- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

Current Chemical Genomics and Translational Medicine
(Discontinued)
ISSN: 2213-9885 ― Volume 12, 2018
High Throughput Screening for Inhibitors of Alpha-Galactosidase
Omid Motabar1, 2, Ke Liu1, Noel Southall1, Juan J Marugan1, Ehud Goldin2, Ellen Sidransky2, Wei Zheng*, 1
Abstract
Fabry disease is a rare X-linked lysosomal storage disorder caused by a deficiency in α-galactosidase A (GLA), which catalyzes the hydrolysis of terminal α-galactosyl groups from glycosphingolipids, such as globotriaosylceramide (Gb3). Many of the mutations in the GLA gene are missense alterations that cause misfolding, decreased stability, and/or mistrafficking of this protein. Small molecule compounds that correct the misfolding and mistrafficking, or activate the mutant enzyme, may be useful in the treatment of Fabry disease. We have screened a library of approximately 230,000 compounds using preparations of human recombinant protein and purified coffee bean enzyme in an effort to find activators and inhibitors of this enzyme. Lansoprazole was identified as a small molecule inhibitor of GLA derived from coffee beans (IC50 = 6.4 μM), but no inhibitors or activators were identified for the human enzyme. The screening results indicate that human GLA is a difficult target for small molecule inhibition or activation.
Article Information
Identifiers and Pagination:
Year: 2010Volume: 4
First Page: 67
Last Page: 73
Publisher Id: CCGTM-4-67
DOI: 10.2174/1875397301004010067
Article History:
Received Date: 3/8/2010Revision Received Date: 9/9/2010
Acceptance Date: 13/10/2010
Electronic publication date: 3/12/2010
Collection year: 2010
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the 9800 Medical Center Drive, MSC 3370, Bethesda, MD 20892-3370, Tel: 301-217-5720; Fax: 301-217-5728; E-mail: wzheng@mail.nih.gov
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 3-8-2010 |
Original Manuscript | High Throughput Screening for Inhibitors of Alpha-Galactosidase | |
INTRODUCTION
Alpha-galactosidase A (GLA) is a lysosomal enzyme that catalyzes the hydrolysis of terminal α-galactosyl moieties from glycolipids and glycoproteins. Mutations in the GLA gene can result in the synthesis of misfolded proteins that are retained in the endoplasmic reticulum and degraded prematurely. In Fabry disease, a deficiency in GLA results in the accumulation of globotriaosylceramide (Gb3), a glycosphingolipid, in many cells and organs of the body, including endothelial cells and the smooth muscle cells of blood vessels [1Desnick RJ, Ioannou YA, Eng CM. Alpha-galactosidase a deficiency: Fabry disease In: New York: McGraw-Hill Professional 2001.]. Patients with Fabry disease present clinically with chronic neuronopathic pain, gastrointestinal disturbances, angiokeratomata, progressive renal impairment, cardiomyopathy, premature myocardial infarctions, and stroke, and both life expectancy and quality of life are severely compromised. Enzyme replacement therapy (ERT) has been available for the treatment of Fabry disease since 2001, and is effective in treating the symptoms, but has drawbacks as well [2Brady RO. Enzyme replacement for lysosomal diseases Annu Rev Med 2006; 57: 283-96., 3Lidove O, Joly D, Barbey F, et al. Clinical results of enzyme replacement therapy in Fabry disease: a comprehensive review of literature Int J Clin Pract 2007; 61: 293-302.]. In addition to ERT, there is great interest in correcting enzyme misfolding with small molecules, a strategy referred to as chemical chaperone therapy (CCT). Chemical chaperones are small molecules that bind to mutant proteins and assist in their correct folding, maturation, and trafficking to the functional site, such as the lysosomes. The effects of chemical chaperones have been explored in various lysosomal storage disorders, including Gaucher disease [4Sawkar AR, Cheng WC, Beutler E, Wong CH, Balch WE, Kelly JW. Chemical chaperones increase the cellular activity of N370S beta -glucosidase: a therapeutic strategy for Gaucher disease Proc Natl Acad Sci USA 2002; 99: 15428-33.