- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Laboratory Detection of Respiratory Viruses by Automated Techniques
Mercedes Pérez-Ruiz*, Irene Pedrosa-Corral, Sara Sanbonmatsu-Gámez , José-María Navarro-Marí
Abstract
Advances in clinical virology for detecting respiratory viruses have been focused on nucleic acids amplification techniques, which have converted in the reference method for the diagnosis of acute respiratory infections of viral aetiology. Improvements of current commercial molecular assays to reduce hands-on-time rely on two strategies, a stepwise automation (semi-automation) and the complete automation of the whole procedure. Contributions to the former strategy have been the use of automated nucleic acids extractors, multiplex PCR, real-time PCR and/or DNA arrays for detection of amplicons. Commercial fully-automated molecular systems are now available for the detection of respiratory viruses. Some of them could convert in point-of-care methods substituting antigen tests for detection of respiratory syncytial virus and influenza A and B viruses. This article describes laboratory methods for detection of respiratory viruses. A cost-effective and rational diagnostic algorithm is proposed, considering technical aspects of the available assays, infrastructure possibilities of each laboratory and clinic-epidemiologic factors of the infection
Article Information
Identifiers and Pagination:
Year: 2012Volume: 6
Issue: Suppl 1
First Page: 151
Last Page: 159
Publisher Id: TOVJ-6-151
DOI: 10.2174/1874357901206010151
Article History:
Received Date: 4/7/2012Revision Received Date: 11/9/2012
Acceptance Date: 20/9/2012
Electronic publication date: 30/11/2012
Collection year: 2012
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http: //creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Servicio de Microbiología. Hospital Universitario Virgen de las Nieves, Avda. Fuerzas Armadas, 2, 18014 Granada, Spain; Tel: +34 958 020 717; E-mail: mercedes.perez.ruiz.sspa@juntadeandalucia.es
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 4-7-2012 |
Original Manuscript | Laboratory Detection of Respiratory Viruses by Automated Techniques | |
1. INTRODUCTION
Acute respiratory infection (ARI) of viral aetiology is one of the main causes of morbidity and mortality worldwide [1Hall MJ, DeFrances CJ, Williams SN, Golosinskiy A, Schwartzman A. National Hospital Discharge Survey: 2007 summary Natl Health Stat Report 2010; 29: 1-20-24.-4Niska R, Bhuiya F, Xu J. National hospital ambulatory medical care survey: 2007 emergency department summary Natl Health Stat Report 2010; 26: 1-31.]. Besides classical respiratory viruses (RV), i.e. influenza viruses (Flu), respiratory syncytial virus (RSV), human rhinovirus (HRV), parainfluenza virus (PIV) 1-4, adenovirus (ADV), enterovirus (EV) and human coronavirus (HCoV) OC43 and 229E [5Henderson FW, Clyde WA Jr, Collier AM, et al. The etiologic and epidemiologic spectrum of bronchiolitis in pediatric practice J Pediatr 1979; 95: 183-90.-9Atmar RL, Piedra PA, Patel SM, Greenberg SB, Couch RB, Glezen WP. Picornavirus the most common respiratory virus causing infection among patients of all ages hospitalized with acute respiratory illness J Clin Microbiol 2012; 50: 506-8.], other “new” RV have currently been added to this list, such as human metapneumovirus (hMPV) [10Van den Hoogen BG, Osterhaus DM, Fouchier RA. Clinical impact and diagnosis of human metapneumovirus infection Pediatr Infect Dis J 2004; 23: S25-32.], SARS-CoV [11Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome N Engl J Med 2003; 348: 1953-66.], HCoV NL63 and HKU1 [12Van der Hoek L, Pyrc K, Jebbink MF, et al. Identification of a new human coronavirus Nat Med 2004; 10: 368-73.,13Woo PC, Lau SK, Huang Y, Tsoi HW, Chan KH, Yuen KY. Phylogenetic and recombination analysis of coronavirus HKU1, a novel coronavirus from patients with pneumonia Arch Virol 2005; 150: 2299-311.], human bocavirus (hBoV) [14Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples Proc Natl Acad Sci USA 2005; 102: 12891-6.] and others. Most RV cause benign and self-limited infections in immunocompetent adults. However, clinical impact of ARI increases when they affect children, elderly, immunocompromised individuals and/or subjects with chronic underlying diseases, in whom RV constitute an important cause of hospitalisations, mainly during the cold months of the year [15Monto AS. Epidemiology of viral respiratory infections Am J Med 2002; 112(Suppl 6A): 4S-12S.,16Navarro-Marí JM, Perez-Ruiz M. Virus respiratorios: viejos y nuevos virus. Revisión de métodos diagnósticos Enferm Infecc Microbiol Clin 2007; 25(Suppl 3): 60-5.]. They involve important costs for health systems and constitute a frequent cause of school and work absenteeism. In certain circumstances, emerging RV cause a great impact in health systems worldwide, as occurred with SARS-CoV and Flu pandemics, the last one taking place in 2009.
Aetiological diagnosis of viral ARI is crucial in order to avoid unnecessary antibiotic use, to establish the appropriate use of antiviral drugs and a comprehensive cohort of hospitalised patients that minimizes the risk of nosocomial transmission, and to provide epidemiological information for an early release of recommendations for the prevention and treatment and, finally, to reduce the overall costs derived from patients´ management.
Several diagnostic methods have been described for RV. They can be classified in direct detection methods, which detect the virus or a part of it in the clinical sample, including viral culture, antigen detection methods and molecular techniques; and indirect methods, based on the detection of immune response to the virus by serologic assays. In clinical practice, serology is not useful for the diagnosis of viral infections, such as ARI, that cause frequent recurrences and are mainly restricted to the respiratory mucosa. Thus, in these cases, direct detection of the virus in the site of infection has to be a priority [16Navarro-Marí JM, Perez-Ruiz M. Virus respiratorios: viejos y nuevos virus. Revisión de métodos diagnósticos Enferm Infecc Microbiol Clin 2007; 25(Suppl 3): 60-5.,17Loeffelholz M, Chonmaitree T. Advances in diagnosis of respiratory virus infections Int J Microbiol 2010; 2010: 126049.].
Advances in diagnostic virology for detecting RV have been focussed on nucleic acids amplification techniques (NAAT), since they are more sensitive than traditional methods such as viral culture or antigen detection assays. Moreover, NAAT are the only available method for most new RV because they do not grow or grow poorly in cell culture and for which no efficient antigen detection methods have been commercialised. NAAT offer additional advantages with respect to other classical methods as less turnaround time and less effect of virus viability on the efficiency of the assay [18Mahony JB. Detection of respiratory viruses by molecular methods Clin Microbiol Rev 2008; 21: 716-47.].
On the contrary, some disadvantages have been attributed to NAAT: increased reagents’ costs, higher hands-on time, the need of knowing at least a part of the target genome to design appropriate primers and/or probes. Indeed, many RV are RNA viruses, subjected to a greater genetic variability that may affect the sensitivity and reliability of the molecular method.
Currently, many commercial molecular assays have been developed that allow partial or complete automation of the procedure. Semi-automated or automated assays have solved in part one disadvantage of NAAT, which is the hands-on time. Many commercial NAAT require minimal management of the specimen, taking no more than few minutes.
