- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

Current Chemical Genomics and Translational Medicine
(Discontinued)
ISSN: 2213-9885 ― Volume 12, 2018
The HaloTag: Improving Soluble Expression and Applications in Protein Functional Analysis
Scott N Peterson , Keehwan Kwon*
Abstract
Technological and methodological advances have been critical for the rapidly evolving field of proteomics. The development of fusion tag systems is essential for purification and analysis of recombinant proteins. The HaloTag is a 34 KDa monomeric protein derived from a bacterial haloalkane dehalogenase. The majority of fusion tags in use today utilize a reversible binding interaction with a specific ligand. The HaloTag system is unique in that it forms a covalent linkage to its chloroalkane ligand. This linkage permits attachment of the HaloTag to a variety of functional reporters, which can be used to label and immobilize recombinant proteins. The success rate for HaloTag expression of soluble proteins is very high and comparable to maltose binding protein (MBP) tag. Furthermore, cleavage of the HaloTag does not result in protein insolubility that often is observed with the MBP tag. In the present report, we describe applications of the HaloTag system in our ongoing investigation of protein-protein interactions of the Y. pestis Type 3 secretion system on a custom protein microarray. We also describe the utilization of affinity purification/mass spectroscopy (AP/MS) to evaluate the utility of the Halo Tag system to characterize DNA binding activity and protein specificity.
Article Information
Identifiers and Pagination:
Year: 2012Volume: 6
Issue: Suppl 1
First Page: 8
Last Page: 17
Publisher Id: CCGTM-6-8
DOI: 10.2174/1875397301206010008
Article History:
Received Date: 17/5/2011Revision Received Date: 13/7/2012
Acceptance Date: 18/7/2012
Electronic publication date: 20/9/2012
Collection year: 2012
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the J. Craig Venter Institute, 9704 Medical Center Drive, Rockville, Maryland 20850: Tel: 301-795-7647; Fax: (301)294-3142; E-mail: kkwon@jcvi.org
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 17-5-2011 |
Original Manuscript | The HaloTag: Improving Soluble Expression and Applications in Protein Functional Analysis | |
INTRODUCTION
Advances in DNA sequencing technology have increased sharply over the past 15 years [1Berglund EC, Kiialainen A, Syvanen AC. Next-generation sequencing technologies and applications for human genetic history and forensics Investig Genet 2011; 2: 23.]. These advances have enabled the sequencing of many large and small genomes, resulting in over 3,000 bacterial genomes including ~150 archaea and nearly 200 eukaryotic and mammalian genome sequences (http://www.ncbi.nlm.nih.gov/sites/genome) to be completed. The access to this massive quantity of data has had a strong ripple effect leading to an increased demand for new technologies that will enable scientists to study the activities and functions of these gene sequences in a high throughput manner. Among the numerous discoveries enabled by genome sequence data, one somewhat unanticipated finding relates to the fact that at least one-third of the open reading frames (ORFs) encoded in genomes has no predicted function based on BLAST analysis [2Siddaramappa S, Challacombe JF, De Castro RE, et al. A comparative genomics perspective on the genetic content of the alkaliphilic haloarchaeon Natrialba magadii ATCC 43099T BMC Genomics 2012; 13(1 ): 165.-4Clamp M, Fry B, Kamal M, et al. Distinguishing protein-coding and noncoding genes in the human genome Proc Natl Acad Sci USA 2007; 104(49 ): 19428-33.]. Interestingly, the number of genes of unknown function increases in a linear manner as we sequence additional genomes [5Raskin DM, Seshadri R, Pukatzki SU, Mekalanos JJ. Bacterial genomics and pathogen evolution Cell 2006; 124(4 ): 703-14.]. One might imagine that as we sequence more genomes, the rate that novel genes are identified would begin to decrease rapidly. This is clearly not the case though and strongly support the view that the number of unique gene sequences and functions encoded on our planet is very large. For most microbial species, 10-30% or more of the ORFs encoded in one strain’s genome are novel compared to another strain belonging to the same species. The gene pool of many bacterial species may exceed several tens of thousands of unique genes. It is likely that by the end of this decade, we will have sequenced over 10 million genes of unknown function!
This humbling realization emphasizes the need for substantial improvements in the area of functional genomics if we are to keep pace with the ever-increasing ease that genes and genomes are sequenced. One phenomenon that have been documented, referred to as non-orthologous gene displacement (NODs) may provide an inroad to tackling the monumental problem of determining the function of uncharacterized genes. NODs represent cases where two proteins perform the same cellular function but do not possess an ancestral relationship. We know of several cases like eukaryotic and prokaryotic DNA polymerases that essentially carry out the same cellular functions, but do not share common ancestral relationships. In other words these functions evolved independently during evolution. The vast majority of the assigned functions of genes are based on BLAST and orthology (conservation of DNA or amino acid sequence). If genes arise independently they by definition do not share ancestry nor do they share amino acid sequence identity. The scientific research community has developed strategies to assay a wide range of known protein functions over the years, it may follow that the screening of novel proteins of unknown function using familiar assay systems will yield a surprising number of experimentally determined gene functions. While this explanation may partially explain the reason we are accumulating more and more genes of unknown function in our databases, we remain highly ignorant as to the frequency of NODs in nature.
Massively parallel technologies have been developed, such as microfluidics and DNA and protein microarrays, which present important vehicles to partially enable the large-scale characterization of gene/protein function [6Fordyce PM, Gerber D, Tran D, et al. De novo identification and biophysical characterization of transcription-factor binding sites with microfluidic affinity analysis Nat Biotechnol 2010; 28(9 ): 970-5.-12Zhang L, Villa NY, Rahman MM, et al. Analysis of vaccinia virus-host protein-protein interactions: validations of yeast two-hybrid screenings J Proteome Res 2009; 8(9 ): 4311-8.]. Our ability to determine the function of genes places strong demands on a variety of disciplines related to recombinant protein technologies. The large-scale characterization of protein function requires very efficient recombinant proteins production in a high-throughput environment and the necessary automation to perform high-throughput functional screens [13Lesley SA. High-throughput proteomics: protein expression and purification in the postgenomic world Protein Expr Purif 2001; 22(2 ): 159-64., 14Kwon K, Pieper R, Shallom S, et al. A correlation analysis of protein characteristics associated with genome-wide high throughput expression and solubility of Streptococcus pneumoniae proteins Protein Expr Purif 2007; 55(2 ): 368-78.]. Likewise, complementary technologies that broaden the use of recombinant proteins such as labeling methods, sub-cellular localization determination, enzymatic activity and substrate specificity will also need to be developed and advanced if we are to make significant progress.