-7Zheng W, Padia J, Urban DJ, et al. Three classes of glucocerebrosidase inhibitors identified by quantitative high-throughput screening are chaperone leads for Gaucher disease Proc Natl Acad Sci USA 2007; 104: 13192-7.], Pompe disease [8Okumiya T, Kroos MA, Vliet LV, Takeuchi H, Van der Ploeg AT, Reuser AJ. Chemical chaperones improve transport and enhance stability of mutant alpha-glucosidases in glycogen storage disease type II Mol Genet Metab 2007; 90: 49-57., 9Parenti G, Zuppaldi A, Gabriela PM, et al. Pharmacological enhancement of mutated alpha-glucosidase activity in fibroblasts from patients with Pompe disease Mol Ther 2007; 15: 508-14.], Tay-Sachs/Sandhoff disease [10Tropak MB, Reid SP, Guiral M, Withers SG, Mahuran D. Pharmacological enhancement of beta-hexosaminidase activity in fibroblasts from adult Tay-Sachs and Sandhoff Patients J Biol Chem 2004; 279: 13478-87.], GM1-gangliosidosis [11Matsuda J, Suzuki O, Oshima A, et al. Chemical chaperone therapy for brain pathology in G(M1)-gangliosidosis Proc Natl Acad Sci USA 2003; 100: 15912-7.], as well as Fabry disease. A GLA inhibitor, 1-deoxygalactonojirimycin (DGJ, marketed as AmigalTM by Amicus Therapeutics, Inc.), is currently being studied in a phase 3 clinical trial as a chaperone therapeutic agent for Fabry disease [12Fan JQ, Ishii S. Active-site-specific chaperone therapy for Fabry disease. Yin and Yang of enzyme inhibitors FEBS J 2007; 274: 4962-71.]. In this chemical chaperone therapy, small molecule inhibitors bind to misfolded proteins and correct their folding, maturation and/or trafficking resulting in increased delivery of GLA to lysosomes. Once in the lysosomes the inhibitors bound to GLA are displaced by the abundant substrates. However, DGJ is an iminosugar analog which may not have the desirable drug selectivity, and may potentially cause adverse effects. Thus, new improved GLA inhibitors with chaperone activity, as well as enzyme activators, are needed for drug development for Fabry disease.
Several enzyme assays are available for the high throughput screening (HTS) of small molecule compound libraries against GLA. Chromogenic assays are available that use p-nitrophenol-α-D-galactopyranoside [13de Groot PG, Hamers MN, Westerveld A, Schram AW, Meera KP, Tager JM. A new immunochemical method for the quantitative measurement of specific gene products in man-rodent somatic cell hybrids Hum Genet 1978; 44: 295-304., 14Yagi F, Eckhardt AE, Goldstein IJ. Glycosidases of Ehrlich ascites tumor cells and ascitic fluid--purification and substrate specificity of alpha-N-acetylgalactosaminidase and alpha-galactosidase: comparison with coffee bean alpha-galactosidase Arch Biochem Biophys 1990; 280: 61-7.] or naphthyl-α-D-galactopyranoside [15Tsou KC, Su HC. A study of yeast alpha-galactosidase with naphthyl alpha-D-galactopyranosides as chromogenic substrates Anal Biochem 1964; 8: 415-23.] as substrates. However, absorbance-based assays are usually less sensitive than fluorescence-based assays, and cannot be easily miniaturized into a high-density 1536-well plate format for HTS. The pro-fluorogenic molecule, 4-methylumbelliferyl-α-D-galactopyr-anoside (4MU-α-Gala), is a substrate for GLA, where the product of the enzyme reaction emits blue fluorescence at a peak of 440 nm upon excitement at 365 nm [16Mapes CA, Sweeley CC. Galactosyl (alpha 1--4)galactosylceramide: galactosyl hydrolase activity in normal and Fabry plasma Biochem Biophys Res Commun 1973; 53: 1317-24., 17Hultberg B, Sjoblad S, Ockerman PA. Glycosidases in human skin fibroblast cultures. Alpha-fucosidase, alpha-galactosidase, alpha-glucosidase, beta-mannosidase, and N-acetyl-alphaglucosaminidase Acta Paediatr Scand 1975; 64: 123-31.]. Resorufinyl α-D-galactopyranoside (res-α-Gala) is a red pro-fluorogenic GLA substrate which excites at 573 nm and emits at 610 nm upon galactose cleavage [18Shi ZD, Motabar O, Goldin E, et al. Synthesis and characterization of a new fluorogenic substrate for alpha-galactosidase Anal Bioanal Chem 2009; 394: 1903-9., 19Motabar O, Shi ZD, Goldin E, et al. A new resorufin-based alpha-glucosidase assay for high-throughput screening Anal Biochem 2009; 390: 79-84.]. We report here the identification of a novel small molecule GLA inhibitor from a compound library screen using these pro-fluorogenic substrates. GLA from green coffee beans, which is readily available, was used in the initial assay development and screen validation. Using that protein, we identified a novel small molecule inhibitor of GLA, lansoprazole. However, in a subsequent screen of 230,000 compounds using the human recombinant GLA, we did not find any hits, indicating the difficulty of targeting the human enzyme.
