- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Leukemia Journal
(Discontinued)
ISSN: 1876-8164 ― Volume 5, 2013
Expression of the FOXP1 Transcription Factor is Post-Transcriptionally Silenced in Normal and Malignant CD138+ Plasma Cells
Philip J. Brown1, Andrew J. Campbell1, Linden Lyne1, Jianxiang Chi1, Charles H. Lawrie1, Rajko Kusec2, Alison H. Banham1, *
Abstract
The FOXP1 transcription factor is heterogeneously expressed in normal B cells and is highly expressed in poor prognosis B-cell lymphoma patients. Double immunohistochemical labelling studies identified the striking absence of FOXP1 protein expression in VS38c+, CD38+ and CD138+ plasma cells; prompting an investigation of FOXP1 mRNA and protein expression in multiple myeloma (MM) and the pre-neoplastic plasma cell proliferation monoclonal gammopathy of undetermined significance (MGUS). FOXP1 mRNA expression was assessed by quantitative RT-PCR in normal CD138+ bone marrow plasma cells, MM cell lines (n=4) and cases of MM, including aspirates of whole BM (n=11) and purified CD138+ cells (n=12). Surprisingly both normal and abnormal CD138+ plasma cells expressed the FOXP1 transcript, some cases of each exhibiting high levels, comparable to those in activated B-cell-like diffuse large B-cell lymphoma. However normal CD138+ bone marrow plasma cells, MM cell lines and CD138+ plasma cells in primary MGUS (n=13) and MM biopsies (n=68) were largely devoid of FOXP1 protein expression. The notable exception was two MM patients in which >30% of the CD138+ population was FOXP1+. Mechanisms which block mRNA translation or de-stabilise the FOXP1 protein may silence its expression in plasma cells.
Article Information
Identifiers and Pagination:
Year: 2010Volume: 3
First Page: 16
Last Page: 23
Publisher Id: TOLEUKEMIAJ-3-16
DOI: 10.2174/1876816401003010016
Article History:
Received Date: 12/10/2009Revision Received Date: 5/11/2009
Acceptance Date: 1/12/2009
Electronic publication date: 22/1/2010
Collection year: 2010
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Nuffield Department of Clinical Laboratory Sciences, University of Oxford, Level 4 Academic Block, John Radcliffe Hospital, Headington, Oxfordshire, OX3 9DU, UK; Tel: +44 (0)1865 220246; Fax: +44 (0)1865 228980; E-mail: alison.banham@ndcls.ox.ac.uk
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 12-10-2009 |
Original Manuscript | Expression of the FOXP1 Transcription Factor is Post-Transcriptionally Silenced in Normal and Malignant CD138+ Plasma Cells | |
INTRODUCTION
Forkhead box P1 (FOXP1) is a member of the forkhead box (FOX) family of transcription factors that are defined by a common DNA-binding domain termed the forkhead box or winged helix domain. The FOXP1 gene maps to a tumour suppressor locus at 3p14.1 and is aberrantly expressed at both the mRNA and protein levels in a variety of carcinomas [1Banham AH, Beasley N, Campo E, et al. The FOXP1 winged helix transcription factor is a novel candidate tumor suppressor gene on chromosome 3p Cancer Res 2001; 61: 8820-9.]. In breast cancer patients the loss of FOXP1 protein expression correlates with a poor prognosis, consistent with a role as a tumour suppressor [2Fox SB, Brown P, Han C, et al. Expression of the forkhead transcription factor FOXP1 is associated with estrogen receptor-a and improved survival in primary human breast carcinomas Clin Cancer Res 2004; 10: 3521-7.]. In contrast, the high level expression of the FOXP1 protein in large B-cell Non Hodgkin’s lymphomas (NHL) correlates with a poor prognosis [3Barrans SL, Fenton JA, Banham A, et al. Strong expression of FOXP1 identifies a distinct subset of diffuse large B-cell lymphoma patients with poor outcome Blood 2004; 104: 2933-5.-5Sagaert X, de Paepe P, Libbrecht L, et al. Forkhead box protein P1 expression in mucosa-associated lymphoid tissue lymphomas predicts poor prognosis and transformation to diffuse large B-cell lymphoma J Clin Oncol 2006; 24(16 ): 2490-7.]. While this might initially appear to be counter intuitive there are plenty of examples in the published literature of transcription factors, such as Maf [6Pouponnot C, Sii-Felice K, Hmitou I, et al. Cell context reveals a dual role for Maf in oncogenesis Oncogene 2006; 25(9 ): 1299-310.] and NF-κB [7Perkins ND. NF-kappaB: tumor promoter or suppressor? Trends Cell Biol 2004; 14(2 ): 64-9.], that can display dual roles as either tumour suppressors or oncogenes, depending on the cellular context. Certainly the alterations in apoptosis and cellular proliferation observed in cardiac tissues from Foxp1 knockout mice have indicated that the affects on these processes are regulated both temporally and spatially during development, suggesting that there is complex context dependent regulation of Foxp1 function in vivo [8Wang B, Weidenfeld J, Lu MM, et al. Foxp1 regulates cardiac outflow tract, endocardial cushion morphogenesis and myocytes proliferation and maturation Development 2004; 131: 4477-87.].