With the increasing availability of easy-to-perform and multiplex NAAT for detecting RV, cost-effective diagnostic algorithms should be used. Whether a more or less complete virological study should be carried out on a certain patient’s sample, depends not only on the resources and type of laboratory, but also on clinic-epidemiologic aspects such as patient’s age, severity of the disease, underlying conditions, etc.
This article describes the molecular methods used to detect respiratory viruses and the contribution that increasing automation, especially on molecular assays, has made to improve laboratory diagnosis. The possibilities of integration of these new assays on each laboratory portfolio are also discussed.
2. TRADITIONAL DETECTION METHODS
Traditional direct methods for RV are viral culture and antigen detection assays. Among indirect methods, many serologic assays, such as EIA, haemmagglutination inhibition, complement fixation, etc. have been used for RV. However, serology is not useful in clinical practice since diagnosis must be achieved by demonstration of seroconversion or seroincrease of IgG antibodies, between acute-phase and convalescent-phase serum samples [16Navarro-Marí JM, Perez-Ruiz M. Virus respiratorios: viejos y nuevos virus. Revisión de métodos diagnósticos Enferm Infecc Microbiol Clin 2007; 25(Suppl 3): 60-5.,18Mahony JB. Detection of respiratory viruses by molecular methods Clin Microbiol Rev 2008; 21: 716-47.].
In past decades, viral culture was the gold standard for the aetiological diagnosis of RV. No automation has been reported for this technology. Rather, advances in this field of clinical virology have been focused on shortening detection times and on increasing sensitivity. Currently, this procedure is limited to reference laboratories that are provided with the appropriate infrastructure, with the main objective of recovering the viral strain for further genetic and/or antigenic characterisation.
Traditional viral culture requires at least 7 days for yielding a result. To solve this problem, the next step in the viral culture methodology was the introduction of the shell-vial assay, based on centrifugation-enhanced adsorption of the sample onto the cells monolayer reproduced on cover slips. Detection is commonly achieved by fluorescent assay with monoclonal antibodies on the cell monolayer. This method reduces the time to obtain a result to 24-48 h without a deleterious effect on sensitivity and specificity.
Although viral culture may represent an open system for detecting multiple viral pathogens, there is no single cell line that allows the growth of all RV, and there is no RV that may grow in any cell line. Thus, to detect the most prevalent RV, multiple cell lines must be used. The combination of shell-vial assay with the inclusion of different cell lines in the same vial has represented a subsequent step that has facilitated the recovery of RV isolates from cell cultures [19Navarro-Marí JM, Sanbonmatsu-Gámez S, Pérez-Ruiz M, De La Rosa-Fraile M. Rapid detection of respiratory viruses by shell vial assay using simultaneous cultura of HEp-2, LLC-MK2, and MDCK cells in a single vial J Clin Microbiol 1999; 37: 2346-7.-21Weinberg A, Brewster L, Clark J, Simoes E. ARIVAC consortium. Evaluation of R-Mix shell vials for the diagnosis of viral respiratory tract infections J Clin Virol 2004; 30: 100-5.]. NAAT have recently contributed to increase the sensitivity of viral culture. PCR assays can be carried out on the cell culture supernatant to detect the isolates. This approach has demonstrated the highest sensitive reading procedure, and has allowed identifying viral isolates in cell monolayers with no cytopathic effect [22Pérez-Ruiz M, Yeste R, Ruiz-Pérez MJ, Ruiz-Bravo A, de la Rosa-Fraile M, Navarro-Marí JM. Testing of diagnostic methods for detection of influenza virus for optimal performance in the context of an influenza surveillance network J Clin Microbiol 2007; 45: 3109-10.]. However, viral culture is not useful for detection of new RV, since they do not grow or grow poorly in cell culture. For this purpose, the best alternative is NAAT.
The availability of monoclonal antibodies has allowed the development of different technologies for detecting viral antigens from clinical samples. Fluorescent and chromatographic immunoassays are rapid, easy to perform and have proved their utility, mainly for RSV and Flu. The minimum (hands-on and hands-off) turnaround time that these techniques require for obtaining results have converted them as referral point-of-care diagnostic assays. However, sensitivity widely depends on the commercial method and on the type of patient and sample, ranging from 10% to 70% in the case of pandemic Flu [23Centers for Disease Control and Prevention. Evaluation of rapid Influenza diagnostic tests for detection of Novel Influenza A (H1N1) Virus — United States, 2009 MMWR 2009; 58: 826-9.]. Higher viral loads and thus, higher sensitivity of antigen detection methods for RSV and Flu have been observed in the paediatric population versus adults and in nasopharyngeal aspirates versus nasopharyngeal exudates [24Utokaparch S, Marchant D, Gosselink JV, et al. The relationship between respiratory viral loads and diagnosis in children presenting to a pediatric hospital emergency department Pediatr Infect Dis J 2011; 30: e18-23.,25Lieberman D, Lieberman D, Shimoni A, Keren-Naus A, Steinberg R, Shemer-Avni Y. Identification of respiratory viruses in adults: nasopharyngeal versus oropharyngeal sampling J Clin Microbiol 2009; 47: 3439-43.].
Recently, the goal of automation in these type of diagnostic tools has been achieved by a new commercial assay, the mariPOC® respi test (ArcDia, Turku, Finland). This allows rapid detection and differentiation between viral and bacterial respiratory infections (Flu A, Flu B, RSV, hMPV, PIV 1, PIV2, PIV3, ADV and Streptococcus pneumoniae). The test is very simple to operate and hands-on-time is less than a minute per sample. Results for highly positive samples are obtained in 20 minutes whereas low positive and negative samples are reported in 2 hours.
The mariPOC® system uses polystyrene microparticles as solid phase reaction carriers for the immunocomplex formation. Viral antigens are captured by specific antibodies fixed onto the microsphere and bind to a fluorescently-labelled conjugate. The amount of immunocomplexes formed on the microsphere is proportional to the analyte concentration. Detection of the fluorescence signal is carried out by separation-free two-photon excitation fluorescence detection assay, the ArcDiaTM TPX (TPX). In contrast to other antigen detection methods, TPX assay avoids the need for multiple washes, and readout is achieved by on-line monitoring of reaction kinetics [26Koskinen JO, Vainionpää R, Meltola NJ, Soukka J, Hänninen PE, Soini AE. Rapid method for detection of influenza a and B virus antigens by use of a two-photon excitation assay technique and dry-chemistry reagents J Clin Microbiol 2007; 45: 3581-8.]. Preliminary studies have reported a sensitivity and specificity of this method comparable to ultrasensitive fluorescent immunoassays (DELPHIA® method) [27Koskinen JO, Vainionpää R, Soini AE. Novel multianalyte point-of-care test for pathogen-speci?c diagnosing of respiratory infections Clin Microbiol Infect 2009; 15(Suppl S4): S556.,28Toivola H, Koskinen JM, Lappalainen M, Ruonamo R, Antikainen P, Koskinen JO. Performance evaluation of multianalyte mariPOC® respiratory infection test for adenovirus types 1–8, 14, 19 and 21 Clin Microbiol Infect 2012; 18(Suppl S3): 787-8.] and offers the advantage of automated result readout, allowing connections to laboratory information systems.
