis
Out of 18 HIV seropositive samples, the gag heteroduplex mobility assay of all the 18 samples of Nagaland injecting drug users showed 17 samples as subtype C while one as subtype B (nag120) (Table 1). On the other hand, env heteroduplex mobility assay showed 11 samples as subtype C and rest of the 7 samples e.g. nag1, nag12, nag23, nag 57, nag120, nag135 and nag153 as subtype B. (Fig. 1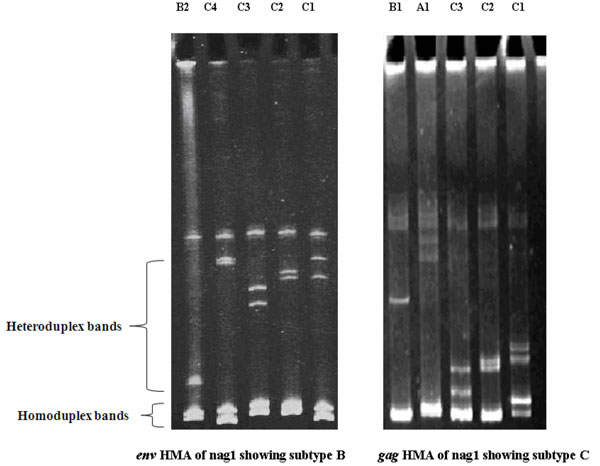 ) shows the subtype specific heteroduplex mobility for env (C2-V3) and gag (p24-p7) genes of “nag 1”.
) shows the subtype specific heteroduplex mobility for env (C2-V3) and gag (p24-p7) genes of “nag 1”.
MHA Analysis
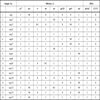
Multiregional hybridization assay was carried out for all the 18 samples (Table 1). The analysis showed that nag 25, nag 33, nag 49, nag 65, nag 81, nag 86, nag 111, nag 113 and nag 157 belonged to subtype C. However, subtype B probe reacted with nag120. Multigenomic recombination was detected for the samples nag 1, nag 12, nag 23, nag 57, nag 88, nag 135, nag 152 and nag 153. The samples nag 23, nag 88 and nag152 showed both dual probe reactivity and multigenomic recombination pattern.
Phylogenetic Analysis
The B/C recombination pattern with respect to gag (p24-p7) and env (C2-V3) of the recombinant samples was confirmed by phylogenetic analysis. Phylogenetic analysis of the gag (p24-p7) gene of Nagaland injecting drug users with the reference subtype C and subtype B sequences available in the database (http://www.hiv.lanl.gov/content/index) clearly showed that the samples nag 1, nag 23, nag 25, nag 33, nag 49, nag 57, nag 65, nag 81, nag 86, nag 88, nag 111, nag 135 clustered with subtype C HIV-1 strains from Africa and nag 12, nag 113, nag 152, nag 153, nag 157 clustered with Indian subtype C. For nag 120, gag (p24-p7) gene clustered with subtype B strains from China (Fig. 2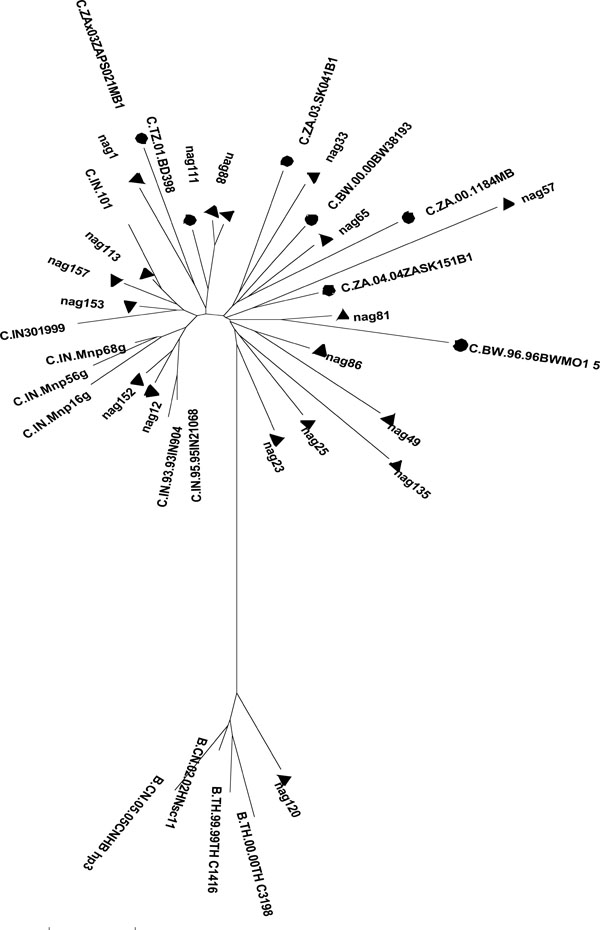 ). On the other hand, phylogenetic analysis of the env C2-V3 gene with other global HIV-1 strains showed that nag 25, nag 33, nag 65, nag 86, nag 88, nag 111, nag 113 formed cluster with subtype C reference sequences from Africa and nag 49, nag 81, nag 152, nag 157 formed cluster with Indian subtype C (Fig. 3
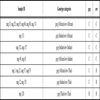