MATERIALS AND METHODS
Enzymes, Substrates, and Other Chemicals
α-Galactosidase A from green coffee beans (G8507, ~10 units/mg protein) and α-glucosidase from rice (G9259, 40-80 units/mg protein) were purchased from Sigma-Aldrich (St. Louis, MO). Glucocerebrosidase was obtained from residual solution after clinical infusions of imiglucerase (Cerezyme®, Genzyme Co., Mr = 60,430), with a specific activity of 42.2 units/mg and 14 units/ml. Recombinant human α-Galactosidase A was also obtained from residual solution after clinical infusions (agalsidase beta, marketed as Fabrazyme® by Genzyme Co.). Glycerol was added to these enzyme solutions to 30%, and small aliquots were stored at -80°C. Enzyme activity was found to be stable in these stock solutions after storage at -80°C for 2 years.
4-methylumbelliferyl-α-D-galactopyranoside (4MU-α-Gala), a blue pro-fluorogenic substrate, and 1-deoxygalactonojirimycin (DGJ), a known inhibitor of α-Galactosidase, were purchased from Sigma-Aldrich, as were citric acid, potassium phosphate trihydrate, tween-20, glycine, and sodium hydroxide. Taurocholic acid sodium salt was obtained from CalbioChem (a subsidiary of EMD Bioscience, San Diego, CA). Four compound libraries, the LOPAC (Library of Pharmacologically Active Compounds) collection, the Prestwick collection, the Spectrum collection, and the Tocris collection were purchased from Sigma-Aldrich, Prestwick Chemicals (Illkirch, France), Micro-Source Discovery Systems (Gaylordsville, CT) and Tocris (Ellisville, MO), respectively.
Buffers
The assay buffer for the α-galactosidase and glucocerebrosidase enzyme assays consisted of 50 mM citric acid, 176 mM K2PO4, and 0.01% Tween-20 at pH 5.9. The assay buffer for α-glucosidase was similar, except that it was titrated with K2PO4 to pH 5.0. The buffers were stored at 4°C for use up to 6 months. The stop solution consisted of 0.5 M sodium hydroxide and 0.5 M glycine at pH 11.6.
Instruments for Liquid Handling and Plate Detection
An FRDTM automated microvolume dispensing station (Aurora Discovery, San Diego, CA) was used to dispense reagents into 1536-well plates at volumes from 1-3 μl. Initially, the compounds were serially diluted in DMSO in 384-well plates using a CyBi®-Well dispensing station with a 384-well head (Cybio Inc., Woburn, MA), and then reformatted into 1536-well plates at 7 μl/well. Nanoliter volumes of these compounds were transferred to 1536-well assay plates using an automated pin-tool station (Kalypsys, Inc., San Diego, CA). A ViewLuxTM CCD-based imaging plate reader (PerkinElmer, Boston, MA) was used for fluorescence detection at a speed of 30 seconds per plate. A Safire2™ monochromator scanning fluorescence plate reader (Tecan Group Ltd., Männedorf, Switzerland) was used for determining the fluorescence excitation and emission spectra.
Enzyme Kinetics Assay
The kinetics assay was carried out in a 384-well plate format using 1 nM enzyme with varying concentrations of substrate. Initially, 10 μl/well of the varying concentrations of substrate were added to the plate. The reaction was initiated by addition of 20 μl/well of enzyme solution. The 4MU-α-Gala stock solution was serially diluted 1:1.5 to give eight concentrations. The final concentrations of substrate used in the assay were 500, 333, 222, 148, 98.8, 65.8, 43.9, and 29.3 μM. 30 μl/well of stop solution was added after 2, 4, 6, 8, 10, and 12 minute incubation times at RT. A standard curve of the free fluorophore, 4-methylumbelliferone (4MU), in the same volume of assay buffer and stop solution was generated for calculating the enzyme product. The plate was read in the ViewLux plate reader at an emission wavelength of 440 nm and an excitation wavelength of 365 nm.
In order to determine the type of inhibition and the inhibition constant of lansoprazole, five enzyme kinetics plots were generated in the presence of 80, 40, 20, 10, and 0 μM of the inhibitor. The final concentrations of substrate were 500, 333, 222, 148, 98.8, 65.8, 43.9, 29.3, and 19.5 μM. The plate was read in the ViewLux at 2, 4, 6, 8, and 10 minute time intervals. As in the above kinetic assay, a standard curve of the free fluorophore, 4MU, was generated. The plate was read at 440 nm emission upon excitation at 365 nm.
qHTS of Compound Collections
Quantitative high-throughput screening (qHTS) is a method for simultaneously screening large compound libraries at multiple concentrations. The Library of Pharmacologically Active Compounds (LOPAC) with 1,280 compounds from Sigma-Aldrich is commonly used to validate the screening assay. Once validated, NCGC’s collection of 230,000 compounds was screened for GLA activity. The compounds in these libraries were serially diluted in DMSO at a ratio of 1:5 for up to seven concentrations in 384-well plates. Four sets of the inter-plate dilution plates were reformatted into one set of 1536-well plates, with compound concentrations ranging from 0.29 μM to 10 mM. The pintool station was used to transfer 23 nl of compounds in DMSO solution to the assay plates with the enzyme solutions. The final compound concentrations in a 3 μl assay volume ranged from 1.9 nM to 66.7 μM.