3. NAAT FOR DETECTION OF RESPIRATORY VIRUSES: FROM THE SIMPLEST TO THE MOST COMPLEX TARGETING APPROACH
NAAT, particularly the polymerase chain reaction (PCR), have expanded the range of pathogens that can be identified in clinical laboratories. They have shown their utility in clinical diagnosis of viral respiratory infections. Molecular assays have greater sensitivity than conventional methods as viral culture or antigen detection and indeed, they are able to detect new viral agents. Another advantage of NAAT compared with viral culture is that they do not require viable viruses. Thus, they have converted in the gold standard for detection of RV. Thus, most advances in clinical diagnostic virology have been made on NAAT.
NAAT are especially useful for detecting “low-positive” patients. Yearly, during periods of RSV and Flu epidemics, the availability of NAAT eases the management of hospitalised patients and the control of outbreaks, since antigen detection tests are not sensitive enough for certain population groups, such as immunosuppressed individuals.
Flu, HRV, PIV 1, PIV 3, PIV 4, ADV and HCoV (except SARS-CoV) usually cause upper respiratory tract infections (URTI), whereas RSV, hMPV and PIV 2 have been frequently associated to lower respiratory tract infections (LRTI) [18Mahony JB. Detection of respiratory viruses by molecular methods Clin Microbiol Rev 2008; 21: 716-47.].
Most RV are RNA viruses except ADV. Two alternatives are feasible for reversetranscription (RT) of viral RNA into its appropriate cDNA PCR target. One approach is the use of random and/or oligo(dT) primers in the RT step. Subsequently, PCR can be directed against any RV with specific primers. The main disadvantage of this procedure is the need for opening the RT tube to carry out the PCR. One-step RT-PCR protocols have been reported elsewhere for most RNA viruses, and solve the problem the former protocol has. These are based on the inclusion in the reaction mix of reversetranscriptases and DNA polymerases for both the RT and the PCR steps, or of enzymes with reversetranscriptase and DNA polymerase activities.
Monoplex PCRs have been published and commercial kits have been developed for detecting the most prevalent RV [18Mahony JB. Detection of respiratory viruses by molecular methods Clin Microbiol Rev 2008; 21: 716-47.]. Whether a simpler or a more complete virological study must be carried out depends on many factors: severity of the disease, type of patient, capacity of the laboratory, availability of validated and optimized trade-marked commercial kits, number of samples that the laboratory can process, etc.
Nevertheless, in many cases, clinical signs and symptoms of ARI are not specific of a single RV, and mild and severe infections have been described for all viruses mentioned above. Currently, monoplex PCR is not useful in a clinical laboratory equipped enough to conduct an extended aetiological diagnosis. However, the advent of the 2009 pandemic allowed many laboratories for equipping with the infrastructure necessary to carry out PCR assays for detection of pandemic Flu. In these circumstances, when a rapid answer is encouraging, monoplex PCRs provide an efficient response.
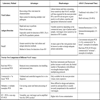
Multiplex PCR assays have been the most widely used molecular methods for RV detection in laboratories of clinical virology capable of developing in-house and/or robust published protocols. The advantage with respect to monoplex PCR is the cost savings compared to multiple monoplex PCRs and an easier processing of runs. Indeed, multiplex PCR extends the portfolio of virological diagnostic methods. Although advantages are strong enough to compensate for any disadvantage, multiplex PCR have intrinsic inconveniences: as the number of targets increase, the possibility of losing sensitivity and specificity increases as well, due to competition among primers and viral targets that consequently yields unspecific PCR products. Indeed, multiplex PCR has to be optimized to ensure the adequate amplification of each viral genome. Multiplex PCR assays have demonstrated that co-detections of RV in a sample are a common event [29Cilla G, Oñate E, Perez-Yarza EG, Montes M, Vicente D, Perez-Trallero E. Viruses in community-acquired pneumonia in children aged less than 3 years old: High rate of viral coinfection J Med Virol 2008; 80: 1843-9.]. Whether these co-detections express true infections by all viruses and/or prolonged shedding of certain agents is difficult to demonstrate, although some authors have pointed out the role of co-detections of RV in the severity of the disease [30Mansbach JM, Piedra PA, Teach SJ, et al. Prospective multicenter study of viral etiology and hospital length of stay in children with severe bronchiolitis Arch Pediatr Adolesc Med 2012; 166(8 ): 700-6.]. Table 1 shows the most important advantages and disadvantages of different methods for detection of RV.
4. AUTOMATION POSSIBILITIES FOR DETECTING RESPIRATORY VIRUSES BY NAAT
Automation in molecular assays, aiming at a continuous reduction of hands-on time, yields higher reproducibility and offers possibility of detecting several pathogens with a minimum processing.
The availability of automated procedures for detecting RV is increasing day by day. This obliges the virologists to choose the best alternative taking into account laboratory throughput and epidemiologic, clinical and cost-effectiveness parameters since, in many instances, each commercial assay requires its own apparatus. Indeed, commercial diagnostic kits are less versatile and often, more expensive than well-optimized in-house PCR.
Molecular procedures for achieving automation goals for RV detection could be divided in two main straightforward phases: procedures and protocols for a stepwise automation (semi-automation) and fully-automated NAAT.
4.1. Stepwise Automation (Semi-Automation)
From nucleic acids extraction to the readout of PCR results, many efficient automated systems covering all steps of the whole procedure have emerged in the last 10-15 years. By chronological order, automated extractors, real-time PCR instruments, DNA arrays (and other systems) for detection of PCR products, and recent platforms for nucleic acids purification and preparation of the PCR master mix, are the main contributions to a semi-automation of molecular methods.
4.1.1. Automated Nucleic Acids Extraction Methods
Several commercial assays offer automated solutions for nucleic acids extraction, isolation and purification that range from low to high throughput. The high quality of nucleic acids obtained with these systems are suitable for a broad range of applications in nucleic acid research, including gene expression analysis (using real-time PCR or conventional PCR on standard block cyclers), microarray analysis, genotyping reactions, and many other technologies.
These instrument purify viral DNA and RNA from a broad range of respiratory samples (nasopharyngeal aspirates and swabs, sputum, bronchoalveolar lavages,…). Contaminations are reduced since the system is closed, which avoids splashing and generation of aerosols and drops. Many of these systems include ready-to-run, prefilled cartridges with all reagents required for the purification procedure. Setup is rapid and saves time. Commercial assays for the simultaneous processing of a maximum of 8 up to 96 samples are available.
Specific studies on the efficiency of these methods in recovery of DNA and RNA from RV have been carried out [31Wilson D, Yen-Lieberman B, Reischl U, Warshawsky I, Procop GW. Comparison of five methods for extraction of Legionella pneumophila from respiratory specimens J Clin Microbiol 2004; 42: 5913-6.-34Yang G, Erdman DE, Kodani M, Kools J, Bowen MD, Fields BS. Comparison of commercial systems for extraction of nucleic acids from DNA/RNA respiratory pathogens J Virol Methods 2011; 171: 195-9.]. Besides its higher reproducibility, the amount of nucleic acids recovered from automated extraction methods is greater than the amount achieved with manual methods [31Wilson D, Yen-Lieberman B, Reischl U, Warshawsky I, Procop GW. Comparison of five methods for extraction of Legionella pneumophila from respiratory specimens J Clin Microbiol 2004; 42: 5913-6.]. Overall, a good recovery of viral RNA and DNA, without significant differences among automated extractors, has been found. No evidence of the presence of inhibitors is observed as well. However, some studies have assessed that, whereas most instruments yield the same render in recovering DNA from respiratory pathogens, they differ in reproducibility, recovery and linearity in recovering viral RNA [34Yang G, Erdman DE, Kodani M, Kools J, Bowen MD, Fields BS. Comparison of commercial systems for extraction of nucleic acids from DNA/RNA respiratory pathogens J Virol Methods 2011; 171: 195-9.].