The identification of recurrent chromosome translocations targeting the FOXP1 gene in both mucosal associated lymphoid tissue (MALT) lymphomas and in diffuse large B-cell lymphomas (DLBCL) suggested that FOXP1 de-regulation might have an important role in lymphomagenesis [9Streubel B, Vinatzer U, Lamprecht A, et al. T(3; 14)(p14.1 q32) involving IGH and FOXP1 is a novel recurrent chromosomal aberration in MALT lymphoma Leukemia 2005; 19: 53-8.-11Fenton JA, Schuuring E, Barrans SL, et al. t (3; 114)(p14. q32) results in aberrant expression of FOXP1 in a case of diffuse large B-cell lymphoma Genes Chromosomes Cancer 2006; 45(2 ): 164-8.]. Interestingly these translocations commonly target the coding region of the FOXP1 gene [12Goatly A, Bacon CM, Nakamura S, et al. FOXP1 abnormalities in lymphoma: translocation breakpoint mapping reveals insights into deregulated transcriptional control Mod Pathol 2008; 21(7 ): 902-11.]. This is consistent with our data suggesting that N-terminal truncation of FOXP1 (by alternative splicing) may be a key event in normal B-cell activation that is retained in the activated B-cell-like (ABC) DLBCL subtype [13Brown PJ, Ashe SL, Leich E, et al. Potentially oncogenic B-cell activation-induced smaller isoforms of FOXP1 are highly expressed in the activated B cell-like subtype of DLBCL Blood 2008; 111(5 ): 2816-4.]. Furthermore, N-terminal truncation of foxP1 is an oncogenic event in a retroviral insertion model identifying novel oncogenes that cause avian nephroblastoma [14Pajer P, Pecenka V, Kralova J, et al. Identification of potential human oncogenes by mapping the common viral integration sites in avian nephroblastoma Cancer Res 2006; 66(1 ): 78-86.]. This raises the possibility that constitutive expression of smaller FOXP1 isoforms may have an oncogenic role in ABC-like DLBCL.
The majority of studies have focussed on FOXP1 in malignancy and thus there is still relatively little information concerning the roles of FOXP1 in normal tissues. Studies using knock out mice have identified an essential role for Foxp1 in embryonic development with phenotypes affecting cardiac [8Wang B, Weidenfeld J, Lu MM, et al. Foxp1 regulates cardiac outflow tract, endocardial cushion morphogenesis and myocytes proliferation and maturation Development 2004; 131: 4477-87.], neuronal [15Dasen JS, De Camilli A, Wang B, et al. Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor FoxP1 Cell 2008; 134(2 ): 304-16., 16Rousso DL, Gaber ZB, Wellik D, et al. Coordinated actions of the forkhead protein Foxp1 and Hox proteins in the columnar organization of spinal motor neurons Neuron 2008; 59(2 ): 226-40.], lung [17Shu W, Lu MM, Zhang Y, et al. Foxp2 and Foxp1 cooperatively regulate lung and esophagus development Development 2007; 134(10 ): 1991-2000.] and B-cell development [18Hu H, Wang B, Borde M, et al. Foxp1 is an essential transcriptional regulator of B cell development Nat Immunol 2006; 7(8 ): 819-26.]. Foxp1 is also required for monocyte differentiation and macrophage function [19Shi C, Zhang X, Chen Z, et al. Integrin engagement regulates monocyte differentiation through the forkhead transcription factor Foxp1 J Clin Invest 2004; 114(3 ): 408-18., 20Shi C, Sakuma M, Mooroka T, et al. Down-regulation of the forkhead transcription factor Foxp1 is required for monocyte differentiation and macrophage function Blood 2008; 112(12 ): 4699-711.], adipocyte differentiation and is implicated in human heart failure. Recently FOXP1 silencing in ex vivo expanded bone marrow mesenchymal stem cells has been reported to suppress their self-renewal capacity and their adipogenic differentiation [21Kubo H, Shimizu M, Taya Y, et al. Identification of mesenchymal stem cell (MSC)-transcription factors by microarray and knockdown analyses, and signature molecule-marked MSC in bone marrow by immunohistochemistry Genes Cells 2009; 14(3 ): 407-24.].