Other Enzyme Assays to Determine Compound Selectivity
The enzyme assays of two other hydrolases, rice α-glucosidase (GAA) and human recombinant glucocerebrosidase (GC) were used for determining the selectivity of the active compounds identified from HTS. Both enzyme assays employed the similar fluorogenic substrates, 4-methylumbelliferyl-α-D-glucopyranoside (4MU-α-glc) for GAA and 4-methylumbelliferyl β-D-glucopyranoside (4MU-β-Glc) for GC, which were purchased from Sigma-Aldrich (St. Louis, MO). The assays were performed in 1536-well black assay plates with 2 μl/well enzyme and 23 nl/well of compound in DMSO solution followed by 1 μl/well substrate solution. After a 30-minute incubation at the room temperature (21 °C), 3 μl/well of stop solution was added and the assay plates were counted in a Viewlux plate reader for fluorescence intensity at an excitation of 365 nm and emission of 440 nm. As described previously [7Zheng W, Padia J, Urban DJ, et al. Three classes of glucocerebrosidase inhibitors identified by quantitative high-throughput screening are chaperone leads for Gaucher disease Proc Natl Acad Sci USA 2007; 104: 13192-7.], the final enzyme concentration was 8 nM and substrate concentration was 160 μM for the GAA assay, while the final enzyme and substrate concentrations in the GC assay were 1.9 nM and 800 μM, respectively.
Data Analysis
The primary screen data was analyzed and concentration responses were fit using a customized software developed internally [20Southall N, Jadhav A, Huang R, Nguyen T, Wang Y. Enabling the large scale analysis of quantitative high throughput screening data In: Seethala R, Zhang L, Eds. Handbook of drug screening. 2nd ed. USA: Taylor and Francis 2009; pp. 442-62., 21Wang Y, Jadhav A, Southall N, Huang R, Nguyen DT. A grid algorithm for high throughput fitting of dose-response curve data Curr Chem Genom 2010; 4: 57-66.]. The results from the assay optimization experiments, enzyme kinetics, confirmation experiments, and selectivity assays were analyzed with Prism® (Graphpad, San Diego, CA).
RESULTS AND DISCUSSION
Screening Assay Optimization
The GLA enzyme assay was initially developed using commercially available purified enzyme from green coffee beans. The classical fluorogenic substrate used, 4MU-α-Gala, forms two products, galactose and fluorescent 4MU, upon cleavage by GLA. Since this enzyme is naturally found in the lysosomes, pH optimization was first performed. A pH titration revealed an optimal pH of 5.9 (Fig. 1a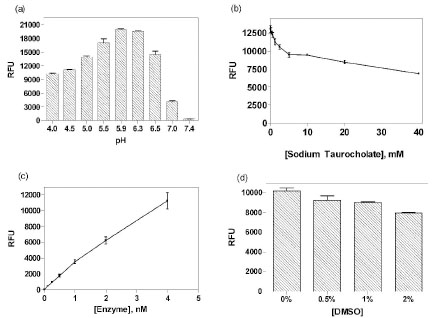 ) for this enzyme assay, and this buffer pH was used for subsequent experiments. An addition of stop solution after the enzyme reaction raised the final pH in assay plates to 10 for optimal fluorescence detection, because the 4MU fluorophore has a pKa of 7.8 and its fluorescence intensity is not favorably detected below this pKa value.
) for this enzyme assay, and this buffer pH was used for subsequent experiments. An addition of stop solution after the enzyme reaction raised the final pH in assay plates to 10 for optimal fluorescence detection, because the 4MU fluorophore has a pKa of 7.8 and its fluorescence intensity is not favorably detected below this pKa value.
