- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

Current Chemical Genomics and Translational Medicine
(Discontinued)
ISSN: 2213-9885 ― Volume 12, 2018
Methods for Activity Analysis of the Proteins that Regulate Histone Methylation
Amy M Quinn#, Anton Simeonov*
Abstract
The enzymes that regulate histone methylation states and the protein domains that recognize methylated histone residues have been implicated in a number of human diseases, including cancer, as a result of their ability to affect transcriptional changes by altering chromatin structure. These proteins are recognized as potential therapeutic targets for the treatment of diseases associated with epigenetic disruption; however, few inhibitors of their activity have been identified. The majority of histone demethylase and methyltransferase enzyme inhibitors have been discovered on the basis of their structural similarity to substrates or known inhibitors of enzymes with analogous mechanisms. The general lack of potency and specificity of these compounds indicates that novel chemotypes are needed to address the large number of recently discovered histone-modifying enzymes. High-throughput screening (HTS) allows rapid testing of chemically diverse small molecule libraries, provided assays amenable to HTS exist. Here we review the biochemical and cellular assays available for testing the proteins and enzymes that regulate histone methylation. Progress in the development of high-throughput, sensitive, and robust assays will enable discovery of small molecules for epigenetic therapy.
Article Information
Identifiers and Pagination:
Year: 2011Volume: 5
Issue: Suppl 1
First Page: 95
Last Page: 105
Publisher Id: CCGTM-5-95
DOI: 10.2174/1875397301005010095
Article History:
Received Date: 9/2/2011Revision Received Date: 23/3/2011
Acceptance Date: 25/4/2011
Electronic publication date: 22/8/2011
Collection year: 2011
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the NIH Chemical Genomics Center, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD 20892-3370, USA; Tel: 301-217-5721; Fax: 301-217-5736; E-mail: asimeono@mail.nih.gov# Present address: Department of Screening and Compound Profiling, GlaxoSmithKline Pharmaceuticals, 1250 South Collegeville Road, Collegeville, PA 19426-0989, USA.
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 9-2-2011 |
Original Manuscript | Methods for Activity Analysis of the Proteins that Regulate Histone Methylation | |
INTRODUCTION
Post-translational modifications (PTMs) of histones, including acetylation, methylation, ubiquitination, and phosphorylation, affect the transcription of associated genes by altering local chromatin structure and the accessibility of transcription factors [1Jenuwein T, Allis CD. Translating the histone code Science 2001; 293(5532): 1074-80.]. These epigenetic marks also act as beacons in the recruitment of enzymes that further modify the landscape of histone PTMs or DNA methylation, or that directly (de)stabilize chromatin structure [2Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers Nat Struct Mol Biol 2007; 14(11): 1025-40.]. Both the enzymes that regulate the addition or removal of histone modifications and the proteins harboring domains that recognize specific PTMs have been implicated in a variety of human diseases, including cancer.
The most thoroughly characterized histone modification is lysine acetylation which generally leads to an increase in transcriptional activity following the loss of lysine’s positive charge with acetylation and decreased electrostatic interaction with the negatively-charged DNA backbone [3Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation Curr Opin Cell Biol 2001; 13(3): 263-73.]. Histone deacetylase (HDAC) inhibitors have been used in the clinic to promote hyperacetylation of histone tails and reactivate aberrantly silenced tumor suppressor genes [4Marks PA, Xu WS. Histone deacetylase inhibitors: potential in cancer therapy J Cell Biochem 2009; 107(4): 600-8.]. The functional consequence of histone methylation is more complex, with methylation of both arginine and lysine possible, and transcriptional changes dependent on the site and degree of methylation. For example, methylation at H3K9, H3K27, and H4K20 typically leads to repressed transcription, whereas methylation at H3K4 and H3K36 results in enhanced transcriptional activation [5Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome Cell 2007; 129(4): 823-37.].
Research on the mechanisms and regulation of histone methylation has intensified in the last decade in response to the discovery that histone methylation is impermanent and the identification of histone demethylases. This surge of activity surrounding epigenetics has resulted in the discovery of greater than 50 methyltransferase enzymes, 30 histone demethylases, and numerous protein domains that recognize and bind methylated histone residues. To study these PTMs and the mechanisms of enzymes that regulate them has necessitated the development of various biochemical and cell-based assays. In addition, validating the involvement of many of these proteins in human disease has generated considerable interest in the discovery of small molecule inhibitors of histone-modifying enzymes and of compounds that disrupt the interactions between histones and the domains that bind specific PTMs, the so-called “readers” of the histone code.
Here we review recent advances in methods to study histone methylation with a focus on the discovery of small molecule modulators of histone methyltransferases, demethylases, and reader domains as potential therapeutic agents in the treatment of cancer and other diseases associated with epigenetic deregulation. To date, no fewer than six high-throughput screens of epigenetic targets have been performed with more than 200,000 compounds and published on the open-access PubChem database (http://pubchem.ncbi. nlm.nih.gov/), demonstrating the need for robust high-throughput assays and techniques for rapid characterization of the downstream effects of compounds that influence histone PTMs. While acknowledging the importance of histone acetylation, phosphorylation, and other PTMs in the development of a spectrum of human diseases, we direct our attention here to the rapidly-evolving field of histone methylation.
METHYLTRANSFERASE ASSAYS
Radiotracer Assays
Radioactivity-based assays to measure histone methyltransferase activity are sensitive, easy to configure, and applicable to a diverse range of enzymes. Filter binding assays measure transfer of tritiated methyl groups from [3H]-SAM to a substrate, followed by removal of unreacted [3H]-SAM and retention of radiolabeled-product by filtration and subsequent scintillation counting (Fig. 1A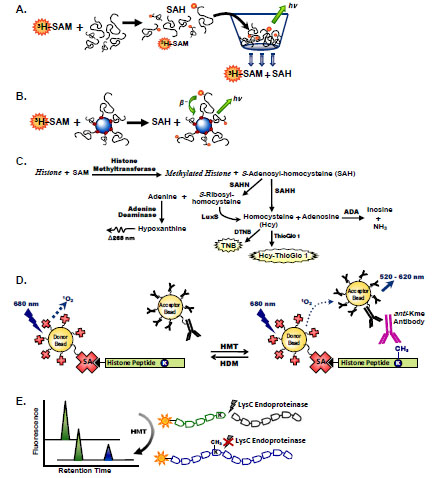 ). The fungal mycotoxin metabolite chaetocin (3,6-epi-dithio-diketopiperazine) was one of the first methyltransferase inhibitors discovered by screening 3,000 compounds against the Drosophila melongaster SU(VAR)3-9 lysine methyltransferase using a radioactive histone H3 peptide filter binding assay (Table 1) [6Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9 Nat Chem Biol 2005; 1(3): 143-5.]. Filters assembled in 96- and 384-well vacuum plates have increased the throughput of this assay. Other plate-based techniques include immobilization of a biotinylated peptide substrate in an avidin/streptavidin-plate and removal of free [3H]-SAM with washing steps. The washing steps may be eliminated with the use of avidin/streptavidin-coated FlashPlates that contain a thin layer of scintillant bound to the walls of the microplate wells where the signal is based on isotope proximity [7Rathert P, Cheng X, Jeltsch A. Continuous enzymatic assay for histone lysine methyltransferases Biotechniques 2007; 43(5): 602-8.].
). The fungal mycotoxin metabolite chaetocin (3,6-epi-dithio-diketopiperazine) was one of the first methyltransferase inhibitors discovered by screening 3,000 compounds against the Drosophila melongaster SU(VAR)3-9 lysine methyltransferase using a radioactive histone H3 peptide filter binding assay (Table 1) [6Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9 Nat Chem Biol 2005; 1(3): 143-5.]. Filters assembled in 96- and 384-well vacuum plates have increased the throughput of this assay. Other plate-based techniques include immobilization of a biotinylated peptide substrate in an avidin/streptavidin-plate and removal of free [3H]-SAM with washing steps. The washing steps may be eliminated with the use of avidin/streptavidin-coated FlashPlates that contain a thin layer of scintillant bound to the walls of the microplate wells where the signal is based on isotope proximity [7Rathert P, Cheng X, Jeltsch A. Continuous enzymatic assay for histone lysine methyltransferases Biotechniques 2007; 43(5): 602-8.].
Transfer of tritiated methyl groups from [3H]-SAM to a bead-bound substrate provides a homogeneous, sensitive method for rapidly measuring methyltransferase activity (Fig. 1B ). Scintillation proximity assays (SPA) utilize microscopic beads containing scintillant. The interaction of these beads with β-particles generated by the radioactive decay of tritium releases photons that may be measured with scintillation counters or charge-coupled device (CCD) imagers. A variety of SPA bead formats allows utilization of various methyltransferase substrates, from biotinylated histone peptides to nucleosomes. The lack of a separation step and intrinsic sensitivity of the assay make SPA techniques amenable for HTS. However, the use of radioactivity is undesirable due to health risks as well as the cost of radiolabeled substrate and waste disposal. Thus the development and use of non-radioactive assays should be utilized whenever possible.