Among the numerous challenges associated with large-scale functional characterization of proteins is the choice of expression systems that are to be employed. Given the fact that several systems offer some discrete advantage, in an ideal world, one would employ many platforms. For practical reasons researchers are forced to make difficult decisions regarding which platform provides the greatest overall utility for the objectives in question. Among the variety of tools being developed that show promise of enabling the functional characterization of protein function, the HaloTag technology developed by scientists at Promega (Madison, WI) is notable [15Los GV, Encell LP, McDougall MG, et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis ACS Chem Biol 2008; 3(6 ): 373-82., 16Ohana RF, Encell LP, Zhao K, et al. HaloTag7: a genetically engineered tag that enhances bacterial expression of soluble proteins and improves protein purification Protein Expr Purif 2009; 68(1 ): 110-20.]. Here we provide an overview of functional assays and experience we have developed in conjunction with the HaloTag technology.
We have used the HaloTag technology for a number of functional studies, including protein microarrays, affinity purification of DNA-protein, protein-protein interactions, and protein complex identification [7Hurst R, Hook B, Slater MR, Hartnett J, Storts DR, Nath N. Protein-protein interaction studies on protein arrays: effect of detection strategies on signal-to-background ratios Anal Biochem 2009; 392(1 ): 45-53., 17Gallo S, Beugnet A, Biffo S. Tagging of functional ribosomes in living cells by HaloTag(R) technology In Vitro Cell Dev Biol Anim 2011; 47(2 ): 132-8.]. The HaloTag is a modified haloalkane dehalogenase designed to covalently bind a series of chloroalkane derivatives such as fluorophore-labeled ligands (Promega). We have observed improved solubility of fusion proteins using this system, comparable to that achieved by the best solubilization fusion partner, the maltose-binding protein MBP [18Kwon K, Hasseman J, Latham S, et al. Recombinant expression and functional analysis of proteases from Streptococcus pneumoniae, Bacillus anthracis, and Yersinia pestis BMC Biochem 2011; 12: 17.]. The HaloTag vector (Promega) adopted a Flexi cloning system that uses traditional restriction site cloning methods. We found this cloning method to be inadequate for high-throughput cloning of genes, and have adapted the cloning platform for compatibility with Gateway and Ligation Independent Cloning (LIC) procedures [19Walhout AJ, Temple GF, Brasch MA, et al. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes Methods Enzymol 2000; 328: 575-92.-22Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res 1990; 18(20 ): 6069-74.]. We have used these vectors in a number of studies including the expression and purification of proteins derived from Influenza virus H1N1, Y. pestis, S. pneumoniae and B. mallei. Genes were expressed using several expression systems including E. coli, a cell-free (wheat germ) system and mammalian cells. The HaloTag supports development of functional assays, such as fluorescence polarization, FRET, on-chip purification in protein microarrays and also allows monitoring sub-cellular protein localization. The rapid covalent attachment of the HaloTag to its specific ligand is a critical feature that separates the HaloTag from any other tags that use reversible interactions [23Ohana RF, Hurst R, Vidugiriene J, Slater MR, Wood KV, Urh M. HaloTag-based purification of functional human kinases from mammalian cells Protein Expr Purif 2011; 76(2 ): 154-64.]. The high affinity covalent interaction is extremely rapid and allows binding reactions to be carried out in minutes. This has proven advantageous in that we observe a dramatic reduction in the background, non-specific binding events that reduce signal to noise assay ratios [16Ohana RF, Encell LP, Zhao K, et al. HaloTag7: a genetically engineered tag that enhances bacterial expression of soluble proteins and improves protein purification Protein Expr Purif 2009; 68(1 ): 110-20., 24Los GV, Wood K. The HaloTag: a novel technology for cell imaging and protein analysis Methods Mol Biol 2007; 356: 195-208.].
HISTORICAL DEVELOPMENT OF HALOTAG
The development of the HaloTag is the result of rational engineering of a bacterially encoded haloalkane dehydrogenase (DhaA) derived from a Rhodococcus spp [25Janssen DB. Evolving haloalkane dehalogenases Curr Opin Chem Biol 2004; 8(2 ): 150-9.], carried out in the laboratories at Promega [15Los GV, Encell LP, McDougall MG, et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis ACS Chem Biol 2008; 3(6 ): 373-82., 16Ohana RF, Encell LP, Zhao K, et al. HaloTag7: a genetically engineered tag that enhances bacterial expression of soluble proteins and improves protein purification Protein Expr Purif 2009; 68(1 ): 110-20., 24Los GV, Wood K. The HaloTag: a novel technology for cell imaging and protein analysis Methods Mol Biol 2007; 356: 195-208.]. The occurrence of this enzyme is phylogenetically restricted to a small number of taxa. The 34 kDa protein cleaves at the carbon-halogen bond of a number of aliphatic halogenated compounds through a mechanism involving a hydrolytic triad within the active site of the enzyme. During the carbon-halogen cleavage reaction, the enzyme forms a transient covalent complex with its substrate, leading to the nucleophilic displacement of the terminal halogen using Asp106. The complex is hydrolyzed in a reaction involving His273 through the activation of a water molecule. In order to stabilize this intermediate, the His273 residue was replaced with a Phe residue that occupies a similar volume in space but does not have the potential as a base to carry out the hydrolysis reaction. Therefore, the covalently linked substrate remains trapped in the active site of the enzyme. Mutagenesis of some residues was made to increase the accessibility of the ligand for the active site and for others to enhance solubility and additional characteristics in the final HaloTag protein. These efforts have resulted in the provision of a novel and robust system for conducting recombinant protein studies in a wide variety of formats.