While sodium taurocholate is routinely used in the GLA assay buffer [22Shin SH, Murray GJ, Kluepfel-Stahl S, et al. Screening for pharmacological chaperones in Fabry disease Biochem Biophys Res Commun 2007; 359: 168-73.], its optimal concentration has not been well-established. It is a bile salt that is required for the activity of certain lysosomal enzymes [23Wenger DA, Clark C, Sattler M, Wharton C. Synthetic substrate beta-glucosidase activity in leukocytes: a reproducible method for the identification of patients and carriers of Gaucher's disease Clin Genet 1978; 13: 145-53.]. A titration of sodium taurocholate was performed in this enzyme assay to establish its optimal concentration. Unexpectedly, we found that sodium taurocholate decreased the GLA activity at all the concentrations tested (ranging from 0.15 to 40 mM) (Fig. 1b ), while enzyme activity was quite high without this bile salt. This result contrasts with the sodium taurocholate dependence of glucocerebrosidase, another lysosomal enzyme, which shows almost no enzyme activity in the absence of this bile salt [7Zheng W, Padia J, Urban DJ, et al. Three classes of glucocerebrosidase inhibitors identified by quantitative high-throughput screening are chaperone leads for Gaucher disease Proc Natl Acad Sci USA 2007; 104: 13192-7.]. Thus, sodium taurocholate was eliminated from the GLA assay buffer.
), while enzyme activity was quite high without this bile salt. This result contrasts with the sodium taurocholate dependence of glucocerebrosidase, another lysosomal enzyme, which shows almost no enzyme activity in the absence of this bile salt [7Zheng W, Padia J, Urban DJ, et al. Three classes of glucocerebrosidase inhibitors identified by quantitative high-throughput screening are chaperone leads for Gaucher disease Proc Natl Acad Sci USA 2007; 104: 13192-7.]. Thus, sodium taurocholate was eliminated from the GLA assay buffer.
The enzyme concentration-response was also measured at a fixed substrate concentration in order to establish the optimal enzyme concentration for compound screening. It showed a nearly linear response from 0.125 nM to 4 nM GLA (Fig. 1c ). Based on this result, 1 nM enzyme was selected for the assay because the fluorescence intensity (~4000 RFU) was adequate, while the substrate consumption was under 10% at this enzyme concentration. An incubation of 20 minutes at RT was selected because the enzyme activity was linear during a 40 minute incubation time (data not shown).
). Based on this result, 1 nM enzyme was selected for the assay because the fluorescence intensity (~4000 RFU) was adequate, while the substrate consumption was under 10% at this enzyme concentration. An incubation of 20 minutes at RT was selected because the enzyme activity was linear during a 40 minute incubation time (data not shown).
DMSO tolerance of this enzyme assay was also evaluated, as it is the solvent used for dissolving library compounds. It was found that the enzyme activity slightly decreased with increasing DMSO concentrations (Fig. 1d ). The enzyme activity was not significantly decreased at the 0.76% DMSO concentration used for the compound screens.
). The enzyme activity was not significantly decreased at the 0.76% DMSO concentration used for the compound screens.
Enzyme Kinetics
The enzyme kinetics experiments were performed using 1 nM enzyme with substrate concentrations ranging from 30 to 500 uM. It was found that the Km was 144 μM and the Vmax 5.74 pmol/min at the optimal pH of 5.9 (Fig. 2a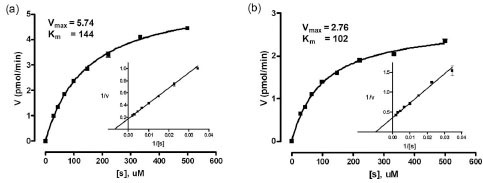 ). This is similar to the value of 239 μM, determined with the same substrate, but at pH 6.5 [24Maranville E, Zhu A. The carboxyl terminus of coffee bean alpha-galactosidase is critical for enzyme activity Arch Biochem Biophys 2000; 373: 225-30.]. In addition, a kinetic analysis was done at the previously used pH of 4.5 [25Maranville E, Zhu A. Assessment of amino-acid substitutions at
tryptophan 16 in alpha-galactosidase Eur J Biochem 2000; 267: 1495-501.], and the Km and Vmax were found to be 102 μM and 2.76 pmol/min, respectively (Fig. 2b
). This is similar to the value of 239 μM, determined with the same substrate, but at pH 6.5 [24Maranville E, Zhu A. The carboxyl terminus of coffee bean alpha-galactosidase is critical for enzyme activity Arch Biochem Biophys 2000; 373: 225-30.]. In addition, a kinetic analysis was done at the previously used pH of 4.5 [25Maranville E, Zhu A. Assessment of amino-acid substitutions at
tryptophan 16 in alpha-galactosidase Eur J Biochem 2000; 267: 1495-501.], and the Km and Vmax were found to be 102 μM and 2.76 pmol/min, respectively (Fig. 2b ). The Km values were similar for the two assays, while the rate of substrate cleavage was about two fold greater at pH 5.9 than at pH 4.5. 40 μM of 4MU-α-Gala was chosen as the substrate concentration for this assay, as it was less than the Km value (important for compound screening sensitivity) and gave sufficient fluorescence.
). The Km values were similar for the two assays, while the rate of substrate cleavage was about two fold greater at pH 5.9 than at pH 4.5. 40 μM of 4MU-α-Gala was chosen as the substrate concentration for this assay, as it was less than the Km value (important for compound screening sensitivity) and gave sufficient fluorescence.