). Scintillation proximity assays (SPA) utilize microscopic beads containing scintillant. The interaction of these beads with β-particles generated by the radioactive decay of tritium releases photons that may be measured with scintillation counters or charge-coupled device (CCD) imagers. A variety of SPA bead formats allows utilization of various methyltransferase substrates, from biotinylated histone peptides to nucleosomes. The lack of a separation step and intrinsic sensitivity of the assay make SPA techniques amenable for HTS. However, the use of radioactivity is undesirable due to health risks as well as the cost of radiolabeled substrate and waste disposal. Thus the development and use of non-radioactive assays should be utilized whenever possible.
Coupled Enzyme Assays
Simple substrate-to-product conversion is difficult to monitor in histone methyltransferase reactions where the spectral differences in SAM and SAH are insufficient for direct monitoring. Several coupled assays have been used to measure the SAH product of methyltransferases (Fig. 1C ). The cysteine-free SAH hydrolase of Sulfolobus solfataricus generates a free thiol in homocysteine that may be measured with thiol-sensitive fluorophores [8Collazo E, Couture JF, Bulfer S, Trievel RC. A coupled fluorescent assay for histone methyltransferases Anal Biochem 2005; 342(1): 86-92.], including the malei-mide derivatives ThioGlo 1 [methyl-10-(2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl)-9-methoxy-3-oxo-3H-benzo[f]chromene-2-carboxylate] and CPM (7-diethylamino-3-(4’-maleimidyl-phenyl)-4-methylcoumarin) [8Collazo E, Couture JF, Bulfer S, Trievel RC. A coupled fluorescent assay for histone methyltransferases Anal Biochem 2005; 342(1): 86-92., 9Chung CC, Ohwaki K, Schneeweis JE, et al. A fluorescence-based thiol quantification assay for ultra-high-throughput screening for inhibitors of coenzyme A production Assay Drug Dev Technol 2008; 6(3): 361-74.]. The free thiol of homocysteine may also be measured by reaction with DTNB (5,5’-dithiobis-2-nitrobenzoic acid) to form the colored product TNB (5-thio-2-nitrobenzoic acid). Homocysteine may also be generated from SAH through the consecutive actions of SAH nucleosidase (SAHN) to generate adenine and S-ribosylhomocysteine and LuxS (S-ribosyl-homocysteinase) to cleave the latter intermediate to form Hcy [10Hendricks CL, Ross JR, Pichersky E, Noel JP, Zhou ZS. An enzyme-coupled colorimetric assay for S-adenosylmethionine-dependent methyltransferases Anal Biochem 2004; 326(1): 100-5.]. Alternatively, the adenine product of SAHN may be utilized by adenine deaminase to form hypoxanthine, where a decrease in UV-absorbance is monitored at 265 nm [11Dorgan KM, Wooderchak WL, Wynn DP, et al. An enzyme-coupled continuous spectrophotometric assay for S-adenosylmethionine-dependent methyltransferases Anal Biochem 2006; 350(2): 249-55.]. The SAHH/ThioGlo assay described by Collazo et al. [8Collazo E, Couture JF, Bulfer S, Trievel RC. A coupled fluorescent assay for histone methyltransferases Anal Biochem 2005; 342(1): 86-92.] is likely the most straightforward and sensitive of these methods to measure SAH production, considering the single coupling enzyme and fluorescence detection. However, assay components must be carefully selected to minimize background fluorescence due to the reaction of free thiols with ThioGlo 1, and chemical blocking of free thiols of the substrate or enzyme may be considered. Coupled enzyme assays are inherently more difficult to optimize and susceptible to false positives when seeking enzyme inhibitors. Nonetheless, the removal of SAH in the coupling assays described here is advantageous for relief of SAH product inhibition, a characteristic common to histone methyltransferases.
). The cysteine-free SAH hydrolase of Sulfolobus solfataricus generates a free thiol in homocysteine that may be measured with thiol-sensitive fluorophores [8Collazo E, Couture JF, Bulfer S, Trievel RC. A coupled fluorescent assay for histone methyltransferases Anal Biochem 2005; 342(1): 86-92.], including the malei-mide derivatives ThioGlo 1 [methyl-10-(2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl)-9-methoxy-3-oxo-3H-benzo[f]chromene-2-carboxylate] and CPM (7-diethylamino-3-(4’-maleimidyl-phenyl)-4-methylcoumarin) [8Collazo E, Couture JF, Bulfer S, Trievel RC. A coupled fluorescent assay for histone methyltransferases Anal Biochem 2005; 342(1): 86-92., 9Chung CC, Ohwaki K, Schneeweis JE, et al. A fluorescence-based thiol quantification assay for ultra-high-throughput screening for inhibitors of coenzyme A production Assay Drug Dev Technol 2008; 6(3): 361-74.]. The free thiol of homocysteine may also be measured by reaction with DTNB (5,5’-dithiobis-2-nitrobenzoic acid) to form the colored product TNB (5-thio-2-nitrobenzoic acid). Homocysteine may also be generated from SAH through the consecutive actions of SAH nucleosidase (SAHN) to generate adenine and S-ribosylhomocysteine and LuxS (S-ribosyl-homocysteinase) to cleave the latter intermediate to form Hcy [10Hendricks CL, Ross JR, Pichersky E, Noel JP, Zhou ZS. An enzyme-coupled colorimetric assay for S-adenosylmethionine-dependent methyltransferases Anal Biochem 2004; 326(1): 100-5.]. Alternatively, the adenine product of SAHN may be utilized by adenine deaminase to form hypoxanthine, where a decrease in UV-absorbance is monitored at 265 nm [11Dorgan KM, Wooderchak WL, Wynn DP, et al. An enzyme-coupled continuous spectrophotometric assay for S-adenosylmethionine-dependent methyltransferases Anal Biochem 2006; 350(2): 249-55.]. The SAHH/ThioGlo assay described by Collazo et al. [8Collazo E, Couture JF, Bulfer S, Trievel RC. A coupled fluorescent assay for histone methyltransferases Anal Biochem 2005; 342(1): 86-92.] is likely the most straightforward and sensitive of these methods to measure SAH production, considering the single coupling enzyme and fluorescence detection. However, assay components must be carefully selected to minimize background fluorescence due to the reaction of free thiols with ThioGlo 1, and chemical blocking of free thiols of the substrate or enzyme may be considered. Coupled enzyme assays are inherently more difficult to optimize and susceptible to false positives when seeking enzyme inhibitors. Nonetheless, the removal of SAH in the coupling assays described here is advantageous for relief of SAH product inhibition, a characteristic common to histone methyltransferases.
Antibody-Based Techniques
Several biochemical assays have been developed that rely on antibody recognition of specific post-translational modifications. Enzyme-linked immunosorbent assays (ELISA) for histone methyltransferases involve immobilization of biotinylated histone peptide substrate on an avidin-coated microplate. Following enzymatic methylation of Lys or Arg residues on the immobilized peptide, a primary antibody specific to the PTM is captured on the plate. A secondary antibody conjugated with horseradish peroxidase (HRP) is used for chemiluminescence detection. The first protein arginine methyltransferase (PRMT) inhibitor AMI-1, a symmetrical sulfonated urea, was identified in an ELISA-based HTS assay of 9,000 diverse compounds using fungal Hmt1p (also known as Rmt1p) and the hnRNP protein Npl3p (Table 1) [12Cheng D, Yadav N, King RW, Swanson MS, Weinstein EJ, Bedford MT. Small molecule regulators of protein arginine methyltransferases J Biol Chem 2004; 279(23): 23892-9.]. AMI-1 is a non-specific arginine methyltransferase inhibitor, although it does not affect methylation of lysine.
A modern version of the ELISA is the dissociation-enhanced lanthanide fluorescent immunoassay (DELFIA) where the secondary antibody is labeled with a lanthanide chelate (typically europium) instead of HRP, and product formation is measured with time-resolved fluorescence (TRF). This technique was used to identify the first inhibitor of a histone lysine methyltransferase that was not competitive with the SAM cosubstrate. BIX-01294 was discovered in a screen of 125,000 compounds as a selective inhibitor of the G9a lysine methyltransferase (IC50 2.7 µM) and was competitive with the histone H3 peptide substrate (Table 1) [13Kubicek S, O'Sullivan RJ, August EM, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase Mol Cell 2007; 25(3): 473-81.]. BIX-01338 was also identified in this screen as a G9a inhibitor (IC50 4.7 µM), but it also inhibited other Lys and Arg methyltransferases and was SAM-competitive. Inhibitors of the arginine methyltransferase PRMT1 were also identified using a DELFIA assay to screen compounds selected by iterative virtual screening of the National Cancer Institute (NCI) diversity collection [14Spannhoff A, Heinke R, Bauer I, et al. Target-based approach to inhibitors of histone arginine methyltransferases J Med Chem 2007; 50(10): 2319-5.]. Two of the compounds active in the PRMT1 in vitro assay, allantodapsone and stilbamidine, were validated by demonstrating hypomethylation of H4R3 in HepG2 cells and inhibition of ERα-transcriptional activation in a cell-based reporter assay.
