- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Cell Development & Biology Journal
(Discontinued)
ISSN: 1874-0855 ― Volume 3, 2011
The Use of Single Base Extension (SBE) for the Assessment of European Mitochondrial DNA Haplogroups in Galician Population
Ignacio Rego1, Mercedes Fernandez-Moreno 1, Sara Relañ1, Joaquin Arenas2, Francisco J. Blanco*, 1
Abstract
Evidences are accumulating on the effects of the mtDNA variants on many complex traits due to the contribution of mtDNA to cellular physiology. More and more studies are being carried out in order to investigate the possible associations between these mtDNA variants or haplogroups and multifactorial complex diseases. Since the required sample size is high, these large-scale epidemiology studies require a suitable method to assign the mtDNA haplogroups in a rapid, accuracy and cost effective manner. In this study we present an assay, based on primer base extension, which permits the rapid genotyping of samples for the assessment of major European mtDNA haplogroups.
Article Information
Identifiers and Pagination:
Year: 2008Volume: 1
First Page: 10
Last Page: 16
Publisher Id: TOCBJ-1-10
DOI: 10.2174/18740855008010010
Article History:
Received Date: 14/11/2007Acceptance Date: 15/12/2007
Electronic publication date: 25/1/2008
Collection year: 2008
* at the Osteoarticular and Aging Research Laboratory, Biomedical Research Center, Complejo Hospitalario Universitario Juan Canalejo, 15006-A Coruña, Spain; Tel: 34-981-178272; Fax: 34-981-178273; E-mail: fblagar@canalejo.org
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 14-11-2007 |
Original Manuscript | The Use of Single Base Extension (SBE) for the Assessment of European Mitochondrial DNA Haplogroups in Galician Population | |
1. INTRODUCTION
It’s well known that Restriction Fragment Length Polymorphisms (RFLPs) studies have revealed a number of stable polymorphic sites in the mitochondrial DNA (mtDNA) coding regions that define related groups of mtDNAs called haplogroups [1]. Most of these haplogroups are continent-specific; so, nine European (H, I, J, K, T, U, V, W and X), seven Asian and three African mtDNA haplogroups have been identified [1], [2]. The specific single nucleotide polymorphisms (SNPs) that characterize the different haplogroups reflect mutations accumulated by a discrete maternal lineage.
Given the several lines of evidence that suggest the contribution of mtDNA to cellular physiology and its critical importance for energy production, an increasing number of studies have been carried out investigating the association between mtDNA haplogroups and multifactorial diseases. Different associations have been found in Leber´s Hereditary Optic Neuropathy (LHON) [3], [4], Alzheimer disease [5], [6], Occipital stroke [7], Sperm mobility [8], Parkinson [9], [10], Type 2 diabetes mellitus [11], multiple sclerosis [12], [13], increased survival after sepsis [14] or hypertrophic cardiomyopathy [15].
Also, mtDNA variants could play a role in successful aging and longevity. In fact, three independent studies have shown that European haplogroup J is over-represented in groups of centenarians with respect to geographically matched younger controls in Northern Italy [16], Northern Ireland [17] and Finland [18]. In the same way, in the Japanese population, the Asian mtDNA haplogroup D was reported to be associated with longevity, being more frequent in centenarians than in a control group of younger subjects [19]. Besides, mtDNA haplogroups have also been exten sively and successfully used as tools for investigating human origin and evolution [20].
Up to date, the method of choice for assessing mtDNA haplogroups relies on RFLP analysis, using up to 14 restriction enzymes [1] or melting curve assays with allele-specific Tm hybridization probes [21]. However, the mtDNA haplogroup association studies with these techniques suffer in several regards: mainly, such studies require a sample size of patients and controls in the several or thousands numbering and, by means of these techniques, a separate PCR amplification for each SNP that comprises the haplogroup is required. Thus, an economic, accuracy and high capacity assay is desirable.