Other parameters have to be considered for the purification of RV genomes. The extractor must be versatile so that it allows extraction from a wide range of respiratory samples and from variable sample volume. Indeed, lysis buffer must be provided independently by the manufacturer to inactivate respiratory specimens in biosafety cabinets if samples are fed in open containers into the instrument.
4.1.2. Real-Time Assays
Currently, real-time PCR assays are widely used in clinical laboratories. The principle of real-time PCR is that amplification of the nucleic acid target is combined with simultaneous detection of amplicons in a single step. There is no need for manipulation of amplified products, which minimises contaminations due to amplicon carry-over and avoids false positive reactions.
The monitoring of accumulating amplicons in real time is achieved by labelling of primers, oligonucleotide probes or amplicons with fluorophores. These emit fluorescent signals that accumulate during the exponential phase of the amplification step, which is recorded by the instrument. The signal is related to the amount of amplicon present during each cycle and will increase as the amount of specific amplicon increases. Various chemistries have been used for real-time PCR assays, i.e. Taqman® probes, FRET probes, intercalating dyes as Sybr Green I, molecular beacons, scorpions oligonucleotides, etc [35Mackay IM. Real-time PCR in the microbiology laboratory Clin Microbiol Infect 2004; 10: 190-212.].
Commercial assays (Prodesse®, Gen-probe), based on real-time PCR for detection of several RV, have received the FDA clearance. Kits for Flu (types and subtypes), hMPV, ADV, RSV and PIV detection are available.
Real-time PCR using dual labelled probes such as Taqman® probes is widely used by virologists in their “in house” protocols, since it is easy to optimize, and despite the higher costs compared to conventional PCR protocols, the reduction in hands-on-time and the high efficiency compensates favourably the use of this approach. Common PCR parameters of 45 cycles of denaturation at 95ºC for 30 s and annealing and extension at 60ºC for 60 s, can be applied to any real-time assay [36Gunson RN, Collins TC, Carman WF. Practical experience of high throughput real time PCR in the routine diagnostic virology setting J Clin Virol 2006; 35: 355-67.]. This protocol is carried out in our laboratory for any RNA virus, including RV such as Flu, RSV, HRV, hMPV, hBoV and HCoV, by using the same instrument and the same protocol (personal communication).
The technical disadvantage of real-time PCR is the limited number of fluorophores that can be included in a single assay, thus, limiting the multiplexing capabilities. In the best case, 3 targets can be detected in a single tube. A published protocol is able to detect 12 RV by a multiplex real-time PCR, including 4 tubes per sample and 3 viral targets per tube [37Gunson RN, Collins TC, Carman WF. Real-time RT-PCR detection of 12 respiratory viral infections in four triplex reactions J Clin Virol 2005; 33: 341-4.].
One of the advantages of real-time PCR compared to other newly described protocols is the possibility of quantitation. The role of high viral loads in ARI have provided in many circumstances valuable information for the optimal management and treatment of infected individuals [38Mahony JB, Petrich A, Smieja M. Molecular diagnosis of respiratory virus infections Crit Rev Clin Lab Sci 2011; 48: 217-49.].
Pre-prepared, frozen real-time PCR reagents reduce turnaround times and increase reproducibility. Frozen aliquots of primers-probes mixes and, on the other hand, of generic PCR reagents (buffers, dNTPs, enzymes, etc) are a good alternative to reduce hands-on-time of the PCR preparation step [36Gunson RN, Collins TC, Carman WF. Practical experience of high throughput real time PCR in the routine diagnostic virology setting J Clin Virol 2006; 35: 355-67.].
Other emerging amplification procedures have been described for RV detection. Examples of these methods are: loop-mediated isothermal amplification, multiplex ligation-dependent probe amplification (MLPA) and padlock probes [39Wu W, Tang YW. Emerging molecular assays for detection and characterization of respiratory viruses Clin Lab Med 2009; 29: 673-93.].
4.1.3. DNA Arrays and Other Systems for Detection of PCR Products
The combination of PCR and hybridisation using DNA arrays is the principle of recently developed commercial assays for RV detection. Depending on the commercial system, it uses generic or multiplex PCR and specific detection relies on the array.
Examples of this technology are: Luminex xTAG® Respiratory Virus Panel (RVP)(Luminex Molecular Diagnostics) and Resplex II (Qiagen).
Luminex xTAG® RVP was the first panel that received the FDA approval. The version RVP Fast has shortened the initial time of 8-10 h required to accomplish the whole procedure. The RVP Fast assay incorporates multiplex RT-PCR using primers with universal tags. The amplified product is hybridized to a bead array conjugated to specific probes. Detection is performed using a streptavidin-R-phycoerythrin conjugate. Each bead population detects a unique viral target by hybridisation to a specific anti-tag. A signal (median fluorescence intensity) is generated for each bead population [40Krunic N, Yager TD, Himsworth D, Merante F, Yaghoubian S, Janeczko R. xTAG RVP assay: analytical and clinical performance J Clin Virol 2007; 40 (Suppl 1): S39-46.]. These fluorescence values are analyzed by using xTAG® data analysis software for RVP Fast (TDAS RVP Fast) to establish the presence or absence of viral targets in each sample. A recent version of the xTAG RVP Fast panel (RVP v2) is CE marked and identifies 19 viral types and subtypes, which now includes Flu A(H1N1)pdm09.
A preliminary evaluation of RVP v2 has demonstrated a higher recovery of RV and 100% specificity compared to “in-house” real-time PCRs. No competition was observed in samples spiked with combinations of 2 RV. Limit of detection for A(H1N1)pdm09, Flu A H3N2 and RSV was calculated with replicates of 10-fold dilutions of positive samples. Semi-quantitation of viral loads was accomplished by measurement of the CT value observed in parallel “in-house” real-time PCRs. For this purpose, mean CT of 30 obtained from the original RT-PCR was chosen. RVP v2 detected RSV and H3N2 down to a 10-2 dilution of the sample, and A(H1N1)pdm09 was detected down to a 10-3 dilution whereas positive “in-house” PCR reached the 10-2 dilution [41Pérez-Ruiz M, Pedrosa-Corral I, Sanbonmatsu-Gámez S, Rodríguez-Granger J, Navarro-Marí JM. Preliminary evaluation of the new version of Luminex® Respiratory Viral Panel (xTAG® RVP Fast v2) In: 15th Annual Meeting of the European Society for Clinical Virology; 4-7 September 2012; Madrid. 105.].