Antibody detection has also been used in a homogeneous assay utilizing AlphaScreen technology to measure histone G9a methyltransferase activity [15Quinn AM, Allali-Hassani A, Vedadi M, Simeonov A. A chemiluminescence-based method for identification of histone lysine methyltransferase inhibitors Mol Biosyst 2010; 6(5): 782-8.]. This dual bead-based technology utilizes laser excitation of donor beads, releasing a flow of singlet oxygen to generate chemiluminescent emission from acceptor beads in close proximity (Fig. 1D ). Acceptor beads coated with secondary antibody are coupled to specific antibody detection of enzyme-modified substrates. AlphaScreen was used in the structural optimization of the BIX-01294 inhibitor of G9a, and IC50 values obtained tracked closely with those measured in a SAHH/ThioGlo 1 coupled assay [16Liu F, Chen X, Allali-Hassani A, et al. Discovery of a 2,4-diamino-7-aminoalkoxyquinazoline as a potent and selective inhibitor of histone lysine methyltransferase G9a J Med Chem 2009; 52(24): 7950-3., 17Liu F, Chen X, Allali-Hassani A, et al. Protein lysine methyltransferase G9a inhibitors: design, synthesis., and structure activity relationships of 2.,4-diamino-7-aminoalkoxy-quinazolines J Med Chem 2010; 53(15): 5844-7.]. These studies led to the discovery of UNC0224 and UNC0321, the most potent G9a inhibitors identified to date (Table 1). The advantage of this assay compared to ELISA-based methods is the homogeneous nature of AlphaScreen where elimination of washing and substrate immobilization steps leads to an increased assay throughput. Importantly, the AlphaScreen assay was readily miniaturized to 1,536-well plate format allowing for its use in screening large compound collections [15Quinn AM, Allali-Hassani A, Vedadi M, Simeonov A. A chemiluminescence-based method for identification of histone lysine methyltransferase inhibitors Mol Biosyst 2010; 6(5): 782-8.].
). Acceptor beads coated with secondary antibody are coupled to specific antibody detection of enzyme-modified substrates. AlphaScreen was used in the structural optimization of the BIX-01294 inhibitor of G9a, and IC50 values obtained tracked closely with those measured in a SAHH/ThioGlo 1 coupled assay [16Liu F, Chen X, Allali-Hassani A, et al. Discovery of a 2,4-diamino-7-aminoalkoxyquinazoline as a potent and selective inhibitor of histone lysine methyltransferase G9a J Med Chem 2009; 52(24): 7950-3., 17Liu F, Chen X, Allali-Hassani A, et al. Protein lysine methyltransferase G9a inhibitors: design, synthesis., and structure activity relationships of 2.,4-diamino-7-aminoalkoxy-quinazolines J Med Chem 2010; 53(15): 5844-7.]. These studies led to the discovery of UNC0224 and UNC0321, the most potent G9a inhibitors identified to date (Table 1). The advantage of this assay compared to ELISA-based methods is the homogeneous nature of AlphaScreen where elimination of washing and substrate immobilization steps leads to an increased assay throughput. Importantly, the AlphaScreen assay was readily miniaturized to 1,536-well plate format allowing for its use in screening large compound collections [15Quinn AM, Allali-Hassani A, Vedadi M, Simeonov A. A chemiluminescence-based method for identification of histone lysine methyltransferase inhibitors Mol Biosyst 2010; 6(5): 782-8.].
AlphaScreen assays to measure histone methyltransferase activity may utilize direct coupling of the acceptor bead to the primary antibody in place of a secondary anti-immunoglobulin donor bead plus primary antibody. Some methyl mark antibodies directly conjugated to acceptor beads are commercially available (http://las.perkinelmer. com/). In vitro AlphaScreen assays most commonly use streptavidin-coated donor beads to bind biotinylated peptide substrates. However, donor beads may be conjugated with antibodies that recognize histone substrates at a site distinct from that modified by the methyltransferase or demethylase enzyme. This sandwich-based approach is suitable for enzymes with no or low activity towards peptide substrates as well as for cell-based quantitation of histone-modifying enzyme activity.
Time-resolved fluorescence resonance energy transfer (TR-FRET), like AlphaScreen, is a proximity based technique that may be used to measure enzymatic activity of histone-modifying enzymes. TR-FRET combines time-resolved fluorescence (TRF) and FRET in a homogeneous format where fluorescence emission of a donor dye overlaps with the excitation spectrum of an acceptor dye. Whereas AlphaScreen donor and acceptor molecules may be separated by up to 200 nm, the distance of interaction is typically limited to 1-10 nm for TR-FRET. To date, FRET has only been used to measure inhibition of HDAC activity in cells using a baculovirus-expressed GFP-p53 fusion protein [18Dudek JM, Horton RA. TR-FRET biochemical assays for detecting posttranslational modifications of p53 J Biomol Screen 2010; 15(5): 569-75.]. A terbium-labeled acetyl-p53 (Lys382) antibody was used to measure activity of the p300 histone acetyltransferase (HAT) using the non-histone GFP-p53 substrate where GFP is the FRET acceptor. We note that if an endpoint assay based on antibody detection is desired, the traditional method of stopping the enzymatic reaction by addition of acid will not be usable: instead, an addition of EDTA at a millimolar concentration can be used as an alternative.
The limitations of these techniques lie in the availability of specific antibodies, particularly in assays that require discrimination between mono- and dimethylated or di- and tri-methylated residues [19Kawamura A, Tumber A, Rose NR, et al. Development of homogeneous luminescence assays for histone demethylase catalysis and binding Anal Biochem 2010; 404(1): 86-93.]. Examination of more than 200 antibodies raised against 57 different histone modifications found that 25% of these failed specificity testing by Western blot and dot blot analysis, and 20% of the specific antibodies were deemed unsuitable for chromatin immunoprecipitation (ChIP) analysis [20Egelhofer TA, Minoda A, Klugman S, et al. An assessment of histone-modification antibody quality Nat Struct Mol Biol 2010; 18(1): 91-3.]. The high failure rate prompted the authors to create a website for posting these and new results of antibody specificity tests for the detection of epigenetic marks (http://compbio.med.harvard.edu/antibodies/). Where specific antibodies are not available, the use of tagged chromatin binding domains to measure changes in levels of specific post-translationally modified residues has been suggested (Wigle, T.J., personal communication), although it is unclear whether the micromolar affinities typical of reader domains for their cognate epigenetic mark would be sufficient for such applications. Additional complications in attempting to quantify PTMs with reader domains in a cellular environment may result from the observation by Fuchs et al. that neighboring PTMs can affect the affinity of chromatin-associated domains for a particular epigenetic mark [21Fuchs SM, Krajewski K, Baker RW, Miller VL, Strahl BD. Influence of combinatorial histone modifications on antibody and effector protein recognition Curr Biol 2011; 21(1): 53-8.]. Using a library of histone peptides with single or multiple PTMs immobilized in a peptide array, both enhanced and diminished binding of several reader domains were observed for combinations of PTMs. Using the same peptide array, the authors also found the specificity of some commercial antibodies raised against a specific epigenetic mark were affected by other PTMs present on the peptide bait [21Fuchs SM, Krajewski K, Baker RW, Miller VL, Strahl BD. Influence of combinatorial histone modifications on antibody and effector protein recognition Curr Biol 2011; 21(1): 53-8.].
Capillary Electrophoresis
The principles of capillary electrophoresis applied to a microfluidic chip (MCE) allow separation of nanoliter-sized samples based on the charge-to-mass ratio of the components of interest. The loss of lysine’s positive charge with acetylation results in facile electrophoretic separation of substrate and product peptides and offers a direct method for measuring HAT or HDAC activity. This type of electrophoretic mobility shift assay has been used to profile inhibitors of HDAC and HAT enzymes, including PCAF and GCN5L2 [22Fanslau C, Pedicord D, Nagulapalli S, et al. An electrophoretic mobility shift assay for the identification and kinetic analysis of acetyl transferase inhibitors Anal Biochem 2010; 402(1): 65-8., 23Blackwell L, Norris J, Suto CM, Janzen WP. The use of diversity profiling to characterize chemical modulators of the histone deacetylases Life Sci 2008; 82(21-22): 1050-8.]. A similar approach may be employed for changes in phosphorylation of histone peptides.
Methylation of lysine, however, does not change the positive charge of this amino acid, rendering a direct electrophoretic separation impossible. To address this limitation, methods originally developed for methyllysine mapping of histones by mass spectrometry (MS) have recently been applied to MCE. These techniques use the endoproteinase LysC to selectively cleave at the carboxyl side of unmodified lysine residues, leaving methylated lysine polypeptides intact (Fig. 1E ). Such a histone mark-dependent cleavage allows for the production of fluorescent peptides of different charge-to-mass ratios. The utility of this method was demonstrated using LysC and fluorescent-labeled histone peptides for inhibitor screening and mechanism of action studies of the G9a methyltransferase and the LSD1 lysine demethylase [24Wigle TJ, Provencher LM, Norris JL, et al. Accessing protein methyltransferase and demethylase enzymology using microfluidic capillary electrophoresis Chem Biol 2010; 17(7): 695-704.]. The G9a MCE assay was used to determine the Morrison Ki (63 pM) of UNC0321, the 2,4-diamino-7-aminoalkoxy-quinazoline compound designed from the BIX-01294 chemical scaffold (Table 1) [17Liu F, Chen X, Allali-Hassani A, et al. Protein lysine methyltransferase G9a inhibitors: design, synthesis., and structure activity relationships of 2.,4-diamino-7-aminoalkoxy-quinazolines J Med Chem 2010; 53(15): 5844-7.]. The multiplicity of Lys residues in the N-terminal tails of histones, however, complicates generation of suitable peptides upon LysC cleavage and may require mutation or chemical modification of non-substrate Lys residues in substrate peptides. In addition to measuring lysine methyltransferase activity, protease-coupled MCE may be applicable to monitoring Arg methylation with the use of the ArgC endoproteinase [24Wigle TJ, Provencher LM, Norris JL, et al. Accessing protein methyltransferase and demethylase enzymology using microfluidic capillary electrophoresis Chem Biol 2010; 17(7): 695-704.].