POTENTIAL ADVANTAGES OF COVALENT LINKAGE
Research objectives focused on high throughput functional characterization of proteins have led to the development of a variety of novel methodological strategies and technologies. Many of these strategies rely on the immobilization of recombinant proteins to matrices with a very large surface area [9Phizicky E, Bastiaens PI, Zhu H, Snyder M, Fields S. Protein analysis on a proteomic scale Nature 2003; 422(6928 ): 208-15., 11Zhu H, Bilgin M, Bangham R, et al. Global analysis of protein activities using proteome chips Science 2001; 293(5537 ): 2101-5., 26Ramachandran N, Hainsworth E, Bhullar B, et al. Self-assembling protein microarrays Science 2004; 305(5680 ): 86-90.-31Labrou NE. Design and selection of ligands for affinity chromatography J Chromatogr B Analyt Technol Biomed Life Sci 2003; 790(1-2 ): 67-78.]. In this regard many of the biochemistry or physical interaction studies being carried out are associated with unique challenges presented by large-scale screening and the immobilization to solid substrates that in some cases may generate significant non-specific binding and high levels of background in the assays performed. The HaloTag technology offers some discrete and potentially important advantages to address these two issues based on the covalent and very high affinity interaction between the HaloTag and its ligand [15Los GV, Encell LP, McDougall MG, et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis ACS Chem Biol 2008; 3(6 ): 373-82., 16Ohana RF, Encell LP, Zhao K, et al. HaloTag7: a genetically engineered tag that enhances bacterial expression of soluble proteins and improves protein purification Protein Expr Purif 2009; 68(1 ): 110-20., 32Urh M, Simpson D, Zhao K. Affinity chromatography: general methods Methods Enzymol 2009; 463: 417-38.]. The covalent linkage of the HaloTag to immobilized surfaces ensures that high stringency washes may be performed without concern of removing the immobilized proteins [33Urh M, Hartzell D, Mendez J, Klaubert DH, Wood K. Methods for detection of protein-protein and protein-DNA interactions using HaloTag Methods Mol Biol 2008; 421: 191-209.]. Perhaps equally important is the high affinity interaction of the HaloTag and its ligand. The on rate of the interaction at typical protein ligand concentrations drives the reaction to near completion very rapidly. In this regard, the functional assays performed with HaloTag recombinant proteins can be conducted in a reduced time frame; thereby decreasing the mass-action, non-specific background signals that may be facilitated by longer incubation times.
ADAPTATION OF HALOTAG TO GATEWAY EXPRESSION VECTORS
One of the essential elements for high-throughput protein production and functional screening is the selection of an expression vector with a specific fusion tag. The trends in high-throughput recombinant protein expression indicate that no single expression system is ideal for all target proteins. Therefore, many expression pipelines include multiple expression vectors which are used in parallel to increase the overall success rate of recovering soluble proteins. However, in order to use multiple expression vectors, efficient cloning methods such as the Gateway recombination cloning method are required [19Walhout AJ, Temple GF, Brasch MA, et al. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes Methods Enzymol 2000; 328: 575-92., 20Reboul J, Vaglio P, Tzellas N, et al. Open-reading-frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans Nat Genet 2001; 27(3 ): 332-6.]. Although the use of multiple expression vectors increases the number of recovered soluble target proteins, for practical purposes, the, use of expression vectors is often limited to one or a few vectors in most high throughput gene cloning pipelines. Therefore, an ideal expression vector possesses excellent fusion tag properties (solubility and purification efficiency) and a high throughput cloning procedure amenable to automation. We have attempted to strike this ideal by constructing a series of expression vectors that merge the qualities associated with the HaloTag to the ease and efficiency associated with the either LIC or Gateway cloning methods. The Gateway compatible expression vector has the added advantage that it allows investigators to utilize existing entry clone sets which have been produced and made available through public repositories (www.beiresources.org) [14Kwon K, Pieper R, Shallom S, et al. A correlation analysis of protein characteristics associated with genome-wide high throughput expression and solubility of Streptococcus pneumoniae proteins Protein Expr Purif 2007; 55(2 ): 368-78., 18Kwon K, Hasseman J, Latham S, et al. Recombinant expression and functional analysis of proteases from Streptococcus pneumoniae, Bacillus anthracis, and Yersinia pestis BMC Biochem 2011; 12: 17.]. We have evaluated the outcomes of a number of protein expression trials using these chimeric vectors.
The vectors, pFN18A, pFN19A, pFC20A and pFC14A were obtained from Promega for expression of various target proteins in E. coli, cell-free lysates and mammalian expression systems (Fig. 1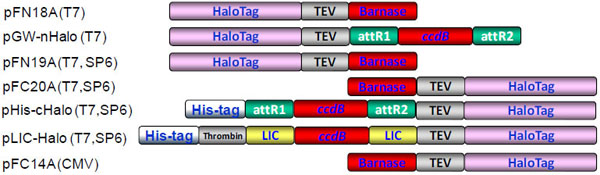 ). We modified these vectors in a variety of ways. Each of the modified vectors contains the E. coliccdB cassette which encodes a product that is toxic to E. coli [34Van Reeth T, Dreze PL, Szpirer J, Szpirer C, Gabant P. Positive selection vectors to generate fused genes for the expression of his-tagged proteins Biotechniques 1998; 25(5
): 898-904.]. We adapted the Gateway cloning method to prepare clones which were easier to use than existing entry clones. The expression vector, pGW-nHalo, is based on the vector pFN18A which replaced the barnase with the attR recombination cloning sites and ccdB cassette. We also constructed pHis-cHalo another Gateway compatible vector based on pFN20A and T02 (pHis) vectors [14Kwon K, Pieper R, Shallom S, et al. A correlation analysis of protein characteristics associated with genome-wide high throughput expression and solubility of Streptococcus pneumoniae proteins Protein Expr Purif 2007; 55(2
): 368-78.] that contains an N-terminal His-tag and a C-terminal HaloTag. We also constructed a ligation independent cloning vector with a C-terminal HaloTag (pLIC-Halo) based on the pMCSG7 vector backbone [35Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site Protein Expr Purif 2002; 25(1
): 8-15.] and consists of an N-terminal His-tag and a C-terminal HaloTag. The His-tag can be removed by thrombin cleavage after purification [21Haun RS, Moss J. Ligation-independent cloning of glutathione S-transferase fusion genes for expression in Escherichia coli Gene 1992; 112(1
): 37-43., 22Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res 1990; 18(20
): 6069-74.]. The addition of the His-tag in the vectors enables the use of the His-tag for purification, when down-stream applications of the purified protein require the HaloTag for fluorophore labeling.