Resplex II (Qiagen) has the same spectrum of aetiological diagnosis than xTAG RVP, and the detection is carried out in a similar instrument based on liquid hybridisation as the former. The main difference relies on the amplification system, the QIAplex technology. It involves two pairs of gene-specific nested primers at extremely low concentration. These primers are used only for the initial enrichment. Efficient amplification is mediated with “SuperPrimers”, universal primers that bind to the inner nested primer pair [42Ritzi-Lehnert M, Himmelreich R, Attig H, et al. On-chip analysis of respiratory viruses from nasopharyngeal samples Biomed Microdevices 2011; 13: 819-27.].
Clinical performance studies in different population groups have demonstrated good results for RV detection using these systems [43Ginocchio CC. Strengths and weaknesses of FDA-approved/cleared diagnostic devices for the molecular detection of respiratory pathogens Clin Infect Dis 2011; 52 Suppl 4: S312-25.,44Balada-Llasat JM, LaRue H, Kelly C, Rigali L, Pancholi P. Evaluation of commercial ResPlex II v2.0, MultiCode-PLx, and xTAG respiratory viral panels for the diagnosis of respiratory viral infections in adults J Clin Virol 2011; 50: 42-5.].
Other automated detection formats have been used for RV NAAT, as Respifinder® 15 assay (PathoFinder) and Seeplex® RV15 (Seegene). Both detect 15 viral pathogens in respiratory samples. The former is based on a multiplex RT-PCR followed a MLPA reaction, which comprises a probe hybridization step, a probe ligation step and a probe amplification step. Only two primers are used in the final PCR. Every target results in a PCR fragment of a specific length [45Reijans M, Dingemans G, Klaassen CH, et al. RespiFinder: a new multiparameter test to differentially identify fifteen respiratory viruses J Clin Microbiol 2008; 46: 1232-40.]. Seeplex® RV15 uses dual-priming oligonucleotides that allow multiplex PCR of high specificity and minimizes the possibility of unspecific DNA products. Detection of amplicons is accomplished by capillary electrophoresis with both assays [46Kim SR, Ki CS, Lee NY. Rapid detection and identification of 12 respiratory viruses using a dual priming oligonucleotide system-based multiplex PCR assay J Virol Methods 2009; 156: 111-6.]. Although these systems do not avoid the potential of contamination with amplicons, they are able to detect a wide range of RV at a lower cost. However, specific instruments for capillary electrophoresis are available only in high-throughput laboratories.
4.1.4. Platforms for Nucleic Acids Purification and Preparation of the PCR Master Mix
In recent years, several automated platforms for nucleic acids purification also enable the preparation of the PCR reaction mix in 96-well microplates ready for amplification-detection steps. VERSANT kPCR Sample Prep System (Siemens), QIAsymphony (Qiagen), m2000sp (Abbott) and MagnaPure 96 (Roche Diagnostics) are examples of these platforms. This improvement reduces even more the hands-on-time by abolishing the manual step associated with preparation of PCR mixes. Evaluations with some of these systems have demonstrated good and comparable performances [33Lee AV, Atkinson C, Manuel RJ, Clark DA. Comparative evaluation of the QIAGEN QIAsymphony® SP system and bioMérieux NucliSens easyMAG automated extraction platforms in a clinical virology laboratory J Clin Virol 2011; 52: 339-43.,47Verheyen J, Kaiser R, Bozic M, Timmen-Wego M, Maier BK, Kessler HH. Extraction of viral nucleic acids: Comparison of five automated nucleic acid extraction platforms J Clin Virol 2012; 54: 255-9.].
4.2. Fully-Automated NAAT Platforms
The availability of fully-automated NAAT for detecting more or less number of RV is continuously increasing. The main advantages associated to the use of these tools are: ease of use, minimisation of hands-on-time, enclosed pouch to perform all reaction steps, minimum contamination risk, high-order multiplex testing, high sensitivity, specificity and reproducibility.
Most of these platforms have focussed on the automated detection of RSV and/or Flu, probably in an attempt to constitute in future a better alternative to antigen detection methods as point-of-care diagnosis.
Some available commercial fully-automated assays that detect Flu A, flu B and/or RSV are: Xpert Flu (Cepheid) that detect Flu A and Flu B and differentiates A(H1N1)pdm09 [48Novak-Weekley SM, Marlowe EM, Poulter M, et al. Evaluation of the Cepheid Xpert Flu Assay for rapid identification and differentiation of influenza A, influenza A 2009 H1N1, and influenza B viruses J Clin Microbiol 2012; 50: 1704-0.] and Simplexa Flu A/B & RSV kit (Focus Diagnostics) for detection and differentiation of Flu A, Flu B and RSV (http://www.focusdx.com/pdfs/pi/US/MOL2600.pdf). The advantage of the first one is that independent modules of the instrument allow continuous workflow, although each added module constitutes a new apparatus and increases costs. The later uses an 8-well cycler (3M® Integrated Cycler), allowing the simultaneous processing of 8 respiratory samples within the same apparatus.
Nanosphere Verigene® Respiratory Virus Nucleic Acids Test (Nanosphere) is a recently commercialised fully-automated assay similar to previous ones, with increased diagnostic capability since it detects and differentiates Flu A, seasonal H1N1, A(H1N1)pdm09 and H3N2 subtypes, Flu B and RSV A and B viruses. Indeed, an added innovation of this system is the detection of the H275Y mutation within the neuraminidase of A(H1N1)pdm09 that confers resistance to oseltamivir. Studies evaluating this commercial test have shown optimal analytical and clinical performances [49Buchan B, Anderson N, Jannetto P, Ledeboer N. Simultaneous detection of influenza A and its subtypes H1 H3 2009 H1N1 influenza B, and RSV A and B in respiratory specimens on an automated random access molecular platform Clin Microbiol Infect 2011; 17 Suppl S4: S3.,50Pedrosa-Corral I, Sanbonmatsu-Gámez S, Cuadros Moronta E, Pérez-Ruiz M, Navarro-Marí JM. Evaluación de una nueva plataforma de nanotecnología para detección de gripe y virus respiratorio sincitial VRS Enferm Infecc Microbiol Clin 2012; 30 Abstract 731. DOI: http: //www.seimc.org/congresos/seimc-EIMC-2012.pdf].
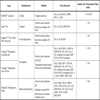
The FilmArray® Respiratory Panel (Idaho Technology Inc.) is commercialised in Europe to fully automate the detection and identification of 21 respiratory pathogens from a single sample in about one hour, including 18 viruses and 3 bacteria. An unprocessed clinical sample is subjected to nucleic acid purification, RT, a high-order nested multiplex PCR and amplicon melting curve analysis in a closed disposable pouch [51Poritz A, Blaschke AJ, Byington CL, et al. FilmArray an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection PLoS One 2011; 6: e26047.]. The larger spectrum of this panel that allows the additional detection of other respiratory pathogens may be an advantage for diagnosing ARI in some population groups [52Loeffelholz MJ, Pong DL, Pyles RB, et al. Comparison of the FilmArray Respiratory Panel and Prodesse real-time PCR assays for detection of respiratory pathogens J Clin Microbiol 2011; 49: 4083-8.]. However, detection of more pathogens leads to a more expensive system. This assay is probably cost-effective for rapid detection of respiratory agents from certain clinically severe patients. Table 2 displays a list of commercial fully-automated assays for detection and identification of RV.