). Such a histone mark-dependent cleavage allows for the production of fluorescent peptides of different charge-to-mass ratios. The utility of this method was demonstrated using LysC and fluorescent-labeled histone peptides for inhibitor screening and mechanism of action studies of the G9a methyltransferase and the LSD1 lysine demethylase [24Wigle TJ, Provencher LM, Norris JL, et al. Accessing protein methyltransferase and demethylase enzymology using microfluidic capillary electrophoresis Chem Biol 2010; 17(7): 695-704.]. The G9a MCE assay was used to determine the Morrison Ki (63 pM) of UNC0321, the 2,4-diamino-7-aminoalkoxy-quinazoline compound designed from the BIX-01294 chemical scaffold (Table 1) [17Liu F, Chen X, Allali-Hassani A, et al. Protein lysine methyltransferase G9a inhibitors: design, synthesis., and structure activity relationships of 2.,4-diamino-7-aminoalkoxy-quinazolines J Med Chem 2010; 53(15): 5844-7.]. The multiplicity of Lys residues in the N-terminal tails of histones, however, complicates generation of suitable peptides upon LysC cleavage and may require mutation or chemical modification of non-substrate Lys residues in substrate peptides. In addition to measuring lysine methyltransferase activity, protease-coupled MCE may be applicable to monitoring Arg methylation with the use of the ArgC endoproteinase [24Wigle TJ, Provencher LM, Norris JL, et al. Accessing protein methyltransferase and demethylase enzymology using microfluidic capillary electrophoresis Chem Biol 2010; 17(7): 695-704.].
Mass Spectrometry
Mass spectrometry (MS) has been an important tool in identifying novel PTMs as well as characterizing activities of histone-modifying enzymes [25Eberl HC, Mann M, Vermeulen M. Quantitative proteomics for epigenetics Chembiochem 2011; 12(2): 224-34.]. The majority of this work has been performed using a bottom-up approach where proteins are digested into small peptides prior to MS analysis. Tandem mass spectrometry (MS/MS) is used to map the site of modification and reconstruct protein identity using sequence information. The major disadvantage to this approach is that information on the combinatorial nature of histone modifications is lost. A top-down approach, on the other hand, refers to MS analysis of intact proteins to yield information on combinations of modifications and their relative abundance [26Garcia BA. What does the future hold for top down mass spectrometry? J Am Soc Mass Spectrom 2010; 21(2): 193-202.]. This is particularly useful in the analysis of heterogeneous mixtures of histones, for example in tissues and cell culture.
An intermediate approach to these techniques was constructed to mitigate the need for a high-resolution mass analyzer in top-down MS while retaining information on the interplay between PTMs on a single histone polypeptide. In middle-down MS, limited proteolysis of histones with endoproteinases such as GluC or AspN generates polypeptides of up to 50 residues [27Taverna SD, Ueberheide BM, Liu Y, et al. Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini Proc Natl Acad Sci USA 2007; 104(7): 2086-91.]. As the majority of PTMs are located on the N-terminal tails of these proteins, information on the complex spectrum of histone PTMs is gained with middle-down MS. This MS proteomics method has been used for global assessment of combinatorial PTMs and their tissue-specific abundance [28Garcia BA, Thomas CE, Kelleher NL, Mizzen CA. Tissue-specific expression and post-translational modification of histone H3 variants J Proteome Res 2008; 7(10): 4225-36.].
Quantitative MS has been used to measure enzymatic activity of histone-modifying enzymes. In vitro approaches generally target specific PTMs and use MS peak intensities of high resolution, well-defined peptides for relative quantitation of product formation (Fig. 2A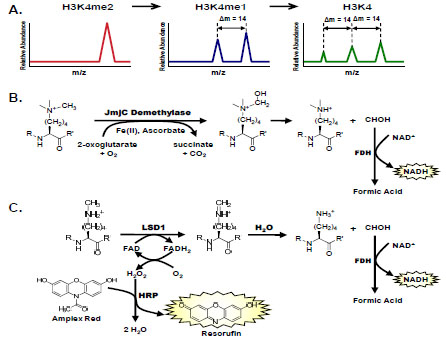 ). This label-free method has been used to measure activities of numerous histone-modifying enzymes [29Tsukada Y, Fang J, Erdjument-Bromage H, et al. Histone demethylation by a family of JmjC domain-containing proteins Nature 2006; 439(7078): 811-6.-31Osborne TC, Obianyo O, Zhang X, Cheng X, Thompson PR. Protein arginine methyltransferase 1: positively charged residues in substrate peptides distal to the site of methylation are important for substrate binding and catalysis Biochemistry 2007; 46(46): 13370-81.] and to determine inhibitor potency [32Chang Y, Zhang X, Horton JR, et al. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294 Nat Struct Mol Biol 2009; 16(3): 312-7., 33Rose NR, Ng SS, Mecinovic J, et al. Inhibitor scaffolds for 2-oxoglutarate-dependent histone lysine demethylases J Med Chem 2008; 51(22): 7053-6.]. An internal standard for normalization that consists of a nonvarying, unmodified peptide may be used in conjunction with peptide intensity profiling to measure changes in histone modifications. Advances in the throughput of MS-based quantitation assays have increased the capacity for inhibitor screening in these sensitive and selective assays [34Roddy TP, Horvath CR, Stout SJ, et al. Mass spectrometric techniques for label-free high-throughput screening in drug discovery Anal Chem 2007; 79(21): 8207-13.]. For example, inhibition of the LSD1 histone demethylase has been measured using an eight-way parallel, staggered flow injection MS from samples in 384-well plate format [35Rye PT, Frick LE, LaMarr WA, Özbal CC. High-throughput Mass Spectrometric Detection of Histone 3 Demethylation Available from: http://www.biocius.com/pdfs/RapidFireHTMS_EpigeneticTargets_Poster.pdf [cited 2011 Jan];]. Other quantitative methods for measuring histone modifying enzymes utilize various labeling techniques, including stable isotope labeling with amino acids in cell culture (SILAC) [36Ong SE, Blagoev B, Kratchmarova I, et al. Stable isotope labeling by amino acids in cell culture, SILAC., as a simple and accurate approach to expression proteomics Mol Cell Proteomics 2002; 1(5): 376-86.], enzymatic incorporation of 18O [37Bantscheff M, Dumpelfeld B, Kuster B. Femtomol sensitivity post-digest (18)O labeling for relative quantification of differential protein complex composition Rapid Commun Mass Spectrom 2004; 18(8): 869-76.], iTRAQ [38Ross PL, Huang YN, Marchese JN, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents Mol Cell Proteomics 2004; 3(12): 1154-69.], and chemical tagging of reactive side chains or groups with stable isotopes [39Smith CM, Gafken PR, Zhang Z, Gottschling DE, Smith JB, Smith DL. Mass spectrometric quantification of acetylation at specific lysines within the amino-terminal tail of histone H4 Anal Biochem 2003; 316(1): 23-33.]. Stable isotopes may also be incorporated into methylated histone residues using [13CD3]-labeled SAM in place of the SAM methyl donor [40Ong SE, Mittler G, Mann M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC Nat Methods 2004; 1(2): 119-26.].
). This label-free method has been used to measure activities of numerous histone-modifying enzymes [29Tsukada Y, Fang J, Erdjument-Bromage H, et al. Histone demethylation by a family of JmjC domain-containing proteins Nature 2006; 439(7078): 811-6.-31Osborne TC, Obianyo O, Zhang X, Cheng X, Thompson PR. Protein arginine methyltransferase 1: positively charged residues in substrate peptides distal to the site of methylation are important for substrate binding and catalysis Biochemistry 2007; 46(46): 13370-81.] and to determine inhibitor potency [32Chang Y, Zhang X, Horton JR, et al. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294 Nat Struct Mol Biol 2009; 16(3): 312-7., 33Rose NR, Ng SS, Mecinovic J, et al. Inhibitor scaffolds for 2-oxoglutarate-dependent histone lysine demethylases J Med Chem 2008; 51(22): 7053-6.]. An internal standard for normalization that consists of a nonvarying, unmodified peptide may be used in conjunction with peptide intensity profiling to measure changes in histone modifications. Advances in the throughput of MS-based quantitation assays have increased the capacity for inhibitor screening in these sensitive and selective assays [34Roddy TP, Horvath CR, Stout SJ, et al. Mass spectrometric techniques for label-free high-throughput screening in drug discovery Anal Chem 2007; 79(21): 8207-13.]. For example, inhibition of the LSD1 histone demethylase has been measured using an eight-way parallel, staggered flow injection MS from samples in 384-well plate format [35Rye PT, Frick LE, LaMarr WA, Özbal CC. High-throughput Mass Spectrometric Detection of Histone 3 Demethylation Available from: http://www.biocius.com/pdfs/RapidFireHTMS_EpigeneticTargets_Poster.pdf [cited 2011 Jan];]. Other quantitative methods for measuring histone modifying enzymes utilize various labeling techniques, including stable isotope labeling with amino acids in cell culture (SILAC) [36Ong SE, Blagoev B, Kratchmarova I, et al. Stable isotope labeling by amino acids in cell culture, SILAC., as a simple and accurate approach to expression proteomics Mol Cell Proteomics 2002; 1(5): 376-86.], enzymatic incorporation of 18O [37Bantscheff M, Dumpelfeld B, Kuster B. Femtomol sensitivity post-digest (18)O labeling for relative quantification of differential protein complex composition Rapid Commun Mass Spectrom 2004; 18(8): 869-76.], iTRAQ [38Ross PL, Huang YN, Marchese JN, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents Mol Cell Proteomics 2004; 3(12): 1154-69.], and chemical tagging of reactive side chains or groups with stable isotopes [39Smith CM, Gafken PR, Zhang Z, Gottschling DE, Smith JB, Smith DL. Mass spectrometric quantification of acetylation at specific lysines within the amino-terminal tail of histone H4 Anal Biochem 2003; 316(1): 23-33.]. Stable isotopes may also be incorporated into methylated histone residues using [13CD3]-labeled SAM in place of the SAM methyl donor [40Ong SE, Mittler G, Mann M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC Nat Methods 2004; 1(2): 119-26.].