In the present study we combine the classical PCR-RFLP technique and the more new Single Base Extension (SBE) assay in order to assess the most common European mtDNA haplogroups (H, V, K, U, T and J) in Galician population (NW Spain). This technique has been successfully used, among others, by Vallone et al. [22] in order to analyze 11 forensically informative SNPs distributed throughout the mitochondrial genome. Also, a similar approach has been carried out by Brandstätter et al. [23] and Wiesbauer et al. [24] to assign mtDNA haplogroups in Austrian populations.
2. MATERIALS AND METHODS
SBE assay consists in the annealing of a single primer to a sequence of the mtDNA template that contains the SNP we want to interrogate, such that the 3´-end of this primer falls one base short of the SNP site present on the template. Since we are using only dideoxynucleotides (ddNTPs) in the reaction, when the complementary base is incorporated by the Taq DNA polymerase, the elongation stops and, depending on the fluorescence emitted, the SNP site will be identified. In contrast, those minor samples that did not belong to any of these 6 mtDNA haplogroups, were analyzed by PCR-RFLP.
Six specific primers were designed in order to amplify the mtDNA fragments that contain each of the informative SNPs, which characterize the 6 major European mtDNA haplogroups, in one multiplex reaction. The polymorphic sites (Table1) analyzed in the present study (7025, 14766, 10394, 4577, 12308 and 4216) were previously reported [1], [25]. Besides, another 6 specific SBE primers were also designed to interrogate each SNP site. In order to design the SBE primers, several considerations were taken into account: (i) the minimum length of each primer consisted of 20 nt, (ii) each primer had a Tm of ~60ºC and (iii) the minimum difference in length among the 6 primers was of 6nt in order to avoid overlapping. In this latest case, is possible to add unspecific nucleotides at 5´-end of the primer for increasing its length. The sequence of PCR, PCR-RFLP and SBE primers are listed in Table2.
Multiplex PCR mixture consisted of a final concentration of 1X Reaction Buffer (Bioline), 0.2mM of each deoxynucleotide (dNTP) (Bioline), 1.5mM MgCl2 (Bioline), 0.025 U/µL of BioTaq DNA polymerase (Bioline) and 0.3 µM of each primer in a volume of 50 µL. Blood isolated (Invisorb Spin Blood Mini Kit from Invitek) genomic DNA (75 ng) was added to the mixture and amplified as follows: 94ºC for 5 minutes, 35 cycles at 95ºC for 60 seconds, 55ºC for 60 seconds and 72ºC for 60 seconds, and a final extension at 70ºC for 10 minutes. After amplification, the PCR products were optionally electrophoresed in a agarose gel and uv-visualized after ethidium bromide treatment.
To remove primers and unincorporated dNTPs, multiplex PCR products were treated with ExoSap-IT (Amersham) following manufacturer´s recommendations: adding 2µL of enzyme to 5 µL of PCR product (or a proportional ratio), followed by an activation of the enzyme at 37ºC for 15 minutes and a further deactivation by incubating at 80ºC for 15 minutes. Later, the samples were placed on ice.
Multiplex SBE reactions were carried out following the recommendations from Applied Biosystems with some modifications. So, in a final volume of 10µL, we added 1.5µL of SNaPshot® Multiplex Kit, 2.5µL of purified PCR product and a final concentration of 0.2µM of the SBE primers mixture. To reach the final volume of 10µL, ddH2O was added. Thermal cycling conditions for SBE were as follows: 96ºC for 60 seconds and 25 cycles at 96ºC for 10 seconds, 60ºC for 5 seconds and 60ºC for 30 seconds. To remove unincorporated ddNTPs, the SBE reaction products were treated with Shrimp Alkaline Phosphatase (SAP) (Amserham) by adding to the mixture 1µL of enzyme, 2µL of SAP reaction buffer and 7µL ddH2O, followed by an activation of the enzyme at 37ºC for 1 hour and a further deactivation by incubating at 75ºC for 15 minutes. Subsequently, the samples were placed on ice.