Thus, for compound screening, 1 nM GLA and 40 μM 4MU-α-Gala were chosen as the enzyme and substrate concentrations, respectively. The assay was performed at pH 5.9 with 20 minutes incubation at RT.
IC50 Determination of a Known GLA Inhibitor
For further assay validation, the activity of a known inhibitor, 1-deoxygalactonojirimycin (DGJ), was evaluated using this enzyme assay. The IC50 value of this known inhibitor was 5.6 nM in this enzyme assay (Fig. 3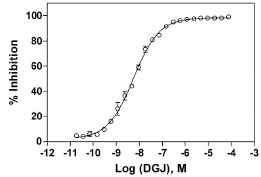 ), which is similar to the reported IC50 value of 3 nM [26Asano N, Ishii S, Kizu H, et al. In vitro inhibition and intracellular
enhancement of lysosomal alpha-galactosidase A activity in Fabry
lymphoblasts by 1-deoxygalactonojirimycin and its derivatives Eur J Biochem 2000; 267: 4179-86.]. This compound was used as an internal control in the subsequent compound screening experiments.
), which is similar to the reported IC50 value of 3 nM [26Asano N, Ishii S, Kizu H, et al. In vitro inhibition and intracellular
enhancement of lysosomal alpha-galactosidase A activity in Fabry
lymphoblasts by 1-deoxygalactonojirimycin and its derivatives Eur J Biochem 2000; 267: 4179-86.]. This compound was used as an internal control in the subsequent compound screening experiments.
 |
Fig. (3) Concentration response of the known GLA inhibitor, DGJ. The substrate concentration was 40 µM and the enzyme concentration was 1 nM. The IC50 was found to be 5.6 nM. |
Screen Validation
In order to test the performance of this enzyme assay for compound screening, we used several small collections of known bioactive compounds, including the LOPAC, Prestwick, Spectrum, and the Tocris-Timtec libraries. In a DMSO plate test without compounds, the signal-to-basal ratio from the control plate was 32.6 fold, and the CV and Z’ factor were 8.1% and 0.76, respectively (Fig. 4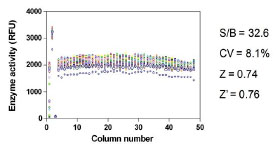 ). These results indicated that this GLA enzyme assay in the 1536-well plate format was robust and suitable for HTS. The qHTS results using the bioactive 6,160 compound set revealed 31 initial “hits”, with a hit rate of approximately 0.50%. This is in the range of the commonly desired 0.1 to 1% hit rate for HTS. However, most of these hits were weak, and were not pursued (Pubchem assay ID: 998). Three of these compounds showed relatively high inhibitory activity against GLA, and were chosen for follow-up experiments. These compounds were lansoprazole, merbromin, and phenylmercuric acetate.
). These results indicated that this GLA enzyme assay in the 1536-well plate format was robust and suitable for HTS. The qHTS results using the bioactive 6,160 compound set revealed 31 initial “hits”, with a hit rate of approximately 0.50%. This is in the range of the commonly desired 0.1 to 1% hit rate for HTS. However, most of these hits were weak, and were not pursued (Pubchem assay ID: 998). Three of these compounds showed relatively high inhibitory activity against GLA, and were chosen for follow-up experiments. These compounds were lansoprazole, merbromin, and phenylmercuric acetate.
 |
Fig. (4) Scatter plot of the results from the control DMSO plate screen. The signal-to-basal ratio was 32.6 with a CV of 8.1% and the Z’ factor was 0.76. |
Hit Confirmation and Compound Characterization
Fresh samples of the three hits from the primary screen were obtained, and their inhibitory activities against GLA were confirmed in the same enzyme assay. To characterize the selectivity of these compounds, they were tested using other lysosomal enzyme assays. Rice α-glucosidase (GAA) and human recombinant glucocerebrosidase (GC) were tested in similar fluorogenic enzyme assay formats. While all three compounds had little effect on GAA, they exhibited inhibitory effects against GC, as well as GLA. Among the three compounds, only lansoprazole showed selectivity for GLA over GC (Fig. 5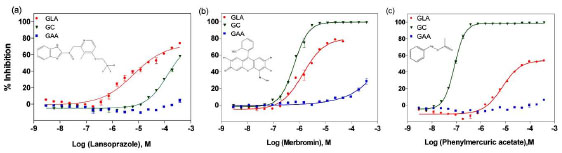 ), as its IC50 for GLA was about 19 times more potent than that for GC (IC50 for GLA was 6.4 μM and IC50 for GC was 122 μM). The other two compounds were not selective for GLA, as the activity of merbromin was 0.63 μM in the GC assay vs. 1.38 μM in the GLA assay, and the IC50 of phenylmercuric acetate was 0.082 μM in the GC assay vs. 7.42 μM in the GLA assay (Fig. 5
), as its IC50 for GLA was about 19 times more potent than that for GC (IC50 for GLA was 6.4 μM and IC50 for GC was 122 μM). The other two compounds were not selective for GLA, as the activity of merbromin was 0.63 μM in the GC assay vs. 1.38 μM in the GLA assay, and the IC50 of phenylmercuric acetate was 0.082 μM in the GC assay vs. 7.42 μM in the GLA assay (Fig. 5 ).