Binding Assays
Given the low turnover of many histone methyltransferases, assays that measure displacement of SAM or a peptide substrate may be used to further characterize inhibitors or to select potential inhibitors. Fluorescence polarization (FP) assays measure changes in fluorescence anisotropy in the presence of compounds that displace fluorescently-labeled cognate peptides from the methyltransferase active site. Exploration of the SAR around the G9a inhibitor BIX-01294 was aided by measuring displacement of a fluorescein-labeled histone H3 peptide upon compound binding [16Liu F, Chen X, Allali-Hassani A, et al. Discovery of a 2,4-diamino-7-aminoalkoxyquinazoline as a potent and selective inhibitor of histone lysine methyltransferase G9a J Med Chem 2009; 52(24): 7950-3.]. A similar FP assay was developed to measure binding of histone H4 peptides to PRMT1 [41Feng Y, Xie N, Wu J, Yang C, Zheng YG. Inhibitory study of protein arginine methyltransferase 1 using a fluorescent approach Biochem Biophys Res Commun 2009; 379(2): 567-72.], and was used for the mechanistic analysis and comparison of binding affinities for published PRMT1 inhibitors (Table 1) [42Feng Y, Li M, Wang B, Zheng YG. Discovery and mechanistic study of a class of protein arginine methylation inhibitors J Med Chem 2010; 53(16): 6028-39.]. FP assays, however, require large amounts of enzyme and may not be appropriate for large-scale discovery of peptide-competitive inhibitors.
Cell-Based Methyltransferase Assays
The options for cell-based assays to measure changes in posttranslational modifications of histone tails are limited and primarily semi-quantitative. The multiplicity of methyltransferases and a lack of characterization of their substrates make it difficult to assign activity to a particular enzyme. Transcriptional changes of genes known to be affected by histone methylation at a particular site are frequently used as surrogate markers of methyltransferase activity in cells [14Spannhoff A, Heinke R, Bauer I, et al. Target-based approach to inhibitors of histone arginine methyltransferases J Med Chem 2007; 50(10): 2319-5.]. ChIP at methyltransferase target genes may also be used to measure methylation changes at promoter-proximal sites. Measurements compared before and after knockdown of the methyltransferase by genetic or RNAi-based methods assist in validation of the inhibitor’s target.
Cell-based assays to measure changes in a particular epigenetic mark include fluorescence immunostaining with specific antibodies in live cells [43King ON, Li XS, Sakurai M, et al. Quantitative high-throughput screening identifies 8-hydroxyquinolines as cell-active histone demethylase inhibitors PLoS One 2010; 5(11): e15535.]. A similar technique is standard ELISA with fixed, permeabilized cells using a primary antibody directed against a specific PTM and a europium-labeled secondary antibody for TRF detection [14Spannhoff A, Heinke R, Bauer I, et al. Target-based approach to inhibitors of histone arginine methyltransferases J Med Chem 2007; 50(10): 2319-5.]. As discussed above, quantitation of methylation at a specific site is complicated by the availability of specific antibodies.
HISTONE DEMETHYLASES
The JmjC domain demethylases utilize non-heme iron (II) in a 2-oxoglutarate (2-OG)–dependent oxygenation mechanism to demethylate lysine residues of histone tails [29Tsukada Y, Fang J, Erdjument-Bromage H, et al. Histone demethylation by a family of JmjC domain-containing proteins Nature 2006; 439(7078): 811-6.]. Their aberrant activities are associated with cancer, inflammation, and X-linked mental retardation [44Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease Nat Rev Genet 2007; 8(11): 829-33.]. Reported inhibitors of the JmjC demethylases include 2,4-pyridinedicarboxylic acid (PDCA) and hydroxamic acid derivatives, small compounds of micromolar potency that are competitive with 2-OG (Table 1) [33Rose NR, Ng SS, Mecinovic J, et al. Inhibitor scaffolds for 2-oxoglutarate-dependent histone lysine demethylases J Med Chem 2008; 51(22): 7053-6.]. The coupling enzyme formaldehyde dehydrogenase (FDH) has been used to oxidize the demethylase product formaldehyde with concomitant reduction of its NAD+ cofactor to monitor the change in UV absorbance or fluorescence of NADH (Fig. 2B ), thus producing a convenient real time kinetic assay for JmjC activity [45Sakurai M, Rose NR, Schultz L, et al. A miniaturized screen for inhibitors of Jumonji histone demethylases Mol Biosyst 2010; 6(2): 357-64., 46Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase Nat Struct Mol Biol 2007; 14(8): 689-95.]. AlphaScreen has also been used to measure JMJD2E H3K9me3 demethylase activity in vitro. Using a panel of 2,4-PDCA analogs, the AlphaScreen assay yielded similar IC50 values for JMJD2E inhibition compared to those obtained with the FDH-coupled assay [19Kawamura A, Tumber A, Rose NR, et al. Development of homogeneous luminescence assays for histone demethylase catalysis and binding Anal Biochem 2010; 404(1): 86-93.].
), thus producing a convenient real time kinetic assay for JmjC activity [45Sakurai M, Rose NR, Schultz L, et al. A miniaturized screen for inhibitors of Jumonji histone demethylases Mol Biosyst 2010; 6(2): 357-64., 46Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase Nat Struct Mol Biol 2007; 14(8): 689-95.]. AlphaScreen has also been used to measure JMJD2E H3K9me3 demethylase activity in vitro. Using a panel of 2,4-PDCA analogs, the AlphaScreen assay yielded similar IC50 values for JMJD2E inhibition compared to those obtained with the FDH-coupled assay [19Kawamura A, Tumber A, Rose NR, et al. Development of homogeneous luminescence assays for histone demethylase catalysis and binding Anal Biochem 2010; 404(1): 86-93.].
The continuous and homogenous nature of the FDH-coupled assay allowed its straightforward miniaturization to 1,536-well plate format [45Sakurai M, Rose NR, Schultz L, et al. A miniaturized screen for inhibitors of Jumonji histone demethylases Mol Biosyst 2010; 6(2): 357-64.] and its subsequent use in a high-throughput screen of 236,000 compounds for inhibitors of the JMJD2E enzyme using an H3K9me3 peptide substrate [43King ON, Li XS, Sakurai M, et al. Quantitative high-throughput screening identifies 8-hydroxyquinolines as cell-active histone demethylase inhibitors PLoS One 2010; 5(11): e15535.]. Of the chemical scaffolds designated as active in the screen, optimization of the 8-hyroxyquinolines was performed to generate a 5-carboxy-8HQ cell-permeable compound with 200 nM potency in the enzymatic assay [43King ON, Li XS, Sakurai M, et al. Quantitative high-throughput screening identifies 8-hydroxyquinolines as cell-active histone demethylase inhibitors PLoS One 2010; 5(11): e15535.]. Compared to 2,4-PDCA, the 5-carboxy-8HQ was more selective for JMJD2 enzymes than the 2-OG–dependent hypoxia inducible factor prolyl hydroxylases PHD2 and FIH. The crystal structure of this compound with the homologous JMJD2A protein indicates that 5-carboxy-8HQ disrupts the binding of the active site Fe(II).
A second family of histone demethylases is the lysine specific demethylase (LSD) family, comprised of LSD1 and LSD2. These are flavin adenine dinucleotide (FAD)-dependent amine oxidases that demethylate di- or mono-methylated H3K4 [47Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1 Cell 2004; 119(7): 941-53.]. The rapid turnover of LSD enzymes compared to JMJ enzymes enables easier detection of demethylase activity. While LSD enzymes generate formaldehyde and may be coupled to FDH, as outlined above, more robust assays exist that measure the hydrogen peroxide product (Fig. 2B ). Horseradish peroxidase (HRP)-coupled assays use H2O2 to oxidize a number of HRP substrates including 4-aminoantipyrine, AmplexRed (10-acetyl-3,7-dihydroxyphenoxazine), and luminol for detection using absorbance, fluorescence, or chemiluminescence, respectively.
). Horseradish peroxidase (HRP)-coupled assays use H2O2 to oxidize a number of HRP substrates including 4-aminoantipyrine, AmplexRed (10-acetyl-3,7-dihydroxyphenoxazine), and luminol for detection using absorbance, fluorescence, or chemiluminescence, respectively.