A brief description of FOXP1 expression in B cells in reactive tonsil has already reported that this protein is expressed in the majority of mantle zone B cells, a variable proportion of germinal centre (GC) cells and in many cells in the interfollicular area [1Banham AH, Beasley N, Campo E, et al. The FOXP1 winged helix transcription factor is a novel candidate tumor suppressor gene on chromosome 3p Cancer Res 2001; 61: 8820-9.]. In addition to staining haematopoietic cells, FOXP1 expression was also observed in both tonsillar epithelium and endothelium. In the current study we have employed double immunoenzymatic labelling techniques to identify the absence of FOXP1 protein expression in normal CD138+ tonsillar plasma cells. Despite the presence of FOXP1 transcripts, the vast majority of both normal and abnormal bone marrow plasma cells also lacked FOXP1 protein expression. Silencing of FOXP1 protein expression may have an important biological role during terminal B-cell differentiation to plasma cells and represents a novel mechanism by which FOXP1 expression is regulated.
METHODS
Patients and Tissues
Patients’ bone marrow samples and normal reactive tonsils from routine tonsillectomy were obtained with informed consent from the John Radcliffe Hospital, Oxford and the Dubrava University hospital, Zagreb, Croatia. This study was conducted under ethical approval from the Oxfordshire Clinical Research Ethics Committee.
Immunohistochemistry
Four micron sections of FFPE tonsil or cell pellets were captured on charged slides and dried at 37°C over night. Sections were dewaxed then antigen retrieved by microwaving in 50mM Tris/2mM EDTA, pH 9.0 [1Banham AH, Beasley N, Campo E, et al. The FOXP1 winged helix transcription factor is a novel candidate tumor suppressor gene on chromosome 3p Cancer Res 2001; 61: 8820-9.]. The anti-FOXP1 monoclonal antibody, JC12 (in house hybridoma supernatant, 1/80 dilution) was applied for 30 minutes at room temperature and binding was detected using the Envision-HRP Kit and DAB+ substrate (DakoCytomation). For single labelling, sections were counterstained with hematoxylin (Gill’s No. 2; Sigma-Aldrich) and mounted in Aquatex (VWR International). For double immunoenzymatic labelling, similar sequential rounds of staining were conducted with DAB+ substrate used in the first round and Vector SG substrate (Vector Laboratories) in the second, without the nuclear counterstain. The second primary antibodies were as follows: VS38c, undiluted monoclonal hybridoma supernatant; CD138, clone MI15, diluted 1/100 (DakoCytomation); CD38, clone SPC32, diluted 1/100 (Novocastra). After detection, slides were washed in water, dehydrated in xylene and mounted in VectaMount Permanent Mounting Medium (Vector Laboratories).
Cell Lines
MM cell lines included in the study were JJN3, NCIH929, RPMI8226 and THIEL. DLBCL cell lines OCI-Ly3, OCI-Ly10 (ABC-derived), SUDHL6, SUDHL10 and DB (GC-derived) were a kind gift from Dr Eric Davis (National Cancer Institute, Bethesda, MD), RIVA (ABC-derived) was a kind gift from Professor Martin Dyer (Leicester University, United Kingdom) and HLY-1 (ABC-derived) was generously provided by Dr Talal Al Saati (Purpan Hospital, Toulouse, France). Cell lines were maintained in RPMI 1640 media supplemented with 10% FCS, 2mM glutamine and antibiotics [streptomycin (50μg/ml) and penicillin (50U/ml)] at 37oC and 5% CO2. For each line, approximately 1x107 cells were formalin-fixed and paraffin embedded, additional cell pellets were snap frozen and used for Western blotting or RNA extraction.
qRT-PCR
CD138+ cells were isolated from bone marrow (BM) aspirates using MACS microbeads, according to the manufacturer’s instructions (Miltenyi Biotec). Total RNA was extracted from CD138+ cells and cell lines using the RNeasy Mini Kit (Qiagen). 100ng of total RNA was reverse transcribed using random primers (Promega) and Superscript III reverse transcriptase (Invitrogen). RNA from BM aspirates was isolated using QIAamp RNA Blood Mini Kit (Qiagen) and one microgram of total RNA was used for cDNA synthesis using GeneAmp Gold RNA PCR Core Kit (Applied BioSystems). 20 μl multiplex qPCR reactions were setup in triplicate using 1x EXPRESS qPCR SuperMix with ROX reference dye (Invitrogen), 1μl FOXP1-FAM TaqMan probe (Hs00212860_m1), 1μl TBP-VIC TaqMan probe (TBP; 4326322E) (Applied Biosystems), 1μl GAPDH-CY5 custom probe (150 nM primers, 250nM probe, final concentration) (Eurogentec) and 1 μl of cDNA template. Real-Time PCR amplification was performed in triplicate with a Chromo 4 continuous fluorescence detector (MJ Research). FOXP1 mRNA expression is presented as relative to that in the DB DLBCL cell line which was assigned a value of 1.