A recent revision of molecular assays for RV carried out by Ginocchio records other available automated and semi-automated commercial tests [43Ginocchio CC. Strengths and weaknesses of FDA-approved/cleared diagnostic devices for the molecular detection of respiratory pathogens Clin Infect Dis 2011; 52 Suppl 4: S312-25.].
5. COST-EFFECTIVENESS OF LABORATORY PROCEDURES: INTEGRATION OF AUTOMATED NAAT TO RATIONAL DIAGNOSTIC ALGORITHMS
The optimal use of the diagnostic tools for RV detection depends on the correct choice of the method in combination with other clinic-epidemiologic criteria. The origin of the sample, patient’s age, predisposing factors for severe disease and the prevalence of a specific RV in a certain area and within a study period must be considered for an appropriate diagnostic algorithm. The overwhelming demand for RV detection in clinical samples has been evident and continuous since the advent of the 2009 influenza pandemic, when clinicians and health institutions have realised that, in many circumstances, an aetiological diagnosis of ARI is desirable. Thus, clinical laboratories must take into account all these points and the availability of infrastructure and resources, to organise their workflow and portfolio related to the diagnosis of viral ARI.
A rational algorithm for viral diagnosis of ARI could be divided in four levels of increasing use of diagnostic tools depending on the type of patient [53Navarro-Marí JM, Pérez-Ruiz M, Galán JC, et al. Circulation of other respiratory viruses and viral co-infection during the 2009 pandemic influenza Enferm Infecc Microbiol Clin 2012; 30(Supl 4): 25-31.].
Overall, viral diagnosis in outpatients with mild ARI is not probably cost-effective, and should not be routinely performed, except for studies from surveillance programs or of epidemiological interest.
Antigen detection methods for RSV and/or Flu may be enough for patients attended at emergency units that do not fulfil severity clinical criteria for hospitalisation. Some individuals should constitute an exception of this protocol as pregnant women, patients with severe underlying conditions and other special situations (immunosupression, obesity, residents in chronic-care facilities, etc) for which a more extended aetiological study and/or the use of NAAT for the most prevalent RV is encouraging [54Fiore AE, Fry A, Shay D, Gubareva L, Bresee JS, Uyeki TM. Centers for Disease Control and Prevention (CDC). Antiviral agents for the treatment and chemoprophylaxis of influenza. Recommendations of the Advisory Committee on Immunization Practices (ACIP) MMWR Recomm Rep 2011; 60: 1-24.-57Van Leeuwen JC, Goossens LK, Hendrix RM, Van Der Palen J, Lusthusz A, Thio BJ. Equal virulence of rhinovirus and respiratory syncytial virus in infants hospitalized for lower respiratory tract infection Pediatr Infect Dis J 2012; 31: 84-6.].
Accurate and timely diagnosis of viral ARI has additional benefits as preventing unnecessary use of antibiotics, thus, decreasing the overall costs of treatments and minimizing the selection of resistant organisms. Moreover, specific antiviral therapy is recommended for treating certain risk groups with Flu and RSV infection. Neuraminidase inhibitors are recommended in patients with suspected or confirmed Flu who require hospitalisation and/or in high-risk groups. Early treatment with neuraminidase inhibitors has demonstrated to decrease the risk of complications, to reduce the duration of the disease and to improve outcome [54Fiore AE, Fry A, Shay D, Gubareva L, Bresee JS, Uyeki TM. Centers for Disease Control and Prevention (CDC). Antiviral agents for the treatment and chemoprophylaxis of influenza. Recommendations of the Advisory Committee on Immunization Practices (ACIP) MMWR Recomm Rep 2011; 60: 1-24.]. Ribavirin is approved for the treatment of hospitalised infants, young children and immunocompromised adults with severe lower respiratory tract infections due to RSV, despite the limited evidence of benefit [58Ventre K, Randolph A. WITHDRAWN: Ribavirin for respiratory syncytial virus infection of the lower respiratory tract in infants and young children Cochrane Database Syst Rev 2010; 5CD000181,59Chemaly RF, Torres HA, Munsell MF, et al. An adaptive randomized trial of intermittent dosing schedule of aerosolized ribavirin in cancer patients with respiratory syncytial virus infections J Infect Dis 2012; 206(9 ): 1367-91.]. New automated assays may improve point-of-care diagnosis of RV since they include a wider range of pathogens within the same kit. In future, automated NAAT may be the alternative of antigen detection methods if costs become comparable and a reduction of turnaround time is achieved.
Samples from patients requiring hospitalisation with negative RSV or Flu antigen results should be subjected to extended virological study that includes viral culture assays and/or NAAT targeted at more or less RV depending on the severity of the disease and the need for a rapid result. With this premise, NAAT for detection of RSV and Flu (at the type and subtype level) in patients with less severe ARI, that do not require admission at intensive care units (ICU), should be carried out first of all. Otherwise, other molecular multiplexed methods of low complexity could be performed in certain patients that included hMPV, HRV and PIV detection, depending on patient’s age and/or the existence of underlying diseases. More complex NAAT that detect other less frequent RV should be reserved to certain situations such as more severe cases (ICU patients) in which previous NAAT for the most prevalent viruses have yielded negative results [60Kehl SC, Kumar S. Utilization of nucleic acid amplification assays for the detection of respiratory viruses Clin Lab Med 2009; 29: 661-71.-62Dundas NE, Ziadie MS, Revell PA, et al. A lean laboratory: operational simplicity and cost effectiveness of the LuminexTAG™ respiratory viral panel J Mol Diagn 2011; 13: 175-9.]. At this level, automated and semi-automated NAAT may provide higher throughput than in-house protocols.
In the future, the feasibility of introducing fully-automated NAAT in a wider range of health institutions can constitute an improvement in viral diagnosis of ARI. Basic and first-level hospitals may provide, as well, with an efficient point-of-care assay to ensure the appropriate management of their in-patients.
6. CONCLUSIONS
Automation in the field of clinical virology for diagnosis of ARI relies on molecular assays. New approaches have yielded point-of-care methods, optimal for a rapid detection of several RV.
Cost-effective laboratory methods must take into account the benefits of aetiological diagnosis depending on technical characteristics of the assays, on the availability of the appropriate resources and on clinic-epidemiologic criteria. Automated NAAT are expensive but probably the most cost-effective tools for severely ill hospitalised patients. Many automated commercial assays require their own instrument; thus, each laboratory should make its forecast for future to provide with the most feasible automation strategies. Since no expertise is required to carry out automated molecular assays, extended aetiological diagnosis of ARI can be achieved if first-level hospitals implement these techniques in their portfolio.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
ACKNOWLEDGEMENTS
Declared none.