Of the reported LSD inhibitors, the majority are monoamine oxidase (MAO) inhibitors used in the clinic for antidepressant therapy, including pargyline and tranylcypromine [48Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications Chem Biol 2006; 13(6): 563-7., 49Karytinos A, Forneris F, Profumo A, et al. A novel mammalian flavin-dependent histone demethylase J Biol Chem 2009; 284(26): 17775-82.]. These are mechanism-based inhibitors that form covalent adducts with the FAD cofactor of LSD1/2 [50Culhane JC, Szewczuk LM, Liu X, Da G, Marmorstein R, Cole PA. A mechanism-based inactivator for histone demethylase LSD1 J Am Chem Soc 2006; 128(14): 4536-7., 51Binda C, Valente S, Romanenghi M, et al. Biochemical, structural., and biological evaluation of tranylcypromine derivatives as inhibitors of histone demethylases LSD1 and LSD2 J Am Chem Soc 2010; 132(19): 6827-33.]. Libraries constructed around these MAO inhibitor scaffolds have improved inhibitor potency and selectivity for LSD over MAO enzymes [51Binda C, Valente S, Romanenghi M, et al. Biochemical, structural., and biological evaluation of tranylcypromine derivatives as inhibitors of histone demethylases LSD1 and LSD2 J Am Chem Soc 2010; 132(19): 6827-33., 52Culhane JC, Wang D, Yen PM, Cole PA. Comparative analysis of small molecules and histone substrate analogues as LSD1 lysine demethylase inhibitors J Am Chem Soc 2010; 132(9): 3164-76.]. Biguanide and bisguanidine polyamine analogues have also been described as LSD1 inhibitors and result in reexpression of aberrantly silenced tumor suppressor genes in a human colon cancer cell line [53Huang Y, Stewart TM, Wu Y, et al. Novel oligoamine analogues inhibit lysine-specific demethylase 1 and induce reexpression of epigenetically silenced genes Clin Cancer Res 2009; 15(23): 7217-28., 54Huang Y, Greene E, Murray ST, et al. Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes Proc Natl Acad Sci USA 2007; 104(19): 8023-.].
HISTONE CODE READER DOMAINS
Protein domains that bind to and recognize posttranslational modifications of histones are referred to as readers of the histone code. These domains include the Royal family of proteins (chromo-, tudor-, malignant brain tumor-domains, others) that recognize PTMs of Lys residues and bromodomains that recognize acetylated Lys [1Jenuwein T, Allis CD. Translating the histone code Science 2001; 293(5532): 1074-80.]. Histone code readers function as adaptors to recruit proteins for further modification of chromatin structure [2Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers Nat Struct Mol Biol 2007; 14(11): 1025-40.]. In addition to recruitment of histone-modifying enzymes, domains that bind specific epigenetic marks link histone modification to changes in DNA methylation [55Zhao Q, Rank G, Tan YT, et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing Nat Struct Mol Biol 2009; 16(3): 304-11.]. The important role of these recognition motifs and their increasing association with disease make compounds that disrupt these protein-protein interactions a novel strategy for targeting epigenetic diseases.
The multiplicity of histone code readers suggests diversity in substrate specificity and, indeed, the interactions of these protein domains with the large spectrum of histone tail PTMs demonstrate a high level of specificity [56Kim J, Daniel J, Espejo A, et al. Tudor, MBT and chromo domains gauge the degree of lysine methylation EMBO Rep 2006; 7(4): 397-403.]. Crystal structures of bromodomains, a recognition domain family comprised of at least 57 members, indicate diversity in loop regions that dictate substrate specificity despite a conserved binding channel [57Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains Nature 2010; 468(7327): 1067-73.]. Thus, characterization of disease-associated epigenetic marks and reader domain specificity is crucial in targeting these binding domains. Peptide arrays coupled with mass spectrometry are used to bait, capture, and identify chromatin-associated domain binding partners [58Liu H, Galka M, Iberg A, et al. Systematic identification of methyllysine-driven interactions for histone and nonhistone targets J Proteome Res 2010; 9(11): 5827-36.]. Combining this technique with SILAC to expose “heavy” cell extracts to post-translationally modified peptide bait and “light” cell extracts to unmodified peptide, specific binders are distinguished from background binders [36Ong SE, Blagoev B, Kratchmarova I, et al. Stable isotope labeling by amino acids in cell culture, SILAC., as a simple and accurate approach to expression proteomics Mol Cell Proteomics 2002; 1(5): 376-86.]. Characterization of transcriptional target genes may be achieved with ChIP-Seq and MS-based sequencing, where the histone marks bound by these reader domains should overlap with the target gene mapping [59Vermeulen M, Eberl HC, Matarese F, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers Cell 2010; 142(6): 967-80.].
Two small molecule compounds that disrupt binding of a BRD4 bromodomain to acetylated histones were recently published in parallel, providing an excellent proof of concept for targeting bromodomains [57Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains Nature 2010; 468(7327): 1067-73., 60Nicodeme E, Jeffrey KL, Schaefer U, et al. Suppression of inflammation by a synthetic histone mimic Nature 2010; 468(7327): 1119-23.]. The very similar triazole-diazepine-fused ring compounds, (+)-JQ1 and I-BET, bind stereoselectively to the bromodomain in the acetyllysine binding channel with equilibrium affinities in the range of 50 – 100 nM. Filippakopoulos et al. demonstrated (+)-JQ1-dependent cellular displacement of BRD4 from chromatin and antitumor activity in a mouse xenograft model of midline carcinoma [57Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains Nature 2010; 468(7327): 1067-73.]. Nicodeme et al. investigated the immunomodulatory effects of I-BET and found that it inhibited transcription of inflammatory genes in activated macrophages and attenuated the death of mice induced by septic shock [60Nicodeme E, Jeffrey KL, Schaefer U, et al. Suppression of inflammation by a synthetic histone mimic Nature 2010; 468(7327): 1119-23.]. Despite the remarkable success in developing these small molecule inhibitors of a bromodomain protein-protein interaction, very little is known about the chemical space surrounding other histone code readers, particularly regarding methyllysine or methylarginine marks, and high-throughput assays are needed to identify novel chemical scaffolds.
Several approaches have been taken to identify small molecule disruptors of histone code readers. Fluorescence polarization has been used to measure binding affinities of histone code readers to fluorescein-labeled peptides (Fig. 3A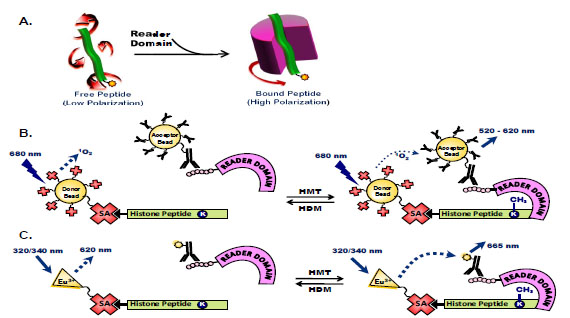 ) [58Liu H, Galka M, Iberg A, et al. Systematic identification of methyllysine-driven interactions for histone and nonhistone targets J Proteome Res 2010; 9(11): 5827-36., 61Fischle W, Tseng BS, Dormann HL, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation Nature 2005; 438(7071): 1116-22., 62Kalakonda N, Fischle W, Boccuni P, et al. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1 Oncogene 2008; 27(31): 4293-304.]. However, these low affinity micromolar interactions make an FP assay format unsuitable for high-throughput identification of small molecule inhibitors of peptide-protein binding. An AlphaScreen assay was developed using biotinylated histone peptides synthesized with an epigenetic mark of interest incorporated to quantify binding to an epitope-tagged reader domain protein (Fig. 3B
) [58Liu H, Galka M, Iberg A, et al. Systematic identification of methyllysine-driven interactions for histone and nonhistone targets J Proteome Res 2010; 9(11): 5827-36., 61Fischle W, Tseng BS, Dormann HL, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation Nature 2005; 438(7071): 1116-22., 62Kalakonda N, Fischle W, Boccuni P, et al. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1 Oncogene 2008; 27(31): 4293-304.]. However, these low affinity micromolar interactions make an FP assay format unsuitable for high-throughput identification of small molecule inhibitors of peptide-protein binding. An AlphaScreen assay was developed using biotinylated histone peptides synthesized with an epigenetic mark of interest incorporated to quantify binding to an epitope-tagged reader domain protein (Fig. 3B ) [63Quinn AM, Bedford MT, Espejo A, et al. A homogeneous method for investigation of methylation-dependent protein-protein interactions in epigenetics Nucleic Acids Res 2010; 38(2): e11., 64Wigle TJ, Herold JM, Senisterra GA, et al. Screening for inhibitors of low-affinity epigenetic peptide-protein interactions: an AlphaScreen-based assay for antagonists of methyl-lysine binding proteins J Biomol Screen 2010; 15(1): 62-71.]. This assay format allows facile miniaturization to 1,536-well plate density and is modular in nature with simple substitution of the reader domain or peptide modification of interest. For example, an AlphaScreen assay measuring binding of the MBT domain protein L3MBTL1 to a monomethylated histone peptide was successfully used to assess inhibitor potency of virtual screening hits identified through pharmacophore modeling and a methyllysine similarity search [65Kireev D, Wigle TJ, Norris-Drouin J, Herold JM, Janzen WP, Frye SV. Identification of non-peptide malignant brain tumor (MBT) repeat antagonists by virtual screening of commercially available compounds J Med Chem 2010; 53(21): 7625-31.]. A similar approach to measuring these peptide/protein interactions may be taken using a TR-FRET readout, for example using europium-labeled streptavidin and an acceptor fluorophore-conjugated antibody raised against the epitope tag of a reader domain fusion protein (Fig. 3C
) [63Quinn AM, Bedford MT, Espejo A, et al. A homogeneous method for investigation of methylation-dependent protein-protein interactions in epigenetics Nucleic Acids Res 2010; 38(2): e11., 64Wigle TJ, Herold JM, Senisterra GA, et al. Screening for inhibitors of low-affinity epigenetic peptide-protein interactions: an AlphaScreen-based assay for antagonists of methyl-lysine binding proteins J Biomol Screen 2010; 15(1): 62-71.]. This assay format allows facile miniaturization to 1,536-well plate density and is modular in nature with simple substitution of the reader domain or peptide modification of interest. For example, an AlphaScreen assay measuring binding of the MBT domain protein L3MBTL1 to a monomethylated histone peptide was successfully used to assess inhibitor potency of virtual screening hits identified through pharmacophore modeling and a methyllysine similarity search [65Kireev D, Wigle TJ, Norris-Drouin J, Herold JM, Janzen WP, Frye SV. Identification of non-peptide malignant brain tumor (MBT) repeat antagonists by virtual screening of commercially available compounds J Med Chem 2010; 53(21): 7625-31.]. A similar approach to measuring these peptide/protein interactions may be taken using a TR-FRET readout, for example using europium-labeled streptavidin and an acceptor fluorophore-conjugated antibody raised against the epitope tag of a reader domain fusion protein (Fig. 3C ) [60Nicodeme E, Jeffrey KL, Schaefer U, et al. Suppression of inflammation by a synthetic histone mimic Nature 2010; 468(7327): 1119-23.].