Western Blotting
Nuclear extracts were prepared from approximately 1x107 cells using the Nuclear Extraction Kit (Panomics). Extracts were resolved on 10% SDSPAGE gels and transferred to Hybond nitrocellulose membrane (GE Healthcare) using semidry apparatus. Membranes were blocked for 1 hour with PBS/5% dried skimmed milk powder, 0.02% Tween. The JC12 antibody was used to probe the membrane at 1/10 dilution in blocking buffer on a rocking platform at 4°C overnight. The membrane was washed (3 x 10 minutes) in PBS/0.05% Tween, then the secondary antibody, goat anti-mouse Ig-HRP (DakoCytomation) was applied at a 1/5000 dilution in blocking buffer for 1 hour at room temperature. The wash steps were repeated and the membrane was developed using enhanced chemiluminescence (ECL) reagent (GE Healthcare). The membrane was subsequently reprobed with the anti-TBP antibody (clone 1TBP18, Abcam) used at 1/1000 and detected as above to confirm adequate sample loading.
RESULTS
Using double immunoenzymatic labelling studies we have identified the absence of FOXP1 protein expression in terminally differentiated CD38+, CD138+ or VS38c+ plasma cells from tonsil (Fig. 1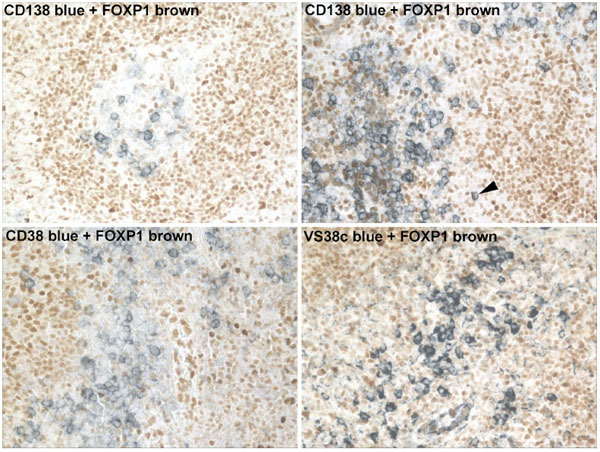 ). The occasional CD38+/FOXP1+ and CD138+/FOXP1+ lymphocytes were noted, indicating that a relatively rare population of terminally differentiated B cells do express FOXP1.
). The occasional CD38+/FOXP1+ and CD138+/FOXP1+ lymphocytes were noted, indicating that a relatively rare population of terminally differentiated B cells do express FOXP1.
While analysing FOXP1 expression in lymphoma cell lines we were surprised to find that the JJN3 MM cell line expressed relatively high levels of the FOXP1 mRNA (Fig. 2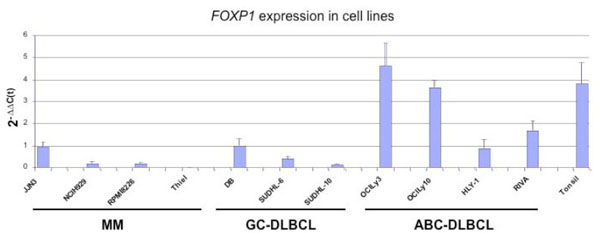 ). The levels were comparable to those in the HLY1 and DB DLBCL cell lines, which we have previously shown to strongly express the FOXP1 mRNA and protein [13Brown PJ, Ashe SL, Leich E, et al. Potentially oncogenic B-cell activation-induced smaller isoforms of FOXP1 are highly expressed in the activated B cell-like subtype of DLBCL Blood 2008; 111(5
): 2816-4.]. The NCIH929 and RPMI8226 cell lines expressed low levels of FOXP1 mRNA, while none was detectable in the THIEL cell line. Western blotting studies showed that FOXP1 protein expression was barely detectable in the majority of MM cell lines, in contrast to the highly FOXP1-positive OCI-Ly3 and DB DLBCL cell lines used as controls (Fig. 3