). We modified these vectors in a variety of ways. Each of the modified vectors contains the E. coliccdB cassette which encodes a product that is toxic to E. coli [34Van Reeth T, Dreze PL, Szpirer J, Szpirer C, Gabant P. Positive selection vectors to generate fused genes for the expression of his-tagged proteins Biotechniques 1998; 25(5
): 898-904.]. We adapted the Gateway cloning method to prepare clones which were easier to use than existing entry clones. The expression vector, pGW-nHalo, is based on the vector pFN18A which replaced the barnase with the attR recombination cloning sites and ccdB cassette. We also constructed pHis-cHalo another Gateway compatible vector based on pFN20A and T02 (pHis) vectors [14Kwon K, Pieper R, Shallom S, et al. A correlation analysis of protein characteristics associated with genome-wide high throughput expression and solubility of Streptococcus pneumoniae proteins Protein Expr Purif 2007; 55(2
): 368-78.] that contains an N-terminal His-tag and a C-terminal HaloTag. We also constructed a ligation independent cloning vector with a C-terminal HaloTag (pLIC-Halo) based on the pMCSG7 vector backbone [35Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site Protein Expr Purif 2002; 25(1
): 8-15.] and consists of an N-terminal His-tag and a C-terminal HaloTag. The His-tag can be removed by thrombin cleavage after purification [21Haun RS, Moss J. Ligation-independent cloning of glutathione S-transferase fusion genes for expression in Escherichia coli Gene 1992; 112(1
): 37-43., 22Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res 1990; 18(20
): 6069-74.]. The addition of the His-tag in the vectors enables the use of the His-tag for purification, when down-stream applications of the purified protein require the HaloTag for fluorophore labeling.
 |
Fig. (1) The HaloTag expression vectors used for protein expression and functional studies Various expression vector systems were used for HaloTag recombinant protein expression. The vectors, pFN18A, pFN19A, pFC20A and pFC14A were obtained from Promega for expression of target proteins in E. coli, cell-free and mammalian expression systems. In order to modify applicable cloning methods, we further modified these vectors to contain the ccdB cassette for positive selection of cloned plasmids. The expression vector, pGW-nHalo is based on pFN18A and ccdB cassette was incorporated into the vector. The pHis-cHalo was based on pFN20A and T02 (pHis) vector [14Kwon K, Pieper R, Shallom S, et al. A correlation analysis of protein characteristics associated with genome-wide high throughput expression and solubility of Streptococcus pneumoniae proteins Protein Expr Purif 2007; 55(2 ): 368-78.]. The expression vector, pLIC-Halo was also based on pFC20A and LIC cloning site was incorporated with ccdB cassette. These vectors were used for expression and solubility studies of proteins in S. pneumoniae TIGR4, Y. pestis KIM 10, B. mallei ATCC 23344 and H1N, and for functional analysis. |
COMPARISON OF EXPRESSION VECTORS
Success rates in recovering solubly expressed target proteins using the various HaloTag vectors (Fig. 1 ) were evaluated in E. coli, cell-free expression system and mammalian cells and compared with previous expression studies that employed fusion proteins such as: His-tag, MBP, DsbA and GST (Table 1 and Supplementary Table 1) [14Kwon K, Pieper R, Shallom S, et al. A correlation analysis of protein characteristics associated with genome-wide high throughput expression and solubility of Streptococcus pneumoniae proteins Protein Expr Purif 2007; 55(2
): 368-78., 18Kwon K, Hasseman J, Latham S, et al. Recombinant expression and functional analysis of proteases from Streptococcus pneumoniae, Bacillus anthracis, and Yersinia pestis BMC Biochem 2011; 12: 17.]. As depicted in Fig. (1
) were evaluated in E. coli, cell-free expression system and mammalian cells and compared with previous expression studies that employed fusion proteins such as: His-tag, MBP, DsbA and GST (Table 1 and Supplementary Table 1) [14Kwon K, Pieper R, Shallom S, et al. A correlation analysis of protein characteristics associated with genome-wide high throughput expression and solubility of Streptococcus pneumoniae proteins Protein Expr Purif 2007; 55(2
): 368-78., 18Kwon K, Hasseman J, Latham S, et al. Recombinant expression and functional analysis of proteases from Streptococcus pneumoniae, Bacillus anthracis, and Yersinia pestis BMC Biochem 2011; 12: 17.]. As depicted in Fig. (1 ), each HaloTag vector has specific characteristics such as the location of the HaloTag, drug resistance markers and cloning strategies. Four of those vectors, pFN19A, pFC20A, pHis-cHalo and pLIC-Halo all contain dual promoters, T7 and SP6, which express proteins in either E. coli or wheat germ in vitro expression systems. As a contrast, vectors, pFN18A and pGW-nHalo, allow the expression of proteins in E. coli expression system with the T7 promoter alone.
), each HaloTag vector has specific characteristics such as the location of the HaloTag, drug resistance markers and cloning strategies. Four of those vectors, pFN19A, pFC20A, pHis-cHalo and pLIC-Halo all contain dual promoters, T7 and SP6, which express proteins in either E. coli or wheat germ in vitro expression systems. As a contrast, vectors, pFN18A and pGW-nHalo, allow the expression of proteins in E. coli expression system with the T7 promoter alone.