) [60Nicodeme E, Jeffrey KL, Schaefer U, et al. Suppression of inflammation by a synthetic histone mimic Nature 2010; 468(7327): 1119-23.].
Variations of FRET assays have been used in a cell-based context to measure bromodomain binding to acetylated histone tails [66Kanno T, Kanno Y, Siegel RM, Jang MK, Lenardo MJ, Ozato K. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells Mol Cell 2004; 13(1): 33-43., 67Padilla-Parra S, Auduge N, Coppey-Moisan M, Tramier M. Quantitative FRET analysis by fast acquisition time domain FLIM at high spatial resolution in living cells Biophys J 2008; 95(6): 2976-88.]. Fluorescence recovery after photobleaching (FRAP) assays in living cells have also been used to show binding of chromatin-associated domains to epigenetic marks and a decreased BRD4/chromatin association in the presence of (+)-JQ1 [57Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains Nature 2010; 468(7327): 1067-73.]. While being effective tools for demonstrating a compound’s on-target mode of action, these cellular assays to measure binding of epigenetic readers to chromatin are lacking in the throughput and quantitative nature necessary for identification of novel small molecules to target these protein-protein interactions.
SUMMARY
We have described a number of methods to measure histone methyltransferase and demethylase activity, as well as techniques to quantify binding of chromatin-associated domains to post-translationally modified peptides. While radioactivity-based assays remain the gold standard for evaluating histone-modifying enzymes, a number of robust and sensitive assays have been developed to address the large number of recently-identified histone methyltransferases and demethylases. Despite intense efforts to identify small molecule modulators of these enzymes, few inhibitors have been described. Of these, the majority are non-specific SAM competitors or metal ion chelators of methyltransferase or demethylase enzymes, respectively. The need exists for identification of novel scaffolds as a starting point for drug discovery related to epigenetic therapeutic targets. Robust high-throughput assays are necessary for screening large compound collections to enable detection of these chemical chemotypes and their downstream cellular effects. Continued improvements in assay technologies and novel methodologies will enable understanding of the biological role of histone methylation and our ability to mediate the epigenetic changes that affect human disease.
CONFLICT OF INTEREST
None declared.
ACKNOWLEDGEMENTS
This research was supported in part by the Molecular Libraries Initiative of the NIH Roadmap for Medical Research and the Intramural Research Program of NHGRI, NIH.
ABBREVIATIONS
REFERENCES
| [1] | Jenuwein T, Allis CD. Translating the histone code Science 2001; 293(5532): 1074-80. |
| [2] | Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers Nat Struct Mol Biol 2007; 14(11): 1025-40. |
| [3] | Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation Curr Opin Cell Biol 2001; 13(3): 263-73. |
| [4] | Marks PA, Xu WS. Histone deacetylase inhibitors: potential in cancer therapy J Cell Biochem 2009; 107(4): 600-8. |
| [5] | Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome Cell 2007; 129(4): 823-37. |
| [6] | Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9 Nat Chem Biol 2005; 1(3): 143-5. |
| [7] | Rathert P, Cheng X, Jeltsch A. Continuous enzymatic assay for histone lysine methyltransferases Biotechniques 2007; 43(5): 602-8. |
| [8] | Collazo E, Couture JF, Bulfer S, Trievel RC. A coupled fluorescent assay for histone methyltransferases Anal Biochem 2005; 342(1): 86-92. |
| [9] | Chung CC, Ohwaki K, Schneeweis JE, et al. A fluorescence-based thiol quantification assay for ultra-high-throughput screening for inhibitors of coenzyme A production Assay Drug Dev Technol 2008; 6(3): 361-74. |
| [10] | Hendricks CL, Ross JR, Pichersky E, Noel JP, Zhou ZS. An enzyme-coupled colorimetric assay for S-adenosylmethionine-dependent methyltransferases Anal Biochem 2004; 326(1): 100-5. |
| [11] | Dorgan KM, Wooderchak WL, Wynn DP, et al. An enzyme-coupled continuous spectrophotometric assay for S-adenosylmethionine-dependent methyltransferases Anal Biochem 2006; 350(2): 249-55. |
| [12] | Cheng D, Yadav N, King RW, Swanson MS, Weinstein EJ, Bedford MT. Small molecule regulators of protein arginine methyltransferases J Biol Chem 2004; 279(23): 23892-9. |
| [13] | Kubicek S, O'Sullivan RJ, August EM, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase Mol Cell 2007; 25(3): 473-81. |
| [14] | Spannhoff A, Heinke R, Bauer I, et al. Target-based approach to inhibitors of histone arginine methyltransferases J Med Chem 2007; 50(10): 2319-5. |
| [15] | Quinn AM, Allali-Hassani A, Vedadi M, Simeonov A. A chemiluminescence-based method for identification of histone lysine methyltransferase inhibitors Mol Biosyst 2010; 6(5): 782-8. |
| [16] | Liu F, Chen X, Allali-Hassani A, et al. Discovery of a 2,4-diamino-7-aminoalkoxyquinazoline as a potent and selective inhibitor of histone lysine methyltransferase G9a J Med Chem 2009; 52(24): 7950-3. |
| [17] | Liu F, Chen X, Allali-Hassani A, et al. Protein lysine methyltransferase G9a inhibitors: design, synthesis., and structure activity relationships of 2.,4-diamino-7-aminoalkoxy-quinazolines J Med Chem 2010; 53(15): 5844-7. |
| [18] | Dudek JM, Horton RA. TR-FRET biochemical assays for detecting posttranslational modifications of p53 J Biomol Screen 2010; 15(5): 569-75. |
| [19] | Kawamura A, Tumber A, Rose NR, et al. Development of homogeneous luminescence assays for histone demethylase catalysis and binding Anal Biochem 2010; 404(1): 86-93. |
| [20] | Egelhofer TA, Minoda A, Klugman S, et al. An assessment of histone-modification antibody quality Nat Struct Mol Biol 2010; 18(1): 91-3. |
| [21] | Fuchs SM, Krajewski K, Baker RW, Miller VL, Strahl BD. Influence of combinatorial histone modifications on antibody and effector protein recognition Curr Biol 2011; 21(1): 53-8. |
| [22] | Fanslau C, Pedicord D, Nagulapalli S, et al. An electrophoretic mobility shift assay for the identification and kinetic analysis of acetyl transferase inhibitors Anal Biochem 2010; 402(1): 65-8. |
| [23] | Blackwell L, Norris J, Suto CM, Janzen WP. The use of diversity profiling to characterize chemical modulators of the histone deacetylases Life Sci 2008; 82(21-22): 1050-8. |
| [24] | Wigle TJ, Provencher LM, Norris JL, et al. Accessing protein methyltransferase and demethylase enzymology using microfluidic capillary electrophoresis Chem Biol 2010; 17(7): 695-704. |
| [25] | Eberl HC, Mann M, Vermeulen M. Quantitative proteomics for epigenetics Chembiochem 2011; 12(2): 224-34. |
| [26] | Garcia BA. What does the future hold for top down mass spectrometry? J Am Soc Mass Spectrom 2010; 21(2): 193-202. |
| [27] | Taverna SD, Ueberheide BM, Liu Y, et al. Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini Proc Natl Acad Sci USA 2007; 104(7): 2086-91. |
| [28] | Garcia BA, Thomas CE, Kelleher NL, Mizzen CA. Tissue-specific expression and post-translational modification of histone H3 variants J Proteome Res 2008; 7(10): 4225-36. |
| [29] | Tsukada Y, Fang J, Erdjument-Bromage H, et al. Histone demethylation by a family of JmjC domain-containing proteins Nature 2006; 439(7078): 811-6. |
| [30] | Whetstine JR, Nottke A, Lan F, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases Cell 2006; 125(3): 467-81. |
| [31] | Osborne TC, Obianyo O, Zhang X, Cheng X, Thompson PR. Protein arginine methyltransferase 1: positively charged residues in substrate peptides distal to the site of methylation are important for substrate binding and catalysis Biochemistry 2007; 46(46): 13370-81. |