). The levels were comparable to those in the HLY1 and DB DLBCL cell lines, which we have previously shown to strongly express the FOXP1 mRNA and protein [13Brown PJ, Ashe SL, Leich E, et al. Potentially oncogenic B-cell activation-induced smaller isoforms of FOXP1 are highly expressed in the activated B cell-like subtype of DLBCL Blood 2008; 111(5
): 2816-4.]. The NCIH929 and RPMI8226 cell lines expressed low levels of FOXP1 mRNA, while none was detectable in the THIEL cell line. Western blotting studies showed that FOXP1 protein expression was barely detectable in the majority of MM cell lines, in contrast to the highly FOXP1-positive OCI-Ly3 and DB DLBCL cell lines used as controls (Fig. 3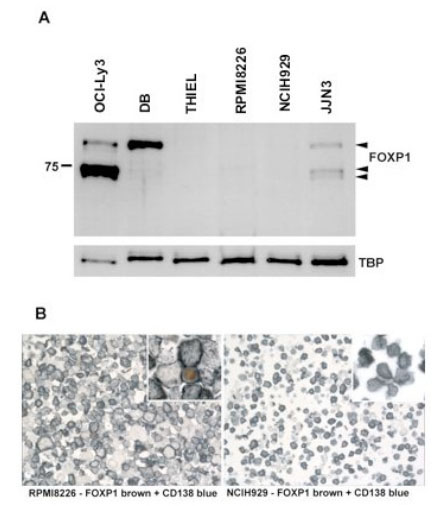 ). Interestingly the JJN3 cell line weakly expressed both the full-length FOXP1 protein and similar levels of the two smaller isoforms that are also present in the OCI-Ly3 DLBCL cell line. Using immunoenzymatic labelling to double stain the MM cell lines for FOXP1 expression (brown) and CD138 (blue) we observed no FOXP1 protein expression in the NCIH929 and THIEL cell lines, while the occasional FOXP1 positive nucleus was detected in the RPMI8226 and JJN3 cell lines.
). Interestingly the JJN3 cell line weakly expressed both the full-length FOXP1 protein and similar levels of the two smaller isoforms that are also present in the OCI-Ly3 DLBCL cell line. Using immunoenzymatic labelling to double stain the MM cell lines for FOXP1 expression (brown) and CD138 (blue) we observed no FOXP1 protein expression in the NCIH929 and THIEL cell lines, while the occasional FOXP1 positive nucleus was detected in the RPMI8226 and JJN3 cell lines.
This study was extended to investigate, whether abnormal and malignant plasma cells in bone marrow also lacked FOXP1 expression. In whole bone marrow aspirates from Croatian patients without plasma cell malignancy or those with MGUS or MM there was no significant difference in FOXP1 mRNA levels between patient groups (Fig. 4a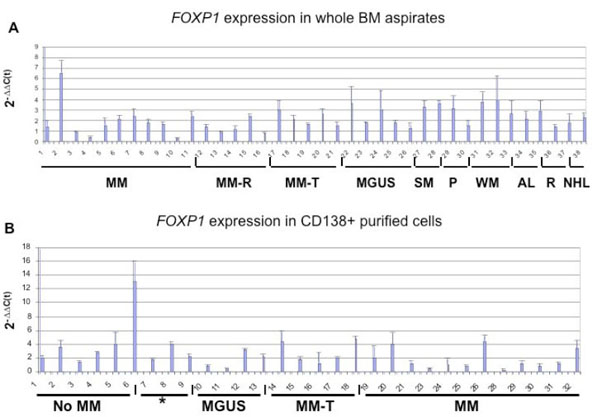 ). Although, most samples expressed relatively high levels of FOXP1 mRNA there was no correlation with the expression levels of CD138 mRNA, which was used to assess the level of plasma cell infiltration (data not shown; Campbell et al., Br J Haem 2010; in press).
). Although, most samples expressed relatively high levels of FOXP1 mRNA there was no correlation with the expression levels of CD138 mRNA, which was used to assess the level of plasma cell infiltration (data not shown; Campbell et al., Br J Haem 2010; in press).