The His-tag expression vector, T02 (pHis) yielded soluble proteins in 43.2 % of attempts when targeting the complete set of ORFs encoded in S. pneumoniae TIGR4 [14Kwon K, Pieper R, Shallom S, et al. A correlation analysis of protein characteristics associated with genome-wide high throughput expression and solubility of Streptococcus pneumoniae proteins Protein Expr Purif 2007; 55(2 ): 368-78.]. A second study focused on expression of proteases resulted in similar outcomes with 39.6% success [18Kwon K, Hasseman J, Latham S, et al. Recombinant expression and functional analysis of proteases from Streptococcus pneumoniae, Bacillus anthracis, and Yersinia pestis BMC Biochem 2011; 12: 17.]. The success frequencies were below 50% for each of the vectors tested in these studies except cases employing the MBP-tag or the HaloTag. The pMBP produced soluble proteins for more than 70% of target proteins. Both the pFN19A, and the pGW-nHalo, which are N-terminal HaloTag vectors, produced soluble proteins in E. coli at very similar frequencies. Our efforts pertaining to the construction of a vector (pHis-cHalo) containing the Gateway attR cloning sites and a C-terminal HaloTag was not generally useful for protein expression for reasons that remain unclear, while pGW-nHalo, Gateway compatible vector with an N-terminal HaloTag, displayed excellent expression and solubility of target proteins, similar to outcomes obtained with pFN19A that also contains an N-terminal HaloTag. Influenza virus (H1N1) proteins were expressed using pFC14A, which contains the CMV promoter and a C-terminal HaloTag, and 8 proteins from this virus were well expressed in HEK293T. These same proteins were expressed in truncated form when using the E. coli expression system. Although target proteins for the expression attempts are not identical and therefore not directly comparable, the proteins in attempts using HaloTag vectors contain a randomly selected set and difficult membrane localized protein sets such as type III and type VI secretion systems. Overall, the body of experience using HaloTag is now large enough to enable comparison to overall outcomes associated with other vector systems and conclude that the HaloTag enhances expression and solubility of target proteins to levels comparable to that of the previously defined “best” solubilization tag, MBP [36Shih YP, Kung WM, Chen JC, Yeh CH, Wang AH, Wang TF. High-throughput screening of soluble recombinant proteins Protein Sci 2002; 11(7 ): 1714-9., 37Hammarstrom M, Hellgren N, van Den Berg S, Berglund H, Hard T. Rapid screening for improved solubility of small human proteins produced as fusion proteins in Escherichia coli Protein Sci 2002; 11(2 ): 313-21.].
INCREASE SUCCESS RATES OF SOLUBLE EXPRESSION
In order to characterize proteins of interest, soluble expression and purification of proteins are essential. Here, we describe two strategies we employed to increase the success rate of soluble expression/purification of proteins of interest. First, a complementary pair of expression vectors containing the same fusion tag (C-terminal and N-terminal) increases the overall recovery of soluble proteins. We have used the expression vectors, pFN19A and pFC20A for this purpose to express a group of E. coli proteins (Fig. 2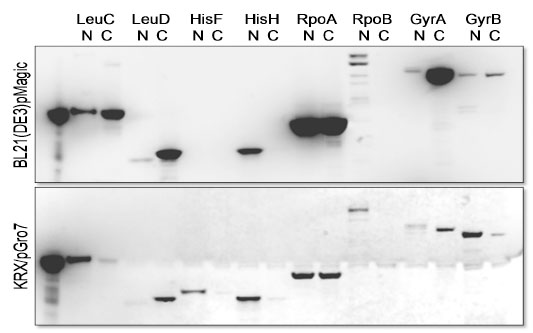 ). Second, we evaluated the success rate of traditional column-based purification procedures to in situ purification and determined that the latter increased overall success and yield of purified proteins (Fig. 3
). Second, we evaluated the success rate of traditional column-based purification procedures to in situ purification and determined that the latter increased overall success and yield of purified proteins (Fig. 3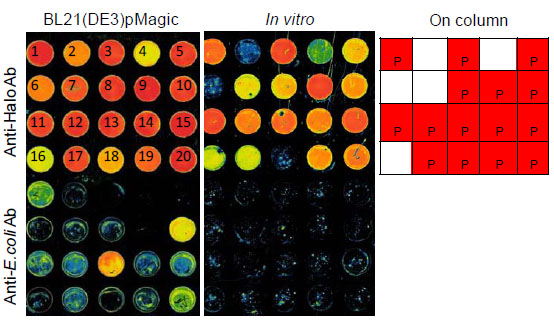 ).
).
 |
Fig. (2) Expression of E. coli proteins for protein-protein interactions. The HaloTag recombinant proteins were visualized with TMR ligand. |
We used pFN19A (N-terminal HaloTag) and pFC20A (C-terminal HaloTag) to increase the overall recovery of soluble proteins of E. coli proteins of interest, LeuC, LeuD, HisF, HisH, RpoA, RpoB, GyrA and GyrB. For these studies we used two E. coli expression strains to enhance the recovery of soluble proteins. BL21(DE3)/pMagic, an E. coli B strain derivative containing the pMagic plasmid that encodes tRNAs that are rare in E. coli and KRX/pGro7, a K-12 derivative containing a plasmid expressing the chaperone complex, GroEL/ES [38Nishihara K, Kanemori M, Kitagawa M, Yanagi H, Yura T. Chaperone coexpression plasmids: differential and synergistic roles of DnaK-DnaJ-GrpE and GroEL-GroES in assisting folding of an allergen of Japanese cedar pollen, Cryj2, in Escherichia coli Appl Environ Microbiol 1998; 64(5
): 1694-9.]. The use of pFN19A and pFC20A vectors displayed similar outcomes in most cases but also displayed complementary outcomes in several instances as shown in Fig. (2 ). For example, LeuD and GyrA displayed higher soluble expression using pFC20A while almost no soluble protein was recovered with pFN19A. In contrast, HisF, HisH and RpoB were recovered as soluble proteins only pFN19A. Similarly, HisF and GyrB were expressed in soluble form at higher levels in vector pFN19A in KRX/pGro7 while soluble LeuC was expressed at higher levels using BL21(DE3)/pMagic. Soluble HisF was obtained solely with N-terminal HaloTag vector in KRX/pGro7. The combination of expression vectors, pF19A and pFC20A and two expression strains allowed the recovery of all targets in soluble form with adequate yield and purity.
). For example, LeuD and GyrA displayed higher soluble expression using pFC20A while almost no soluble protein was recovered with pFN19A. In contrast, HisF, HisH and RpoB were recovered as soluble proteins only pFN19A. Similarly, HisF and GyrB were expressed in soluble form at higher levels in vector pFN19A in KRX/pGro7 while soluble LeuC was expressed at higher levels using BL21(DE3)/pMagic. Soluble HisF was obtained solely with N-terminal HaloTag vector in KRX/pGro7. The combination of expression vectors, pF19A and pFC20A and two expression strains allowed the recovery of all targets in soluble form with adequate yield and purity.