REFERENCES
| [1] | Hall MJ, DeFrances CJ, Williams SN, Golosinskiy A, Schwartzman A. National Hospital Discharge Survey: 2007 summary Natl Health Stat Report 2010; 29: 1-20-24. |
| [2] | Hsiao C-J, Cherry DK, Beatty PC, Rechtsteiner E. National ambulatory medical care survey: 2007 summary Natl Health Stat Report 2010; 27: 1-32. |
| [3] | Hing E, Hall MJ, Ashman JJ, Xu J. National hospital ambulatory medical care survey: 2007 outpatient department summary Natl Health Stat Report 2010; 28: 1-32. |
| [4] | Niska R, Bhuiya F, Xu J. National hospital ambulatory medical care survey: 2007 emergency department summary Natl Health Stat Report 2010; 26: 1-31. |
| [5] | Henderson FW, Clyde WA Jr, Collier AM, et al. The etiologic and epidemiologic spectrum of bronchiolitis in pediatric practice J Pediatr 1979; 95: 183-90. |
| [6] | Yun BY, Kim MR, Park JY, Choi EH, Lee HJ, Yun CK. Viral etiology and epidemiology of acute lower respiratory tract infections in Korean children Pediatr Infect Dis J 1995; 14: 1054-9. |
| [7] | Jennings LC, Anderson TP, Beynon KA, et al. Incidence and characteristics of viral community-acquired pneumonia in adults Thorax 2008; 63: 42-8. |
| [8] | Dimopoulos G, Lerikou M, Tsiodras S, et al. Viral epidemiology of acute exacerbations of chronic obstructive pulmonary disease Pulmonary Pharmacol Ther 2012; 25: 12-8. |
| [9] | Atmar RL, Piedra PA, Patel SM, Greenberg SB, Couch RB, Glezen WP. Picornavirus the most common respiratory virus causing infection among patients of all ages hospitalized with acute respiratory illness J Clin Microbiol 2012; 50: 506-8. |
| [10] | Van den Hoogen BG, Osterhaus DM, Fouchier RA. Clinical impact and diagnosis of human metapneumovirus infection Pediatr Infect Dis J 2004; 23: S25-32. |
| [11] | Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome N Engl J Med 2003; 348: 1953-66. |
| [12] | Van der Hoek L, Pyrc K, Jebbink MF, et al. Identification of a new human coronavirus Nat Med 2004; 10: 368-73. |
| [13] | Woo PC, Lau SK, Huang Y, Tsoi HW, Chan KH, Yuen KY. Phylogenetic and recombination analysis of coronavirus HKU1, a novel coronavirus from patients with pneumonia Arch Virol 2005; 150: 2299-311. |
| [14] | Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples Proc Natl Acad Sci USA 2005; 102: 12891-6. |
| [15] | Monto AS. Epidemiology of viral respiratory infections Am J Med 2002; 112(Suppl 6A): 4S-12S. |
| [16] | Navarro-Marí JM, Perez-Ruiz M. Virus respiratorios: viejos y nuevos virus. Revisión de métodos diagnósticos Enferm Infecc Microbiol Clin 2007; 25(Suppl 3): 60-5. |
| [17] | Loeffelholz M, Chonmaitree T. Advances in diagnosis of respiratory virus infections Int J Microbiol 2010; 2010: 126049. |
| [18] | Mahony JB. Detection of respiratory viruses by molecular methods Clin Microbiol Rev 2008; 21: 716-47. |
| [19] | Navarro-Marí JM, Sanbonmatsu-Gámez S, Pérez-Ruiz M, De La Rosa-Fraile M. Rapid detection of respiratory viruses by shell vial assay using simultaneous cultura of HEp-2, LLC-MK2, and MDCK cells in a single vial J Clin Microbiol 1999; 37: 2346-7. |
| [20] | Dunn JJ, Woolstenhulme RD, Langer J, Carroll KC. Sensitivity of respiratory virus culture when screening with R-mix fresh cells J Clin Microbiol 2004; 42: 79-82. |
| [21] | Weinberg A, Brewster L, Clark J, Simoes E. ARIVAC consortium. Evaluation of R-Mix shell vials for the diagnosis of viral respiratory tract infections J Clin Virol 2004; 30: 100-5. |
| [22] | Pérez-Ruiz M, Yeste R, Ruiz-Pérez MJ, Ruiz-Bravo A, de la Rosa-Fraile M, Navarro-Marí JM. Testing of diagnostic methods for detection of influenza virus for optimal performance in the context of an influenza surveillance network J Clin Microbiol 2007; 45: 3109-10. |
| [23] | Centers for Disease Control and Prevention. Evaluation of rapid Influenza diagnostic tests for detection of Novel Influenza A (H1N1) Virus — United States, 2009 MMWR 2009; 58: 826-9. |
| [24] | Utokaparch S, Marchant D, Gosselink JV, et al. The relationship between respiratory viral loads and diagnosis in children presenting to a pediatric hospital emergency department Pediatr Infect Dis J 2011; 30: e18-23. |
| [25] | Lieberman D, Lieberman D, Shimoni A, Keren-Naus A, Steinberg R, Shemer-Avni Y. Identification of respiratory viruses in adults: nasopharyngeal versus oropharyngeal sampling J Clin Microbiol 2009; 47: 3439-43. |
| [26] | Koskinen JO, Vainionpää R, Meltola NJ, Soukka J, Hänninen PE, Soini AE. Rapid method for detection of influenza a and B virus antigens by use of a two-photon excitation assay technique and dry-chemistry reagents J Clin Microbiol 2007; 45: 3581-8. |
| [27] | Koskinen JO, Vainionpää R, Soini AE. Novel multianalyte point-of-care test for pathogen-speci?c diagnosing of respiratory infections Clin Microbiol Infect 2009; 15(Suppl S4): S556. |
| [28] | Toivola H, Koskinen JM, Lappalainen M, Ruonamo R, Antikainen P, Koskinen JO. Performance evaluation of multianalyte mariPOC® respiratory infection test for adenovirus types 1–8, 14, 19 and 21 Clin Microbiol Infect 2012; 18(Suppl S3): 787-8. |
| [29] | Cilla G, Oñate E, Perez-Yarza EG, Montes M, Vicente D, Perez-Trallero E. Viruses in community-acquired pneumonia in children aged less than 3 years old: High rate of viral coinfection J Med Virol 2008; 80: 1843-9. |
| [30] | Mansbach JM, Piedra PA, Teach SJ, et al. Prospective multicenter study of viral etiology and hospital length of stay in children with severe bronchiolitis Arch Pediatr Adolesc Med 2012; 166(8 ): 700-6. |
| [31] | Wilson D, Yen-Lieberman B, Reischl U, Warshawsky I, Procop GW. Comparison of five methods for extraction of Legionella pneumophila from respiratory specimens J Clin Microbiol 2004; 42: 5913-6. |
| [32] | Chan KH, Yam WC, Pang CM, et al. Comparison of the NucliSens easyMAG and Qiagen BioRobot 9604 nucleic acid extraction systems for detection of RNA and DNA respiratory viruses in nasopharyngeal aspirate samples J Clin Microbiol 2008; 46: 2195-9. |
| [33] | Lee AV, Atkinson C, Manuel RJ, Clark DA. Comparative evaluation of the QIAGEN QIAsymphony® SP system and bioMérieux NucliSens easyMAG automated extraction platforms in a clinical virology laboratory J Clin Virol 2011; 52: 339-43. |