).
To characterize the mechanism of inhibition on GLA, varying concentrations of lansoprazole were tested in the GLA kinetics assay. A Lineweaver-burk plot of the results showed that the linear regression curves of the reciprocal data neither converged on the y-axis (competitive inhibition) or the x-axis (non-competitive inhibition) (Fig. 6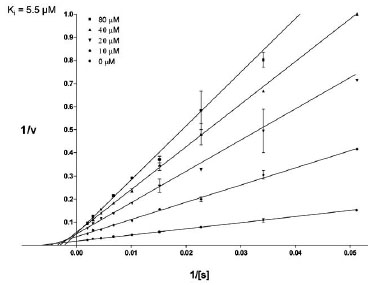 ). The cross point of these linear regression curves was in between the y-axis and x-axis, indicating a mixed type of inhibition. Both the Km and Vmax of GLA were affected in the presence of varying concentrations of this inhibitor. The Ki of lansoprazole was calculated as 5.5 μM.
). The cross point of these linear regression curves was in between the y-axis and x-axis, indicating a mixed type of inhibition. Both the Km and Vmax of GLA were affected in the presence of varying concentrations of this inhibitor. The Ki of lansoprazole was calculated as 5.5 μM.
Lansoprazole is an FDA approved drug used to treat peptic ulcers and gastroesophageal reflux disease. It inhibits the proton-pump of gastric parietal cells, and thus decreases the amount of gastric acid secretion in the stomach. It has not previously been reported to have inhibitory activity on GLA.
HTS of 230,000 Compounds Using the Human Recombinant GLA
Once human recombinant GLA became available for screening, the enzyme assay was optimized once again, as described above. A screen of ~230,000 diverse compounds was carried out using human enzyme in the qHTS format. The primary screen results yielded 136 hits with a hit rate of 0.06%, a rate that was quite low. All of the primary results as well as the assay conditions have been deposited into Pubchem (Assay ID: 1467). Further confirmation and counter screen tests of these primary hits revealed that there were no selective and relatively potent (IC50 < 50 µM) GLA inhibitors against the human enzyme. In an additional effort to find new GLA inhibitors for the human enzyme, we developed an assay using GLA in human spleen homogenate as the enzyme source. This assay used the native enzyme from human tissue, since the GLA enzyme should be in its native conformation and in a physiologically relevant environment with subunits and co-factors. A qHTS was carried out with this assay using the same compound library and 194 primary hits were found, with a hit rate of 0.08% (Assay ID: 2107). No relatively potent GLA inhibitor (IC50 < 50 µM) remained after the confirmation and selectivity tests. In both screens, the previously identified coffee bean GLA inhibitor, lansoprazole, was not active with the human enzyme, and no enzyme activators were found as well. These negative screening results for the human GLA preparation were in contrast to screens with other lysosomal enzymes, including human glucocerebrosidase and α-glucosidase, where potent activators and inhibitors were identified [7Zheng W, Padia J, Urban DJ, et al. Three classes of glucocerebrosidase inhibitors identified by quantitative high-throughput screening are chaperone leads for Gaucher disease Proc Natl Acad Sci USA 2007; 104: 13192-7., 27Marugan JJ, Zheng W, Motabar O, et al. Evaluation of 2-thioxo-2,3,5,6,7,8-hexahydropyrimido[4,5-d]pyrimidin-4(1H)-one analogues as GAA activators Eur J Med Chem 2010; 45: 1880-97.]. The difficulty in identifying activators and inhibitors of human GLA indicates that this enzyme might be a difficult or “undruggable” target for small molecules. It should be noted that the compound libraries had little diversity in chemotypes that mimic sugars such as DGJ.