Immunolabelling studies subsequently identified the widespread expression of the FOXP1 protein in CD138-negative bone marrow cells (Fig. 5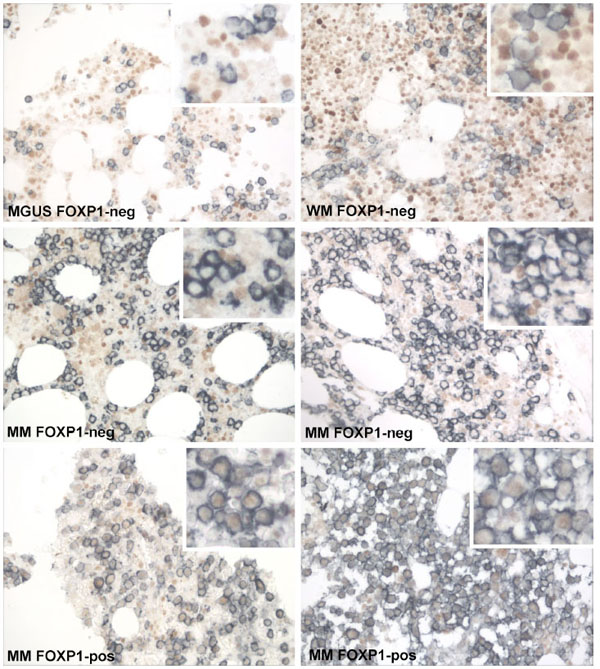 ). Therefore, FOXP1 mRNA expression was also further investigated in CD138+ purified bone marrow plasma cells from a cohort of patients from Oxford to exclude other cell populations from the expression analysis (Fig. 4b
). Therefore, FOXP1 mRNA expression was also further investigated in CD138+ purified bone marrow plasma cells from a cohort of patients from Oxford to exclude other cell populations from the expression analysis (Fig. 4b ). FOXP1 was differentially expressed between the CD138+ bone marrow samples, but there was no evidence for elevated FOXP1 mRNA expression in malignant plasma cells when compared to those from MGUS patients or patients without plasma cell malignancy. However FOXP1 mRNA was detectable in all the plasma cell samples, a significant proportion of which (26/30, 86.6%) expressed levels comparable to that seen in some strongly positive DLBCL cell lines, such as DB and the ABC-DLBCL lines OCI-Ly3 and OCI-Ly10 [13Brown PJ, Ashe SL, Leich E, et al. Potentially oncogenic B-cell activation-induced smaller isoforms of FOXP1 are highly expressed in the activated B cell-like subtype of DLBCL Blood 2008; 111(5
): 2816-4.]. Interestingly, the levels of FOXP1 mRNA in both normal and abnormal CD138+ bone marrow plasma cells was generally higher than those observed in the MM-derived cell lines. These data suggest that FOXP1 mRNA expression is not commonly transcriptionally silenced in normal or abnormal plasma cells.
). FOXP1 was differentially expressed between the CD138+ bone marrow samples, but there was no evidence for elevated FOXP1 mRNA expression in malignant plasma cells when compared to those from MGUS patients or patients without plasma cell malignancy. However FOXP1 mRNA was detectable in all the plasma cell samples, a significant proportion of which (26/30, 86.6%) expressed levels comparable to that seen in some strongly positive DLBCL cell lines, such as DB and the ABC-DLBCL lines OCI-Ly3 and OCI-Ly10 [13Brown PJ, Ashe SL, Leich E, et al. Potentially oncogenic B-cell activation-induced smaller isoforms of FOXP1 are highly expressed in the activated B cell-like subtype of DLBCL Blood 2008; 111(5
): 2816-4.]. Interestingly, the levels of FOXP1 mRNA in both normal and abnormal CD138+ bone marrow plasma cells was generally higher than those observed in the MM-derived cell lines. These data suggest that FOXP1 mRNA expression is not commonly transcriptionally silenced in normal or abnormal plasma cells.
Double immunoenzymatic labelling was used to investigate the expression of FOXP1 at the protein level in CD138+ plasma cells. Paraffin embedded bone marrow trephines were available from fifteen Croatian patients (including nine MM patients and two MGUS patients) of which ten (patients numbered; 1, 2, 3, 10, 12, 22, 23, 27, 31 and 32) corresponded to those where the expression levels of FOXP1 mRNA had been determined in whole bone marrow. There was no FOXP1 protein detected in fourteen of the patients’ biopsies. Samples with occasional rare FOXP1+ plasma cells were scored as negative throughout this study. The exception was patient number 3, where weak/moderate expression of the FOXP1 protein was observed in the nuclei of the majority of the CD138+ population (Fig. 5 , bottom right). This 61 year old male patient was diagnosed with multiple myeloma, IgG kappa paraprotein of 63g/l, albumin 22g/l, Beta2microglobulin 7.0, (stage 3 ISS), with >80% plasma cell infiltrate in the aspirate/trephine and had multiple osteolytic lesions. He initially received VAD and thalidomide that had to be discontinued due to the cardiac side effects (bradycardia, arrhythmia). He was continued on Dexamethasone with which he achieved partial remission. The patient received tandem autologous stem cell transplants and is still alive two years after diagnosis. An unusual feature of this patient was that the diagnostic bone marrow sample was taken while he was recovering from sepsis (Streptococcus pneumoniae). We cannot exclude the possibility that this event may have affected the expression of FOXP1; certainly cytokines such as IL-2, IL-10 and TGF-beta have a key role in the development and function of regulatory T-cells characterised by the expression of the related FOXP3 protein. However, further studies will be needed to determine whether cytokines do indeed also have a role in modulating FOXP1 expression and function. Interestingly, this case had one of the lowest levels of FOXP1 mRNA expression suggesting that FOXP1 mRNA levels do not reflect tumoural FOXP1 protein expression in whole bone marrow aspirates.