Finally, 9µL of Hi-DiTM Formamide (Applied Biosystems), 0.5µL of size internal standard (120 Liz Size Standard from Applied Biosystems) and 0.5µL of purified SBE product were mixed and denatured at 95ºC for 5 minutes prior to load into an ABI 3100 genetic analyzer. The configuration system is based on a 36cm length capillary filled with a Performance Optimized Polymer 4 (POP4) containing urea, from Applied Biosystems. The SBE default run module consisted in 22 seconds of injection time, 16 minutes of run time and a run voltage of 15 KVolts. Once the runs were finished, the whole data was analyzed in GeneMapper v3.5 software (Applied Biosystems), which is able to assign the different alleles (SNPs) in each locus, prior designing of a reference sequence that encompasses all the allelic variants for each locus (Fig.1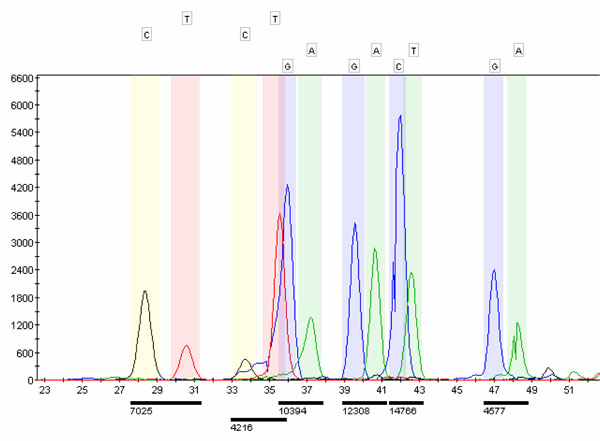 ). In order to verify the results obtained, some samples were analyzed by PCR-RFLP and direct sequencing of the PCR product.
). In order to verify the results obtained, some samples were analyzed by PCR-RFLP and direct sequencing of the PCR product.
 |
Fig. (1) Reference sequence encompassing the allelic variants for each polymorphic site. |
3. RESULTS AND DISCUSSION
The present study consisted of 778 unrelated Europeans from Galicia, in the Northwest of Spain. Of this cohort of samples, 535 were women and 243 were men. The mean age of the samples was 66 years old. All subjects signed an informed consent in order to authorize this study.
When running the PCR multiplex, ~85% of samples were assigned to one of the six most common mtDNA European haplogroups (H, V, K, U, T or J). Typical patterns are shown in Fig.(2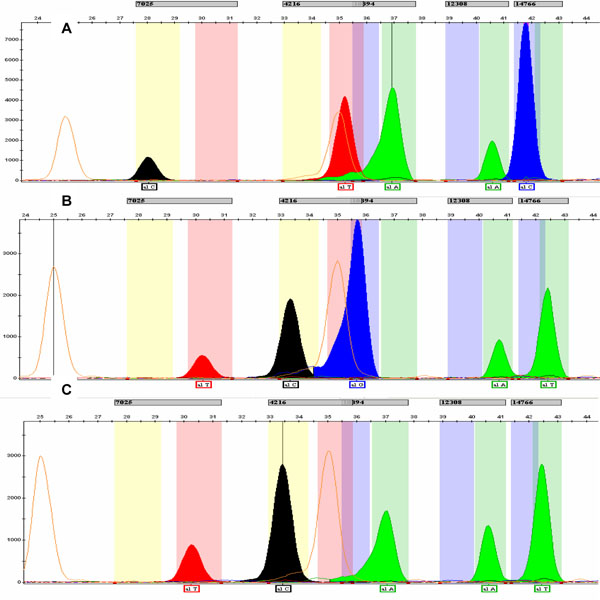 ). The less common haplogroups (W, I and X) were assessed by means of PCR-RFLP according to the hierarchical scheme described by Macaulay et al. [25]. Remaining samples were assigned depending on the polymorphic site 10394 (A10398G): those samples with the 10398G allele were tested for 10032AluI (+10032AluI were assigned to haplogroup I, 10029G allele), whereas those with the 10398A allele were tested for either 14465AccI (+14465AccI were assigned to haplogroup X, 14470C allele) or 8994HaeIII (-14465AccI, -8994HaeIII were assigned to haplogroup W, 14470T and 8994A alleles respectively) (Fig. 3
). The less common haplogroups (W, I and X) were assessed by means of PCR-RFLP according to the hierarchical scheme described by Macaulay et al. [25]. Remaining samples were assigned depending on the polymorphic site 10394 (A10398G): those samples with the 10398G allele were tested for 10032AluI (+10032AluI were assigned to haplogroup I, 10029G allele), whereas those with the 10398A allele were tested for either 14465AccI (+14465AccI were assigned to haplogroup X, 14470C allele) or 8994HaeIII (-14465AccI, -8994HaeIII were assigned to haplogroup W, 14470T and 8994A alleles respectively) (Fig. 3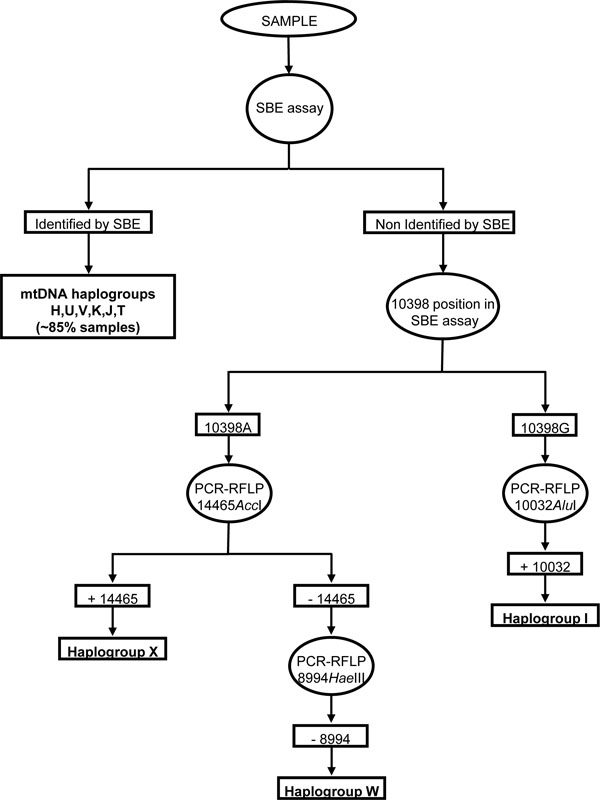 ).
