- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Leukemia Journal
(Discontinued)
ISSN: 1876-8164 ― Volume 5, 2013
Mitochondrial DNA Alterations and Oxidative Stress in Acute Leukemia
Mouna Saadaoui1, 4, Lamia Aissaoui2, Véronique Salaun3, Mohamed Manai4, Stéphane Allouche* , 1
Abstract
Mitochondrial DNA (mtDNA) alterations were reported in many cancers but their roles in oncogenesis are still debated. We aimed to examine qualitative and quantitative mtDNA modifications and oxidative stress in normal and cancer cells. 23 leukemia patients and 18 healthy subjects were recruited from the hospital of Azziza Othmana (Tunisia). Mitochondrial D-loop was sequenced, mtDNA level was determined by Q-PCR, the oxidative stress was assessed by a fluorescent probe and the mitochondrial transcription factor A (mTFA) level was quantified. No somatic mutation was evidenced in leukemia cells compared to non-malignant cells. However, a significant higher level of mtDNA associated with an increase of mTFA expression and reactive oxygen species (ROS) production were measured in patients’ cells compared to non-malignant cells. In conclusion, our results don’t support the role of mtDNA mutations in leukemogenesis but increase of mtDNA and ROS levels would be molecular signatures of leukemia.
Article Information
Identifiers and Pagination:
Year: 2013Volume: 5
First Page: 1
Last Page: 6
Publisher Id: TOLEUKEMIAJ-5-1
DOI: 10.2174/1876816420130418001
Article History:
Received Date: 12/1/2013Revision Received Date: 27/3/2013
Acceptance Date: 4/4/2013
Electronic publication date: 3/5/2013
Collection year: 2013
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/ which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Service de Biochimie, Centre Hospitalier et Universitaire, Avenue côte de nacre, 14033 Caen Cedex, France; Tel: +33 231065419; Fax: +33231065172; E-mail: allouche-s@chu-caen.fr
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 12-1-2013 |
Original Manuscript | Mitochondrial DNA Alterations and Oxidative Stress in Acute Leukemia | |
INTRODUCTION
Since the pioneering works of Otto Warburg in 1920s’ about differences in the mitochondrial metabolism between normal and tumoral tissue, an abundant literature has been published to date pointing out alterations of mitochondria in cancer and their potential roles in carcinogenesis (see for review [1Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism Nat Rev Cancer 2011; 11(5 ): 325-7.]). The human mtDNA contains 16.569 bp [2Anderson S, Bankier AT, Barrell BG, et al. Sequence and organization of the human mitochondrial genome Nature 1981; 290: 457-65.] and plays an important role in energy metabolism. While the majority of mtDNA encodes ribosomal RNAs (rRNAs), transfer RNAs (tRNAs) and proteins, there is a small non-coding region of about 1.1 kb named D-loop which binds regulatory factors such as mitochondrial transcription factor A (mTFA) that plays a major role in the maintenance of mtDNA [3Kang D, Kim SH, Hamasaki N. Mitochondrial transcription factor A (TFAM): roles in maintenance of mtDNA and cellular functions Mitochondrion 2007; 7: 39-44.]. It’s now well admitted that this region is a “hot spot” for mutations of mtDNA associated with various cancers [4Chatterjee A, Dasgupta S, Sidransky D. Mitochondrial subversion in cancer Cancer Prev Res (Phila) 2011; 4: 638-54.] that would result from oxidative stress. Mitochondrion is the major site of reactive oxygen species (ROS) production. Under physiological conditions, ROS participate into various cell functions but their abnormal production or the loss of detoxifying enzyme would cause mtDNA damages and diseases [5Fogg VC, Lanning NJ, Mackeigan JP. Mitochondria in cancer: at the crossroads of life and death Chin J Cancer 2011; 30: 526-39.].
Mitochondria were shown to be altered in hematological disorders. For instance, myelodysplastic syndromes are characterized by an ineffective hematopoiesis caused by mtDNA mutations that would directly be responsible for carcinogenesis [6Wulfert M, Küpper AC, Tapprich C, et al. Analysis of mitochondrial DNA in 104 patients with myelodysplastic syndromes Exp Hematol 2008; 36: 577-86.]. Several laboratories also reported the presence of mtDNA mutations in chronic lymphocytic (CLL) and myeloid (CML) leukemia and in acute lymphoblastic (ALL) and myeloid (AML) leukemia [7Carew JS, Zhou Y, Albitar M, Carew JD, Keating MJ, Huang P. Mitochondrial DNA mutations in primary leukemia cells after chemotherapy clinical significance and therapeutic implications Leukemia 2003; 7: 437-7.-9He L, Luo L, Proctor SJ, et al. Somatic mitochondrial DNA mutations in adult-onset leukaemia Leukemia 2003; 7: 2487-91.]. However, a recent study challenged the potential role of mtDNA mutations in leukemogenesis [10Cerezo M, Bandelt H-J, Martín-Guerrero I, et al. High mitochondrial DNA Stability in B-Cell chronic lymphocytic leukemia PLoS ONE 2009; 4: e7902-0.].
Since the role of mitochondria in cancer is still a matter of debate, our study aimed to evaluate mtDNA instability within the D-loop and mtDNA copy number in Tunisian patients affected by acute leukemia. This was accomplished by PCR/sequencing and quantitative PCR of samples obtained from patients and healthy subjects. The significant increase in mtDNA content observed in tumor cells prompted us to examine mTFA and ROS, two factors known to regulate mitochondrial DNA replication. mTFA was quantified by real-time-RT-PCR and western-blot and ROS production was evaluated by hydroethidine/flow cytometry.