As part of our ongoing efforts to compare a variety of strategies for recombinant protein expression and purification to determine whether any provide a means for achieving higher overall success frequencies in the recovery of soluble recombinant protein. We exploited the covalent linkage of HaloTag recombinant proteins as a means of performing direct protein purification from crude E. coli lysates or from in vitro expression extracts using HaloLink microarray slides (Fig. 3 ). We randomly selected 20 ORFs encoded in the genome of S. pneumoniae and cloned these sequences into pFC20A. Recombinant proteins were either expressed in the BL21(DE3)/pMagic strain or by in vitro expression using the TnT® SP6 Coupled Wheat Germ Extract System (Promega). The over-expressed proteins derived from BL21(DE3)/ pMagic were purified using either HaloLink resin resulting in recovery of 75% of targets as soluble protein. When these proteins were expressed and purified using direct purification on HaloLink glass slides we recovered 100% of the target proteins in soluble form. Finally, when using in vitro transcription and translation systems followed by direct purification using HaloLink slides we recovered 85% of the target proteins in soluble form. Conclusions drawn from these studies must be taken with caution, however it appears that direct purification of recombinant proteins whether expressed in vitro or in E. coli may be more successful than traditional column-based purification schemes. The average purity of recovered proteins over-expressed in the E. coli BL21(DE3)/pMagic strain and purified using in situ purification is estimated to be more than 90% which is adequate for a variety of downstream applications.
). We randomly selected 20 ORFs encoded in the genome of S. pneumoniae and cloned these sequences into pFC20A. Recombinant proteins were either expressed in the BL21(DE3)/pMagic strain or by in vitro expression using the TnT® SP6 Coupled Wheat Germ Extract System (Promega). The over-expressed proteins derived from BL21(DE3)/ pMagic were purified using either HaloLink resin resulting in recovery of 75% of targets as soluble protein. When these proteins were expressed and purified using direct purification on HaloLink glass slides we recovered 100% of the target proteins in soluble form. Finally, when using in vitro transcription and translation systems followed by direct purification using HaloLink slides we recovered 85% of the target proteins in soluble form. Conclusions drawn from these studies must be taken with caution, however it appears that direct purification of recombinant proteins whether expressed in vitro or in E. coli may be more successful than traditional column-based purification schemes. The average purity of recovered proteins over-expressed in the E. coli BL21(DE3)/pMagic strain and purified using in situ purification is estimated to be more than 90% which is adequate for a variety of downstream applications.
USE OF HALOTAG RECOMBINANT PROTEINS TO IDENTIFY PROTEIN-PROTEIN INTERACTIONS
As we learn more about the cellular functions of proteins we see that few proteins operate in isolation of other macromolecules, particularly other proteins. The two-hybrid method and immunoprecipitation “pull down” experiments have contributed to our growing perception that proteins often function via physical interaction with one or more proteins [12Zhang L, Villa NY, Rahman MM, et al. Analysis of vaccinia virus-host protein-protein interactions: validations of yeast two-hybrid screenings J Proteome Res 2009; 8(9 ): 4311-8., 39Bruckner A, Polge C, Lentze N, Auerbach D, Schlattner U. Yeast two-hybrid, a powerful tool for systems biology Int J Mol Sci 2009; 10(6 ): 2763-88.]. Our knowledge of numerous binary interactions between proteins and multi-protein complexes e.g. RNA and DNA polymerase, ribosomal subunits etc is extensive for these examples but fundamentally lacking in others. Independent methods are needed to validate and discover protein-protein interactions [12Zhang L, Villa NY, Rahman MM, et al. Analysis of vaccinia virus-host protein-protein interactions: validations of yeast two-hybrid screenings J Proteome Res 2009; 8(9 ): 4311-8.]. We have used the HaloTag technology in a number of formats as a means of identifying or validating a number of binary protein interactions and also to identify constituents of multi-protein complexes [7Hurst R, Hook B, Slater MR, Hartnett J, Storts DR, Nath N. Protein-protein interaction studies on protein arrays: effect of detection strategies on signal-to-background ratios Anal Biochem 2009; 392(1 ): 45-53., 40Nath N, Hurst R, Hook B, et al. Improving protein array performance: focus on washing and storage conditions J Proteome Res 2008; 7(10 ): 4475-82.].
Protein interactions that occur within the Y. pestis Type 3 secretion system (T3SS) were identified using a protein array-based method in which the labeled HaloTag recombinant proteins were used as prey to detect binary protein interactions with immobilized bait proteins. The T3SS apparatus, also known as an injectisome, functions to directly inject effector proteins expressed by the bacterium into its mammalian host during infection [41Kubori T, Matsushima Y, Nakamura D, et al. Supramolecular structure of the Salmonella typhimurium type III protein secretion system Science 1998; 280(5363
): 602-5.-45Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines Nature 2006; 444(7119
): 567-73.]. To carry out this interrogation we cloned the bait proteins (T3SS) into pMBP (His-MBP tag) previously reported in [18Kwon K, Hasseman J, Latham S, et al. Recombinant expression and functional analysis of proteases from Streptococcus pneumoniae, Bacillus anthracis, and Yersinia pestis BMC Biochem 2011; 12: 17.], that were immobilized to a Cu2+ coated microarray slide surface (Fig. 4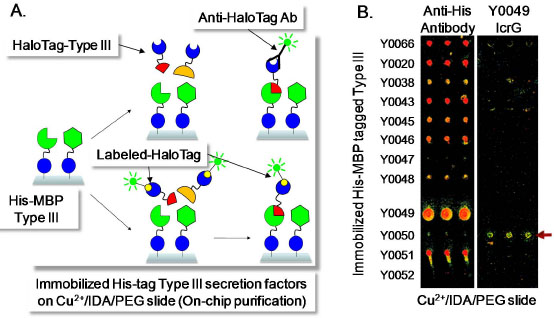 ) [46Kwon K, Grose C, Pieper R, Pandya GA, Fleischmann RD, Peterson SN. High quality protein microarray using in situ protein purification BMC Biotechnol 2009; 9: 72.]. The immobilized bait proteins were challenged with specific HaloTag prey proteins which were derived from pFN18A to establish the specificity of their interactions using indirect detection via an anti-HaloTag antibody or Biotin labeled HaloTag followed by fluorescently labeled streptavidin. The pFN18A vector was used for this study because the HaloTag recombinant T3SS proteins derived from pFN19A and pFC20A were partially degraded when expressed in E. coli. These experiments are particularly challenging since the T3SS is a multi-protein complex involving a number of membrane localized components that are difficult to express as soluble proteins. An example of the results achieved using this strategy is shown in Fig. (4