). On the other hand, phylogenetic analysis of the env C2-V3 gene with other global HIV-1 strains showed that nag 25, nag 33, nag 65, nag 86, nag 88, nag 111, nag 113 formed cluster with subtype C reference sequences from Africa and nag 49, nag 81, nag 152, nag 157 formed cluster with Indian subtype C (Fig. 3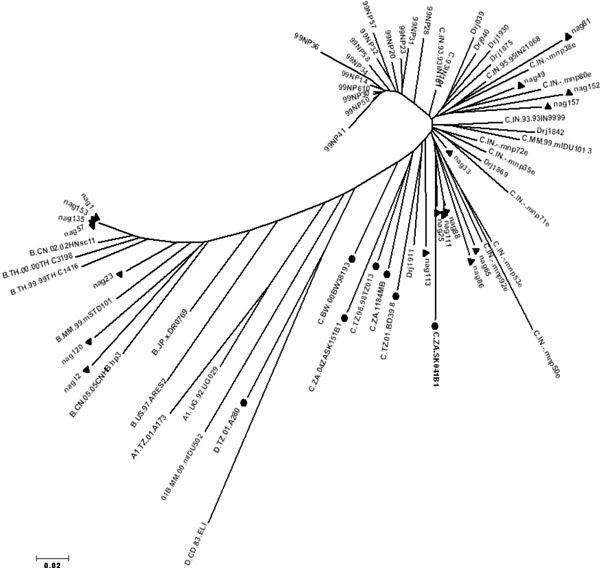 ). However nag 1, nag 57, nag 135, nag 153 formed a unique cluster close to Thai B sequences and nag 12, nag 23 and nag 120 clustered with subtype B sequences from China and Myanmar. The results obtained from the phylogenetic analysis were further validated through Simplot analysis (data not shown). While focusing on individual HIV-1 isolates, the information revealed regarding the genotyping pattern of gag (p24-p7) and env (C2-V3), can be divided into seven categories as shown (Table 2). First, gag B/ env B showed by nag 120; second, gag C (African)/ env C (African) showed by nag 65, nag 86, nag 88, nag 111, nag 33, nag 25; third, gag C (African) / env C (Indian) showed by nag 49, nag 81; fourth gag C (Indian) / env C (Indian) showed by nag 152, nag 157; fifth gag C (Indian) / env C (African) showed by nag 113; sixth gag C (African)/ env (Thai B) showed by nag 1, nag 23, nag 57, nag 135 and the seventh one gag C (Indian) / env (Thai B) showed by nag 12 and nag 153.
). However nag 1, nag 57, nag 135, nag 153 formed a unique cluster close to Thai B sequences and nag 12, nag 23 and nag 120 clustered with subtype B sequences from China and Myanmar. The results obtained from the phylogenetic analysis were further validated through Simplot analysis (data not shown). While focusing on individual HIV-1 isolates, the information revealed regarding the genotyping pattern of gag (p24-p7) and env (C2-V3), can be divided into seven categories as shown (Table 2). First, gag B/ env B showed by nag 120; second, gag C (African)/ env C (African) showed by nag 65, nag 86, nag 88, nag 111, nag 33, nag 25; third, gag C (African) / env C (Indian) showed by nag 49, nag 81; fourth gag C (Indian) / env C (Indian) showed by nag 152, nag 157; fifth gag C (Indian) / env C (African) showed by nag 113; sixth gag C (African)/ env (Thai B) showed by nag 1, nag 23, nag 57, nag 135 and the seventh one gag C (Indian) / env (Thai B) showed by nag 12 and nag 153.