Coffee bean GLA has 59% homology to the recombinant human enzyme [28Zhu A, Goldstein J. Cloning and functional expression of a cDNA encoding coffee bean alpha-galactosidase Gene 1994; 140: 227-31.]. It is known that two enzymes can have similar catalytic mechanisms and active sites despite a modest similarity in primary sequence, as is the case with many other proteins, such as chymotrypsin and subtilisin [29Kraut J. Serine proteases: structure and mechanism of catalysis Annu Rev Biochem 1977; 46: 331-58.]. Despite the functional similarity of coffee bean and human GLAs, species differences in small molecule inhibition of proteins should not be unexpected, as even closely related species often have important activity differences. For example, DT-diaphorase, an enzyme involved in activating antitumor compounds, responded differently to a compound depending on the source of this enzyme. It was found that human DT-diaphorase was not as effective as rat DT-diaphorase in activating cytotoxic antitumor drugs [30Chen S, Knox R, Wu K, et al. Molecular basis of the catalytic differences among DT-diaphorase of human, rat, and mouse J Biol Chem 1997; 272: 1437-9.-32Beall HD, Mulcahy RT, Siegel D, Traver RD, Gibson NW, Ross D. Metabolism of bioreductive antitumor compounds by purified rat and human DT-diaphorases Cancer Res 1994; 54: 3196-201.]. However, replacing one amino acid residue in the human with that present in the rat enhanced the effectiveness of the compound [30Chen S, Knox R, Wu K, et al. Molecular basis of the catalytic differences among DT-diaphorase of human, rat, and mouse J Biol Chem 1997; 272: 1437-9.]. This indicates that a slight difference in amino acid sequence of an enzyme can cause a significant change in compound activity. Conversely, it has been observed that human TRPA1, a cation channel implicated in pain and neurogenic inflammation, is inhibited by certain trichloro(sulfanyl)ethyl benzamides, while these same compounds are inactive or serve as activators for rat TRPA1 [33Chen J, Zhang XF, Kort ME, et al. Molecular determinants of species-specific activation or blockade of TRPA1 channels J Neurosci 2008; 28: 5063-71., 34Klionsky L, Tamir R, Gao B, et al. Species-specific pharmacology of Trichloro(sulfanyl)ethyl benzamides as transient receptor potential ankyrin 1 (TRPA1) antagonists Mol Pain 2007; 3: 39.].
An amino acid sequence analysis shows that coffee bean GLA shares 36% identity and 59% homlogy with human GLA [28Zhu A, Goldstein J. Cloning and functional expression of a cDNA
encoding coffee bean alpha-galactosidase Gene 1994; 140: 227-31.]. There is a conserved CEW sequence in the human, coffee, and rice GLAs, as well as in other organisms (human residues 203-205), which is an important part of the active site in the human protein structure (Fig. 7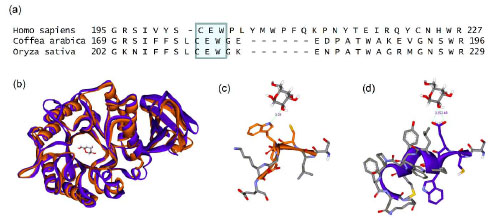 and suppl Fig.
1). However, the crystal structure of rice GLA (PDB code: 1UAS) reveals that this conserved sequence does not support the same structural role as it does in the human protein structure (PDB code: 1R47). Tryptophan 205 is buried in the human protein structure, but its equivalent amino acid in the rice sequence is exposed to solvent. Using the rice template as a model for the coffee bean’s protein structure, it might be hypothesized that this tryptophan provides support (a hot-spot) for lansoprazole binding, while its absence in the human protein active site explains why the observed activity profiles are so different between the two proteins [35NCBI Accession numbers: GI 115482692 (rice), GI 4504009 (human) and GI 504489 (coffee bean) ]. Comparing these two proteins should give better insight into what makes one enzyme “druggable” and another not, and it may be that the difference in hydrophobic surface area provided by the coffee bean’s solvent-exposed tryptophan provides the crucial difference.
and suppl Fig.
1). However, the crystal structure of rice GLA (PDB code: 1UAS) reveals that this conserved sequence does not support the same structural role as it does in the human protein structure (PDB code: 1R47). Tryptophan 205 is buried in the human protein structure, but its equivalent amino acid in the rice sequence is exposed to solvent. Using the rice template as a model for the coffee bean’s protein structure, it might be hypothesized that this tryptophan provides support (a hot-spot) for lansoprazole binding, while its absence in the human protein active site explains why the observed activity profiles are so different between the two proteins [35NCBI Accession numbers: GI 115482692 (rice), GI 4504009 (human) and GI 504489 (coffee bean) ]. Comparing these two proteins should give better insight into what makes one enzyme “druggable” and another not, and it may be that the difference in hydrophobic surface area provided by the coffee bean’s solvent-exposed tryptophan provides the crucial difference.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s website along with the published article Download Supplementary Material.
ACKNOWLEDGEMENTS
This research was supported by the Molecular Libraries Initiative of the NIH Roadmap for Medical Research and the Intramural Research Programs of the National Human Genome Research Institute National Institutes of Health.