) [46Kwon K, Grose C, Pieper R, Pandya GA, Fleischmann RD, Peterson SN. High quality protein microarray using in situ protein purification BMC Biotechnol 2009; 9: 72.]. The immobilized bait proteins were challenged with specific HaloTag prey proteins which were derived from pFN18A to establish the specificity of their interactions using indirect detection via an anti-HaloTag antibody or Biotin labeled HaloTag followed by fluorescently labeled streptavidin. The pFN18A vector was used for this study because the HaloTag recombinant T3SS proteins derived from pFN19A and pFC20A were partially degraded when expressed in E. coli. These experiments are particularly challenging since the T3SS is a multi-protein complex involving a number of membrane localized components that are difficult to express as soluble proteins. An example of the results achieved using this strategy is shown in Fig. (4 ). In this instance, when HaloTag prey protein Y0049 (LcrG) is used to interrogate the protein microarray it interacts specifically with Y0050 (LcrV), an interaction that has been reported previously using independent methods for determining the interaction of these proteins [47Nilles ML, Williams AW, Skrzypek E, Straley SC. Yersinia pestis LcrV forms a stable complex with LcrG and may have a secretion-related regulatory role in the low-Ca2+ response J Bacteriol 1997; 179(4
): 1307-6.-51Matson JS, Nilles ML. Interaction of the Yersinia pestis type III regulatory proteins LcrG and LcrV occurs at a hydrophobic interface BMC Microbiol 2002; 2: 16.].
). In this instance, when HaloTag prey protein Y0049 (LcrG) is used to interrogate the protein microarray it interacts specifically with Y0050 (LcrV), an interaction that has been reported previously using independent methods for determining the interaction of these proteins [47Nilles ML, Williams AW, Skrzypek E, Straley SC. Yersinia pestis LcrV forms a stable complex with LcrG and may have a secretion-related regulatory role in the low-Ca2+ response J Bacteriol 1997; 179(4
): 1307-6.-51Matson JS, Nilles ML. Interaction of the Yersinia pestis type III regulatory proteins LcrG and LcrV occurs at a hydrophobic interface BMC Microbiol 2002; 2: 16.].
We evaluated the use of HaloTag in a more challenging goal to capture the subunits of multi-protein complexes. We selected a well-characterized multi-protein complex, RNA polymerase from E. coli to examine the pull down scheme wherein one suspected member of a protein complex is fused to HaloTag. Based on the work of several studies it is known that RpoA forms direct contacts with itself, AceE, RplA, RpoC, NusA and RpoB, whereas indirect linkages within the complex include the additional proteins TufA and Tig [52Arifuzzaman M, Maeda M, Itoh A, et al. Large-scale identification of protein-protein interaction of Escherichia coli K-12 Genome Res 2006; 16(5
): 686-91.-58Mencia M, Monsalve M, Rojo F, Salas M. Substitution of the C-terminal domain of the Escherichia coli RNA polymerase alpha subunit by that from Bacillus subtilis makes the enzyme responsive to a Bacillus subtilis transcriptional activator J Mol Biol 1998; 275(2
): 177-85.]. We cloned and over-expressed the RpoA subunit as an N-terminal HaloTag (pFN19A) fusion protein in E. coli, BL21(DE3)/pMagic. The assumption made in this experimental procedure is that the fusion protein will retain its ability to interact with the other proteins in the complex with relatively similar efficiency as the endogenously expressed RpoA. The RpoA in the pFN19A vector was over-expressed in 5 mL E. coli culture. The RpoA derived from the whole cell lysate was immobilized onto HaloLink resin and washed extensively to eliminate non-specific interacting proteins. Following recovery of the fusion protein, several protein bands were recovered (Fig. 5A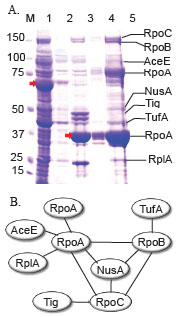 ). These bands were cut from the gel and subjected to MALDI-TOF/TOF-MS to identify those proteins present in the RpoA complex. Our results illustrate the power of the approach as all of the known members of the protein complex were recovered as shown in Fig. 5B
). These bands were cut from the gel and subjected to MALDI-TOF/TOF-MS to identify those proteins present in the RpoA complex. Our results illustrate the power of the approach as all of the known members of the protein complex were recovered as shown in Fig. 5B . This platform can be easily adapted to high throughput platform such as a 96-well format, thus allowing AP/MS to be performed in a high throughput manner.
. This platform can be easily adapted to high throughput platform such as a 96-well format, thus allowing AP/MS to be performed in a high throughput manner.
USE OF HALOTAG RECOMBINANT PROTEINS TO IDENTIFY PROTEIN-DNA INTERACTIONS
The interest in DNA protein interactions, particularly of transcriptional regulatory proteins has been significant for nearly three decades now. There are a variety of methods for studying these interactions but the majority of these are refractory to high throughput characterization. We have evaluated a number of methods including gel mobility shift assays, fluorescence polarization, ChIP-chip and ChIP-Seq analysis and others [8Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions Science 2007; 316(5830 ): 1497-502., 10Pugh BF, Gilmour DS. Genome-wide analysis of protein-DNA interactions in living cells Genome Biol 2001; 2(4 ): REVIEWS1013., 59Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment Methods 1999; 19(3 ): 425-33.-70Hartzell DD, Trinklein ND, Mendez J, et al. A functional analysis of the CREB signaling pathway using HaloCHIP-chip and high throughput reporter assays BMC Genomics 2009; 10: 497.]. Each approach has specific advantages and disadvantages with respect to ease, reproducibility, sensitivity and specificity. The proteomic profiling of transcription factors is often hampered by the low-level expression of these proteins preventing their visualization on 2DE MS/MS based experiments or LC/MS/MS studies. We enriched these proteins from crude lysates derived from Y. pestis by passing the lysate over a DNA cellulose column. The eluted proteins were indeed strongly enriched for transcription factors and other nucleic acid binding proteins. Among the list of recovered proteins was a set of 16 hypothetical proteins. We wished to establish whether these genes of unknown function represented a new class of transcription factors or nucleic acid binding proteins.