DISCUSSION
Due to its geographical proximity with the golden triangle, Manipur and the adjoining states of Nagaland have made the north-eastern region vulnerable to HIV infection [1Sarkar S, Mookerjee P, Roy A, et al. Descriptive epidemiology of intravenous heroin users- a new risk group for transmission of HIV in India J Infect 1991; 23: 201-7., 2Weniger BG, Takebe Y, Ou CY, Yamazaki S. The molecular epidemiology of HIV in Asia AIDS 1994; 8: 13-28.]. Rampant injecting practices have contributed to making these areas as the main abode for HIV. Sharing of injecting equipments frequently occurs among injecting drug users and drug traders as a part of their drug-purchasing behavior, thereby, leading to a possibility of introduction of different HIV- 1 subtypes into circulation. Similar observations have been found in studies from China where HIV epidemic due to different subtypes has been spreading along heroin trafficking routes [11Piyasirisilp S, McCutchan FE, Carr JK, et al. A Recent Outbreak of Human Immunodeficiency Virus Type 1 Infection in Southern China Was Initiated by Two Highly Homogeneous, Geographically Separated Strains, Circulating Recombinant Form AE and a Novel BC Recombinant J Virol 2000; 74: 11286-95.]. Genetic diversity of HIV-1 might also be related to cross-border migration and sharing practices of IDUs of different ethnicity (host factor). The IDUs who had sexual relationship with other IDUs were especially at the brink of acquiring HIV infection. These findings suggest that an IDU having these risk behaviors has multiple chances of getting exposed to different circulating subtypes of HIV-1 thereby facilitating the development of novel recombinant strains. Various other studies have documented the rapid spread of recombinant strains of HIV among IDUs due to unsafe injecting practices [12Kalish ML, Baldwin A, Raktham S, et al. The evolving molecular epidemiology of HIV-1 envelope subtypes in injecting drug users in Bangkok, Thailand: implications for HIV vaccine trials AIDS 1995; 9: 851-7.-14Liitsola K, Tashkinova I, Laukkanen T, et al. HIV-1 genetic subtype A/B recombinant strain causing an explosive epidemic in injecting drug users in Kaliningrad AIDS 1998; 12: 1907-.].
The sequencing and phylogenetic analyses of gag and env genes in this study clearly indicate that the HIV-1 strains from Africa are in circulation along with Indian strains. The introduction of African HIV-1 strains might have occurred in the past and had become stable in the northeastern IDU population. Though it is well known that the B/ C recombinants were evolved from recombination between subtype C and subtype B viruses due to dual infection, however, the recombination pattern in different genes of HIV-1 detected by multiregional hybridization assay, raises the possibility of recombination within recombinant strains and the generation of mosaic recombinant forms of HIV-1. The occurrence of Indian C (gag) /Thai-B (env), African C (gag)/ Thai-B (env), Indian C (gag)/ African C (env), African C (gag)/ Indian C (env) recombinant forms of HIV-1 among the injecting drug users of Nagaland brings about the possibility of incorporation of recombinant HIV-1 strains in Nagaland IDUs. This gives an alarming signal of increasing population with inter and intragenomic recombinant forms of HIV-1 which could lead to serious epidemic among injecting drug users of the north-eastern region. Multiregional hybridization assay along with heteroduplex mobility assay enabled us to scan the whole HIV-1 genome within a short period of time. In Multiregional Hybridization Assay, dual probe reactivity and failure of probe hybridization in a few cases could have been the outcome of genetic diversity of HIV-1 within the clade [9Kijak GH, Tovanabutra S, Sanders-Buell E, et al. Distinguishing molecular forms of HIV-1 in Asia with a high-throughput, fluorescent genotyping assay, MHAbce v.2 Virology 2007; 358: 178-91.]. The full length HIV-1 gene amplification of the samples with B/C recombination is in process to track the possibility of newly circulating recombinant forms among Nagaland injecting drug users. Moreover, it remains to be investigated whether recombination may confer selective advantage over parental viruses.
ACKNOWLEDGEMENTS
We sincerely acknowledge the cooperation of Nagaland State AIDS Control and Prevention in this study. We also thank the Dept. of Biotechnology, Govt. of India and Indian Council of Medical Research for financial support.