, bottom right). This 61 year old male patient was diagnosed with multiple myeloma, IgG kappa paraprotein of 63g/l, albumin 22g/l, Beta2microglobulin 7.0, (stage 3 ISS), with >80% plasma cell infiltrate in the aspirate/trephine and had multiple osteolytic lesions. He initially received VAD and thalidomide that had to be discontinued due to the cardiac side effects (bradycardia, arrhythmia). He was continued on Dexamethasone with which he achieved partial remission. The patient received tandem autologous stem cell transplants and is still alive two years after diagnosis. An unusual feature of this patient was that the diagnostic bone marrow sample was taken while he was recovering from sepsis (Streptococcus pneumoniae). We cannot exclude the possibility that this event may have affected the expression of FOXP1; certainly cytokines such as IL-2, IL-10 and TGF-beta have a key role in the development and function of regulatory T-cells characterised by the expression of the related FOXP3 protein. However, further studies will be needed to determine whether cytokines do indeed also have a role in modulating FOXP1 expression and function. Interestingly, this case had one of the lowest levels of FOXP1 mRNA expression suggesting that FOXP1 mRNA levels do not reflect tumoural FOXP1 protein expression in whole bone marrow aspirates.
FOXP1 protein expression was also investigated by double immunoenzymatic labelling in a larger series of patients from Oxford. Including routinely fixed bone marrow trephines from non-malignant reactive marrows (n=10), patients with MGUS (n=11) and a cohort with MM (n=60). The vast majority of normal and malignant bone marrow CD138+ plasma cells lacked FOXP1 protein expression. The presence of CD138-negative cells expressing FOXP1 served as an internal control for the immunolabelling technique. FOXP1 protein expression in more than 30% of CD138+ cells was only observed in one MM biopsy, taken at time of diagnosis (Fig. 5 , bottom left). The patient had fairly typical clinical features including: age 69 years at diagnosis, 90% plasma cell infiltrate on trephine, no bone marrow involvement, normal calcium, anaemic at diagnosis with haemaglobin of 8.7, normal renal function, IgA lambda paraprotein, albumin 37, Beta2microglobulin 10.8 and was still alive after 7 months follow up. A minor population of FOXP1+/CD138+ plasma cells (<10%) was also observed in two MM cases and one MGUS case. These data suggest that FOXP1 is not frequently expressed at the protein level in MM.
, bottom left). The patient had fairly typical clinical features including: age 69 years at diagnosis, 90% plasma cell infiltrate on trephine, no bone marrow involvement, normal calcium, anaemic at diagnosis with haemaglobin of 8.7, normal renal function, IgA lambda paraprotein, albumin 37, Beta2microglobulin 10.8 and was still alive after 7 months follow up. A minor population of FOXP1+/CD138+ plasma cells (<10%) was also observed in two MM cases and one MGUS case. These data suggest that FOXP1 is not frequently expressed at the protein level in MM.
DISCUSSION
Foxp1 has been shown to function during the pro-B to pre-B cell transition in the early stages of B-cell development (Hu et al., 2006) and regulates Rag gene expression. Our immunolabelling studies have already shown widespread, high level FOXP1 expression in lymphoid tissue, in particular in the naïve B cells of the mantle zone and variable but often strong expression in B cells within the germinal centre [1Banham AH, Beasley N, Campo E, et al. The FOXP1 winged helix transcription factor is a novel candidate tumor suppressor gene on chromosome 3p Cancer Res 2001; 61: 8820-9.]. However, here we show that expression of the FOXP1 protein appears to be tightly regulated in the terminal stages of B-cell development, rarely being expressed in normal plasma cells. However, FOXP1 transcripts were expressed at relatively high levels in CD138+ bone marrow plasma cells from patients without MM.
This pattern of FOXP1 protein expression, with occasional nuclear positivity, was found to be similar in MM cell lines and FOXP1 transcripts were fairly abundantly expressed in the JJN3 myeloma cell line. In the two MM cell lines in which FOXP1 protein was detectable, it appeared that this was restricted to moderate levels of nuclear expression in a very minor population of CD138+ cells. Similarly, we observed that the FOXP1 protein is rarely expressed in malignant plasma cells, yet the FOXP1 transcript is abundant in samples of purified CD138+ cells recovered from MM and MGUS BM aspirates. It was noticeable that both normal and malignant CD138+ primary bone marrow cells tended to expressed considerably higher levels of the FOXP1 mRNA than was observed in three of the four MM-derived cell lines. It is possible that factors in the bone marrow microenvironment may upregulate FOXP1 mRNA expression in bone marrow plasma cells. Certainly expression of the related FOXP3 transcription factor in regulatory T cells is significantly affected by cytokines, such as IL-2 and the signalling molecule TGF-β [22Josefowicz SZ, Rudensky A. Control of regulatory T cell lineage commitment and maintenance Immunity 2009; 30(5 ): 616-25.]. These data suggest that the expression of the FOXP1 protein in both normal and abnormal plasma cells is regulated at a post-transcriptional level.