| [32] | Chang Y, Zhang X, Horton JR, et al. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294 Nat Struct Mol Biol 2009; 16(3): 312-7. |
| [33] | Rose NR, Ng SS, Mecinovic J, et al. Inhibitor scaffolds for 2-oxoglutarate-dependent histone lysine demethylases J Med Chem 2008; 51(22): 7053-6. |
| [34] | Roddy TP, Horvath CR, Stout SJ, et al. Mass spectrometric techniques for label-free high-throughput screening in drug discovery Anal Chem 2007; 79(21): 8207-13. |
| [35] | Rye PT, Frick LE, LaMarr WA, Özbal CC. High-throughput Mass Spectrometric Detection of Histone 3 Demethylation Available from: http://www.biocius.com/pdfs/RapidFireHTMS_EpigeneticTargets_Poster.pdf [cited 2011 Jan]; |
| [36] | Ong SE, Blagoev B, Kratchmarova I, et al. Stable isotope labeling by amino acids in cell culture, SILAC., as a simple and accurate approach to expression proteomics Mol Cell Proteomics 2002; 1(5): 376-86. |
| [37] | Bantscheff M, Dumpelfeld B, Kuster B. Femtomol sensitivity post-digest (18)O labeling for relative quantification of differential protein complex composition Rapid Commun Mass Spectrom 2004; 18(8): 869-76. |
| [38] | Ross PL, Huang YN, Marchese JN, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents Mol Cell Proteomics 2004; 3(12): 1154-69. |
| [39] | Smith CM, Gafken PR, Zhang Z, Gottschling DE, Smith JB, Smith DL. Mass spectrometric quantification of acetylation at specific lysines within the amino-terminal tail of histone H4 Anal Biochem 2003; 316(1): 23-33. |
| [40] | Ong SE, Mittler G, Mann M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC Nat Methods 2004; 1(2): 119-26. |
| [41] | Feng Y, Xie N, Wu J, Yang C, Zheng YG. Inhibitory study of protein arginine methyltransferase 1 using a fluorescent approach Biochem Biophys Res Commun 2009; 379(2): 567-72. |
| [42] | Feng Y, Li M, Wang B, Zheng YG. Discovery and mechanistic study of a class of protein arginine methylation inhibitors J Med Chem 2010; 53(16): 6028-39. |
| [43] | King ON, Li XS, Sakurai M, et al. Quantitative high-throughput screening identifies 8-hydroxyquinolines as cell-active histone demethylase inhibitors PLoS One 2010; 5(11): e15535. |
| [44] | Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease Nat Rev Genet 2007; 8(11): 829-33. |
| [45] | Sakurai M, Rose NR, Schultz L, et al. A miniaturized screen for inhibitors of Jumonji histone demethylases Mol Biosyst 2010; 6(2): 357-64. |
| [46] | Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase Nat Struct Mol Biol 2007; 14(8): 689-95. |
| [47] | Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1 Cell 2004; 119(7): 941-53. |
| [48] | Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications Chem Biol 2006; 13(6): 563-7. |
| [49] | Karytinos A, Forneris F, Profumo A, et al. A novel mammalian flavin-dependent histone demethylase J Biol Chem 2009; 284(26): 17775-82. |
| [50] | Culhane JC, Szewczuk LM, Liu X, Da G, Marmorstein R, Cole PA. A mechanism-based inactivator for histone demethylase LSD1 J Am Chem Soc 2006; 128(14): 4536-7. |
| [51] | Binda C, Valente S, Romanenghi M, et al. Biochemical, structural., and biological evaluation of tranylcypromine derivatives as inhibitors of histone demethylases LSD1 and LSD2 J Am Chem Soc 2010; 132(19): 6827-33. |
| [52] | Culhane JC, Wang D, Yen PM, Cole PA. Comparative analysis of small molecules and histone substrate analogues as LSD1 lysine demethylase inhibitors J Am Chem Soc 2010; 132(9): 3164-76. |
| [53] | Huang Y, Stewart TM, Wu Y, et al. Novel oligoamine analogues inhibit lysine-specific demethylase 1 and induce reexpression of epigenetically silenced genes Clin Cancer Res 2009; 15(23): 7217-28. |
| [54] | Huang Y, Greene E, Murray ST, et al. Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes Proc Natl Acad Sci USA 2007; 104(19): 8023-. |
| [55] | Zhao Q, Rank G, Tan YT, et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing Nat Struct Mol Biol 2009; 16(3): 304-11. |
| [56] | Kim J, Daniel J, Espejo A, et al. Tudor, MBT and chromo domains gauge the degree of lysine methylation EMBO Rep 2006; 7(4): 397-403. |
| [57] | Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains Nature 2010; 468(7327): 1067-73. |
| [58] | Liu H, Galka M, Iberg A, et al. Systematic identification of methyllysine-driven interactions for histone and nonhistone targets J Proteome Res 2010; 9(11): 5827-36. |
| [59] | Vermeulen M, Eberl HC, Matarese F, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers Cell 2010; 142(6): 967-80. |
| [60] | Nicodeme E, Jeffrey KL, Schaefer U, et al. Suppression of inflammation by a synthetic histone mimic Nature 2010; 468(7327): 1119-23. |
| [61] | Fischle W, Tseng BS, Dormann HL, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation Nature 2005; 438(7071): 1116-22. |
| [62] | Kalakonda N, Fischle W, Boccuni P, et al. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1 Oncogene 2008; 27(31): 4293-304. |
| [63] | Quinn AM, Bedford MT, Espejo A, et al. A homogeneous method for investigation of methylation-dependent protein-protein interactions in epigenetics Nucleic Acids Res 2010; 38(2): e11. |
| [64] | Wigle TJ, Herold JM, Senisterra GA, et al. Screening for inhibitors of low-affinity epigenetic peptide-protein interactions: an AlphaScreen-based assay for antagonists of methyl-lysine binding proteins J Biomol Screen 2010; 15(1): 62-71. |
| [65] | Kireev D, Wigle TJ, Norris-Drouin J, Herold JM, Janzen WP, Frye SV. Identification of non-peptide malignant brain tumor (MBT) repeat antagonists by virtual screening of commercially available compounds J Med Chem 2010; 53(21): 7625-31. |
| [66] | Kanno T, Kanno Y, Siegel RM, Jang MK, Lenardo MJ, Ozato K. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells Mol Cell 2004; 13(1): 33-43. |
| [67] | Padilla-Parra S, Auduge N, Coppey-Moisan M, Tramier M. Quantitative FRET analysis by fast acquisition time domain FLIM at high spatial resolution in living cells Biophys J 2008; 95(6): 2976-88. |
| [68] | Purandare AV, Chen Z, Huynh T, et al. Pyrazole inhibitors of coactivator associated arginine methyltransferase 1 (CARM1) Bioorg Med Chem Lett 2008; 18(15): 4438-1. |
| [69] | Vedel M, Lawrence F, Robert-Gero M, Lederer E. The antifungal antibiotic sinefungin as a very active inhibitor of methyltransferases and of the transformation of chick embryo fibroblasts by Rous sarcoma virus Biochem Biophys Res Commun 1978; 85(1): 371-6. |
| [70] | Sekirnik R, Rose NR, Thalhammer A, Seden PT, Mecinovic J, Schofield CJ. Inhibition of the histone lysine demethylase JMJD2A by ejection of structural Zn(II) Chem Commun (Camb) 2009; 14(42): 6376-8. |
| [71] | Spannhoff A, Machmur R, Heinke R, et al. A novel arginine methyltransferase inhibitor with cellular activity Bioorg Med Chem Lett 2007; 17(15): 4150-3. |
| [72] | Chang Y, Ganesh T, Horton JR, et al. Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases J Mol Biol 2010; 400(1): 1-7. |
| [73] | Ueda R, Suzuki T, Mino K, et al. Identification of cell-active lysine specific demethylase 1-selective inhibitors J Am Chem Soc 2009; 131(48): 17536-7. |
| [74] | Hamada S, Kim TD, Suzuki T, et al. Synthesis and activity of N-oxalylglycine and its derivatives as Jumonji C-domain-containing histone lysine demethylase inhibitors Bioorg Med Chem Lett 2009; 19(10): 2852-5. |
| [75] | Rose NR, Woon EC, Kingham GL, et al. Selective inhibitors of the JMJD2 histone demethylases: combined nondenaturing mass spectrometric screening and crystallographic approaches J Med Chem 2010; 53(4): 1810-8. |
| [76] | Cloos PA, Christensen J, Agger K, et al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3 Nature 2006; 442(7100): 307-11. |
| [77] | Smith EH, Janknecht R, Maher LJ 3rd. Succinate inhibition of alpha-ketoglutarate-dependent enzymes in a yeast model of paraganglioma Hum Mol Genet 2007; 16(24): 3136-48. |