We developed an approach to evaluate the DNA binding activity and specificity of these proteins as described below. In this scheme, we cloned each of the putative transcription factors into pFN19A N-terminal HaloTag expression vector. The recombinant proteins were expressed in BL21(DE3)/ pMagic. These proteins were then immobilized onto HaloLink slides. Among the 16 Y. pestis target proteins 12 were expressed in E. coli and 10 of these were recovered as soluble protein. Nine of the soluble proteins were effectively purified by direct purification on HaloLink slides (Fig. 6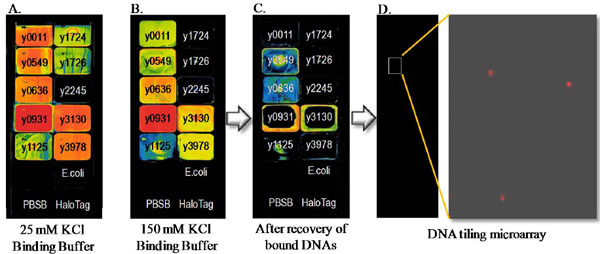 ). We next fluorescently labeled sheared Y. pestis genomic DNA with Cy5. The labeled genomic DNAs were then mixed with each immobilized HaloTag fusion protein in either low or high salt buffer to allow DNA-protein interactions to occur. After appropriate washing of the slide surface, the bound genomic DNA is recovered from the array and used as a hybridization probe of a second DNA oligonucleotide tiling microarray. This microarray represents the entire Y. pestis genome as a series of overlapping 60-mer oligonucleotides alternately covering each strand of DNA and allows the approximation and partial identification of the specific DNA sequences bound by the transcription factor. This straight-forward method is amenable to moderate throughput but can be envisioned as a means of characterizing all annotated transcription factors encoded in a genome of interest. While our experience with this strategy is still limited it is anticipated that the method success will be linked to the affinity of the protein for its cognate DNA sequence motifs and further by our ability to capture growth conditions that permit expression of transcription factors such that they are activated for specific DNA binding such as is expected for the case of two-component regulators that require phosphorylation for DNA binding activity.
). We next fluorescently labeled sheared Y. pestis genomic DNA with Cy5. The labeled genomic DNAs were then mixed with each immobilized HaloTag fusion protein in either low or high salt buffer to allow DNA-protein interactions to occur. After appropriate washing of the slide surface, the bound genomic DNA is recovered from the array and used as a hybridization probe of a second DNA oligonucleotide tiling microarray. This microarray represents the entire Y. pestis genome as a series of overlapping 60-mer oligonucleotides alternately covering each strand of DNA and allows the approximation and partial identification of the specific DNA sequences bound by the transcription factor. This straight-forward method is amenable to moderate throughput but can be envisioned as a means of characterizing all annotated transcription factors encoded in a genome of interest. While our experience with this strategy is still limited it is anticipated that the method success will be linked to the affinity of the protein for its cognate DNA sequence motifs and further by our ability to capture growth conditions that permit expression of transcription factors such that they are activated for specific DNA binding such as is expected for the case of two-component regulators that require phosphorylation for DNA binding activity.
CONCLUSIONS
We have adapted the HaloTag technology to current protein production platforms and examined the enhancement of soluble expression of the proteins of interest. We also examined the use of the HaloTag to high throughput functional studies such as protein-protein interactions and protein-DNA interactions. Several vectors containing HaloTag were made compatible with high throughput cloning strategies and examined for their efficiency in expressing soluble protein. The N-terminal HaloTag Gateway vector (pGW-nHalo) showed that the HaloTag recombinant proteins were solubly expressed with a high success rate and can be used for high throughput cloning using existing entry clone sets. Soluble expression attempts of proteins of interest in E. coli, in vitro and mammalian expression systems were conducted using various HaloTag vectors and the results demonstrated overall high success rates. A combination of N-terminal and C-terminal HaloTag vectors increases overall success rate of soluble protein recovery. We have employed the HaloTag technology in other contexts using protein microarrays for high throughput assay for anti-sera screening and other protein functional analysis. In the protein array schemes, the HaloTag recombinant proteins were successfully used as prey proteins for identification of protein-protein interactions in Y. pestis T3SS with other fusion tagged recombinant proteins, and as bait proteins to identify DNA binding activity of hypothetical proteins. The HaloTag was successfully used for pull-down assays involving E. coli RpoA as part of a multi-protein complex. While we describe here only a limited number of applications of the HaloTag technology, many more strategies are enabled by this versatile technology. In these early days of the post-genomic era, HaloTag and other technologies will be important vehicles for better understanding the breadth of protein functions encoded by the awe inspiring number of unique proteins encoded on our planet.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
SUPPLEMENTARY MATERIAL
Supplementary material (Supplementary Table 1.xlsx) is available on the publisher’s website along with the published article Download Supplementary Material.
ACKNOWLEDGEMENTS
We thank Dr. Marjeta Urh for helpful discussion and review of the manuscript. We also thank Sarah Grimshaw for proofreading of the manuscript. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institute of Health, under contract No. N01-AI15447.
REFERENCES
Endorsements
Browse Contents
Table of Contents
- INTRODUCTION
- HISTORICAL DEVELOPMENT OF HALOTAG
- POTENTIAL ADVANTAGES OF COVALENT LINKAGE
- ADAPTATION OF HALOTAG TO GATEWAY EXPRESSION VECTORS
- COMPARISON OF EXPRESSION VECTORS
- INCREASE SUCCESS RATES OF SOLUBLE EXPRESSION
- USE OF HALOTAG RECOMBINANT PROTEINS TO IDENTIFY PROTEIN-PROTEIN INTERACTIONS
- USE OF HALOTAG RECOMBINANT PROTEINS TO IDENTIFY PROTEIN-DNA INTERACTIONS
- CONCLUSIONS
- CONFLICT OF INTEREST