).
A minimal percentage of samples (5.7%) could not be assigned to any of the European mtDNA haplogroups and were classified as “others”. The obtained haplogroup frequencies were: 45.2% for haplogroup H, 15.6% for haplogroup U, 8% for haplogroup J and K, 7.2% for haplogroup T and 3.7% for haplogroup V. For the less common haplogroups the frequencies were: 3% for haplogroup X and superHV, 0.5% for haplogroup I and 0.3% for haplogroup W. These frequencies were in concordance with those obtained by other authors when analyzed mtDNA haplogroups in different European populations though with slight differences (Table3). A previous study of 686 samples from Spanish populations [26] also showed a similar frequency distribution, with slight differences in haplogroup U. These minimal differences reflected in the studies described above are probably due to both the sample size and the reflecting of geographical distribution of the populations, as described by Samuels et al. [27], and Wiesbauer et al. [24] respectively.
The technique described in this study is rapid and cost effective since ~85% of samples could be assigned to one mtDNA haplogroup in one multiplex reaction by genotyping only 6 SNPs. Besides, its accuracy is also obvious since a few samples (N=20) were also analyzed by both PCR-RFLP and direct sequencing of the PCR product, obtaining the same results (data not shown). A similar approach was
described by Wiesbauer et al. [24], obtaining successfully results. However, it must be taken into account that, in order to perform this assay, an expensive genetic analyzer is required. In our case, the ABI 3100 genetic analyzer, with 16 capillary technology, allowed us a high capacity of processing. In contrast PCR-RFLP, though reliable, is a much less rapid and more laborious technique.
In summary, to our knowledge this is the first study in analyzing, by means of the SBE assay, the frequencies of the European mtDNA haplogroups in the several hundreds numbering of subjects (N=778) from Galician population. These frequencies give high valour when epidemiological association studies for mitochondrial disorders studies in this population is planted. To identify mitochondrial haplogroups in a high number of samples we recommend to follow-up the hierarchical scheme showed in the Fig.(3 ) using the SBE assay as a first approach.
) using the SBE assay as a first approach.