| [34] | Yang G, Erdman DE, Kodani M, Kools J, Bowen MD, Fields BS. Comparison of commercial systems for extraction of nucleic acids from DNA/RNA respiratory pathogens J Virol Methods 2011; 171: 195-9. |
| [35] | Mackay IM. Real-time PCR in the microbiology laboratory Clin Microbiol Infect 2004; 10: 190-212. |
| [36] | Gunson RN, Collins TC, Carman WF. Practical experience of high throughput real time PCR in the routine diagnostic virology setting J Clin Virol 2006; 35: 355-67. |
| [37] | Gunson RN, Collins TC, Carman WF. Real-time RT-PCR detection of 12 respiratory viral infections in four triplex reactions J Clin Virol 2005; 33: 341-4. |
| [38] | Mahony JB, Petrich A, Smieja M. Molecular diagnosis of respiratory virus infections Crit Rev Clin Lab Sci 2011; 48: 217-49. |
| [39] | Wu W, Tang YW. Emerging molecular assays for detection and characterization of respiratory viruses Clin Lab Med 2009; 29: 673-93. |
| [40] | Krunic N, Yager TD, Himsworth D, Merante F, Yaghoubian S, Janeczko R. xTAG RVP assay: analytical and clinical performance J Clin Virol 2007; 40 (Suppl 1): S39-46. |
| [41] | Pérez-Ruiz M, Pedrosa-Corral I, Sanbonmatsu-Gámez S, Rodríguez-Granger J, Navarro-Marí JM. Preliminary evaluation of the new version of Luminex® Respiratory Viral Panel (xTAG® RVP Fast v2) In: 15th Annual Meeting of the European Society for Clinical Virology; 4-7 September 2012; Madrid. 105. |
| [42] | Ritzi-Lehnert M, Himmelreich R, Attig H, et al. On-chip analysis of respiratory viruses from nasopharyngeal samples Biomed Microdevices 2011; 13: 819-27. |
| [43] | Ginocchio CC. Strengths and weaknesses of FDA-approved/cleared diagnostic devices for the molecular detection of respiratory pathogens Clin Infect Dis 2011; 52 Suppl 4: S312-25. |
| [44] | Balada-Llasat JM, LaRue H, Kelly C, Rigali L, Pancholi P. Evaluation of commercial ResPlex II v2.0, MultiCode-PLx, and xTAG respiratory viral panels for the diagnosis of respiratory viral infections in adults J Clin Virol 2011; 50: 42-5. |
| [45] | Reijans M, Dingemans G, Klaassen CH, et al. RespiFinder: a new multiparameter test to differentially identify fifteen respiratory viruses J Clin Microbiol 2008; 46: 1232-40. |
| [46] | Kim SR, Ki CS, Lee NY. Rapid detection and identification of 12 respiratory viruses using a dual priming oligonucleotide system-based multiplex PCR assay J Virol Methods 2009; 156: 111-6. |
| [47] | Verheyen J, Kaiser R, Bozic M, Timmen-Wego M, Maier BK, Kessler HH. Extraction of viral nucleic acids: Comparison of five automated nucleic acid extraction platforms J Clin Virol 2012; 54: 255-9. |
| [48] | Novak-Weekley SM, Marlowe EM, Poulter M, et al. Evaluation of the Cepheid Xpert Flu Assay for rapid identification and differentiation of influenza A, influenza A 2009 H1N1, and influenza B viruses J Clin Microbiol 2012; 50: 1704-0. |
| [49] | Buchan B, Anderson N, Jannetto P, Ledeboer N. Simultaneous detection of influenza A and its subtypes H1 H3 2009 H1N1 influenza B, and RSV A and B in respiratory specimens on an automated random access molecular platform Clin Microbiol Infect 2011; 17 Suppl S4: S3. |
| [50] | Pedrosa-Corral I, Sanbonmatsu-Gámez S, Cuadros Moronta E, Pérez-Ruiz M, Navarro-Marí JM. Evaluación de una nueva plataforma de nanotecnología para detección de gripe y virus respiratorio sincitial VRS Enferm Infecc Microbiol Clin 2012; 30 Abstract 731. DOI: http: //www.seimc.org/congresos/seimc-EIMC-2012.pdf |
| [51] | Poritz A, Blaschke AJ, Byington CL, et al. FilmArray an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection PLoS One 2011; 6: e26047. |
| [52] | Loeffelholz MJ, Pong DL, Pyles RB, et al. Comparison of the FilmArray Respiratory Panel and Prodesse real-time PCR assays for detection of respiratory pathogens J Clin Microbiol 2011; 49: 4083-8. |
| [53] | Navarro-Marí JM, Pérez-Ruiz M, Galán JC, et al. Circulation of other respiratory viruses and viral co-infection during the 2009 pandemic influenza Enferm Infecc Microbiol Clin 2012; 30(Supl 4): 25-31. |
| [54] | Fiore AE, Fry A, Shay D, Gubareva L, Bresee JS, Uyeki TM. Centers for Disease Control and Prevention (CDC). Antiviral agents for the treatment and chemoprophylaxis of influenza. Recommendations of the Advisory Committee on Immunization Practices (ACIP) MMWR Recomm Rep 2011; 60: 1-24. |
| [55] | Kraft CS, Jacob JT, Sears MH, Burd EM, Caliendo AM, Lyon GM. Severity of human rhinovirus infection in immunocompromised adults is similar to that of 2009 H1N1 influenza J Clin Microbiol 2012; 50: 1061-3. |
| [56] | Dubar G, Azria E, Tesnière A, et al. French experience of 2009 A/H1N1v influenza in pregnant women PLoS One 2010; 5(10): pii: e13112. |
| [57] | Van Leeuwen JC, Goossens LK, Hendrix RM, Van Der Palen J, Lusthusz A, Thio BJ. Equal virulence of rhinovirus and respiratory syncytial virus in infants hospitalized for lower respiratory tract infection Pediatr Infect Dis J 2012; 31: 84-6. |
| [58] | Ventre K, Randolph A. WITHDRAWN: Ribavirin for respiratory syncytial virus infection of the lower respiratory tract in infants and young children Cochrane Database Syst Rev 2010; 5CD000181 |
| [59] | Chemaly RF, Torres HA, Munsell MF, et al. An adaptive randomized trial of intermittent dosing schedule of aerosolized ribavirin in cancer patients with respiratory syncytial virus infections J Infect Dis 2012; 206(9 ): 1367-91. |
| [60] | Kehl SC, Kumar S. Utilization of nucleic acid amplification assays for the detection of respiratory viruses Clin Lab Med 2009; 29: 661-71. |
| [61] | Mahony JB, Blackhouse G, Babwah J, et al. Cost analysis of multiplex PCR testing for diagnosing respiratory virus infections J Clin Microbiol 2009; 47: 2812-7. |
| [62] | Dundas NE, Ziadie MS, Revell PA, et al. A lean laboratory: operational simplicity and cost effectiveness of the LuminexTAG™ respiratory viral panel J Mol Diagn 2011; 13: 175-9. |
Endorsements
Browse Contents
Table of Contents
- INTRODUCTION
- TRADITIONAL DETECTION METHODS
- NAAT FOR DETECTION OF RESPIRATORY VIRUSES: FROM THE SIMPLEST TO THE MOST COMPLEX TARGETING APPROACH
- AUTOMATION POSSIBILITIES FOR DETECTING RESPIRATORY VIRUSES BY NAAT
- COST-EFFECTIVENESS OF LABORATORY PROCEDURES: INTEGRATION OF AUTOMATED NAAT TO RATIONAL DIAGNOSTIC ALGORITHMS
- CONCLUSIONS