While we were preparing this manuscript, another group published a paper and a case report describing FOXP1 protein expression in MM; FOXP1 mRNA expression was not analysed in either study [23Korac P, Skrtic A, Peran I, et al. Atypical FOXP1 expression in malignant plasma cells that show several simultaneous translocations Histopathology 2009; 54(6 ): 770-1.]. The case report described a FOXP1+ MM case with rather atypical clinical features, hyperploidy and multiple IGH translocations in which the FOXP1 gene was amplified in CD138+ cells (12-15 copies) [23Korac P, Skrtic A, Peran I, et al. Atypical FOXP1 expression in malignant plasma cells that show several simultaneous translocations Histopathology 2009; 54(6 ): 770-1.]. In a larger series of 13 MGUS and 60 MM patients they identified >30% nuclear FOXP1 protein expression in one MGUS and 12 MM biopsies (20%) [24Korac P, Peran I, Skrtic A, et al. FOXP1 expression in monoclonal gammopathy of undetermined significance and multiple myeloma Pathol Int 2009; 59(5 ): 354-8.]. Korac et al., detected FOXP1 protein expression at a much higher incidence than in our study, as we detected >30% FOXP1 nuclear positivity in only 3% of MM patients (2/68) and none of the MGUS patients (n=13).
The reason(s) for the difference in the findings are unclear. Both groups have used the same monoclonal anti-FOXP1 antibody to detect protein expression in formalin-fixed paraffin-embedded bone marrow trephines by immunohistochemistry (although different labelling kits were used). There was nothing to suggest that FOXP1 expression differed between the cases that we studied from Oxford versus those from Croatia. However, we found that double labelling enabled us to detect weak FOXP1 protein expression levels that would have been difficult to detect in the presence of a nuclear counterstain and to verify that the positive cells were indeed CD138+ plasma cells. As illustrated by the data presented in Fig. (5 ) we observed variable and often significant levels of FOXP1 expression in the CD138-negative population in bone marrow.
) we observed variable and often significant levels of FOXP1 expression in the CD138-negative population in bone marrow.
Korac and colleagues identified FOXP1 copy number changes in 24/60 MM and 5/13 MGUS [24Korac P, Peran I, Skrtic A, et al. FOXP1 expression in monoclonal gammopathy of undetermined significance and multiple myeloma Pathol Int 2009; 59(5 ): 354-8.]. How this relates to the expression of the FOXP1 protein is unclear as the abnormalities were detected at a much higher frequency than was FOXP1 protein expression. The authors did not comment on whether the MM cases reported to express the FOXP1 protein included those with the highest FOXP1 gene copy number. Further study will be necessary to determine whether these genetic changes genuinely contribute to aberrant FOXP1 protein expression in MM. Certainly our data suggest that the FOXP1 mRNA is commonly expressed in both normal and malignant plasma cells in the absence of FOXP1 protein expression.
The rarity of FOXP1 protein expression in normal plasma cells is consistent with the detection of FOXP1 protein expression in only a minority of MM. Other transcription factors with a role in early B-cell development are also involved in terminal differentiation to plasma cells, including PAX5 which is silenced during plasma cell development and IRF4 which is up-regulated during plasma cell differentiation. Thus it is not without precedent to suggest that the absence of FOXP1 protein expression may also be functionally important during terminal B-cell differentiation to plasma cells. It will be interesting in the future to investigate the affects that expressing FOXP1 has on the terminally differentiated phenotype of plasma cells. In particular whether FOXP1 regulates the expression of those cell surface markers and transcription factors whose silencing or induction plays a crucial role in this phenotype. The post-transcriptional regulation of FOXP1 protein expression in both normal and malignant plasma cells provides an explanation as to why its loss of expression would not have been detected by studies of gene expression changes occuring during plasmacytic differentiation. FOXP1 mRNA levels are indicative of the FOXP1 protein expression patterns in lymphomas derived from mature B cells. Therefore, it will be interesting to discover the mechanism of this novel post-transcriptional regulation of FOXP1 expression in terminally differentiated B cells.
ACKNOWLEDGEMENTS
This study was supported by funding from Leukaemia Research. A.J.C has also received funding from the Julian Starmer-Smith Lymphoma Fund, NIHR Biomedical Research Centre Oxford and the Anya Sturdy Clinical Research Fellowship. The authors have no conflict of interest to disclose.