MATERIALS AND METHODOLOGY
Patients
Twenty three Tunisian children or young adults, with a mean age of 8.8 years and a sex ratio male/female of 1.87, affected either by ALL (n=16) or AML (n=7) were studied (Table 1) and compared to eighteen healthy children or young adults from Tunisia with a sex ratio male/female of 1.25 and a mean age of 12.3 years. All patients, except P5, were included without prior chemotherapy. The study was conducted according to the Tunisian biomedical research rules that was approved by the ethics committee of the hospital of Azziza Othmana (Tunisia) and according to the ethical standards formulated in the Helsinki Declaration. All biological samples were collected anonymously.
Peripheral Blood Mononuclear Cells Preparation
Peripheral blood mononuclear cells (PBMC) were obtained from patients and healthy donors using a gradient Ficoll-Paque (Amersham Biosciences, Saclay France) and then were frozen at –192°C into two vials for nucleic acid and protein extraction and measurement of superoxide anion (O2°-) production.
Nucleic Acid Extraction
Total DNA was extracted from isolated PBMC and buccal cells by using the standard phenol/chloroform extraction procedure. Total RNAs were extracted from frozen PBMC cells of patients and healthy donors with TRIzol® reagent (Invitrogen, Saint-Aubin France). 3 ìg of total RNAs were reverse transcribed using the Moloney Murine Leukemia Virus Reverse Transcriptase (Invitrogen) to obtain cDNAs.
mtDNA Sequencing
Detection of polymorphisms/mutations in the D-loop of the mtDNA was performed by PCR using four primer pairs L1R1, L2R2, L3R3 and L4R4 (1) followed by direct sequencing using the Genomelab DTCS (Beckman Coulter, France) and analyzed on a Beckman Coulter CEQ 8000 sequencer. Estimation of mitochondrial haplogroups was determined using the mtDNAmanager interface developed by Hwan Young Lee et al. (2008) using the control region sequence [11Lee HY, Song I, Ha E, Cho S-B, Yang WI, Shin K-J. mtDNAmanager a Web-based tool for the management and quality analysis of mitochondrial DNA control-region sequences BMC Bioinformatics 2008; 9: 483-0.].
Determination of the mtDNA/nDNA Ratio by Real-Time PCR
Quantification of the mtDNA was performed by real-time PCR amplification on a Light Cycler 480 (Roche Diagnostics, Meylan France) using Cycler FastStart DNA master Sybr green I mix (Roche Diagnostics) according to the method developed by May-Panloup et al. [12May-Panloup P, Chrétien M-F, Savagner F, et al. Increased sperm mitochondrial DNA content in male infertility Hum Reprod 2003; 18: 550-6.] except that we amplified the 18S rRNA for nDNA quantification (1). All samples were analyzed in duplicate. Considering that 1 cell contains 10 pg of nDNA, results are expressed as mtDNA copy number per cell.
Real-Time PCR
Real-time quantification of mTFA expression relative to b-actin mRNAs was performed on the Light Cycler 480 system (Roche Diagnostics) using specific set of primers (1). The threshold cycle (Ct) was determined and the mTFA/b-actin ratio was calculated according to the 2-DDCt method [13Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method Methods 2001; 25: 402-8.].
Western-Blot Analysis
mTFA and b-actin expression were studied from PBMC by western-blot with a 1:500 dilution of rabbit anti-human mTFA (Abcam, Paris France) and a 1:1.000 dilution of the monoclonal anti-β-actin (santa-Cruz, Le Perray en Yvelines France) antibodies, respectively. Protein concentration was determined by the BCA Protein assay (Bicinchoninic Acid).
Measurement of Superoxide Anion Production
Superoxide anion radical production was determined by using the fluorescent sensitive probe hydroethidine (HE) (Molecular Probes, Invitrogen, Saint-Aubin France). PBMC (5. 104 cells) obtained from controls or patients were incubated with 78µM HE for 15 min in PBS at 37°C in the dark. Then, fluorescence of oxidized HE was determined by flow cytometry (Becton Dickinson FACS CANTO II, FACS Diva Software version 6.1) with 485 and 510 nm wavelengths for excitation and emission, respectively.
Statistical Analysis
Data presented correspond to mean +/- S.E.M. The Fisher’s exact test and the t-test were used to determine the statistical significance of differences between controls and patients. Differences were considered significant when p values were less than 0.05. For multiple comparison tests, we applied the Bonferroni correction.
RESULTS
mtDNA Sequence Analysis
The D-loop region of the mtDNA was entirely sequenced in leukemia patients and healthy subjects. We observed several homoplasmic and heteroplasmic variations both in patients and controls compared to the rCRS (revised Cambridge Reference Sequence) (2A) but only the heteroplasmic m.343C>T substitution was found significantly under-represented in patients compared to controls (22% vs 89 %, p < 0.001 respectively) (2B). In order to distinguish between somatic and germinal mutation/polymorphism, the m.343C>T substitution was screened in buccal epithelial cells from 11 among 23 patients, an identical distribution of this polymorphism was found in both tissues.
Quantitative Real Time PCR Analysis
As the D-loop plays a major role in the regulation of mtDNA replication, quantification of mtDNA level was achieved from PBMC of patients and healthy donors by quantitative PCR using both mitochondrial and nuclear standards. As depicted in the Fig. (1A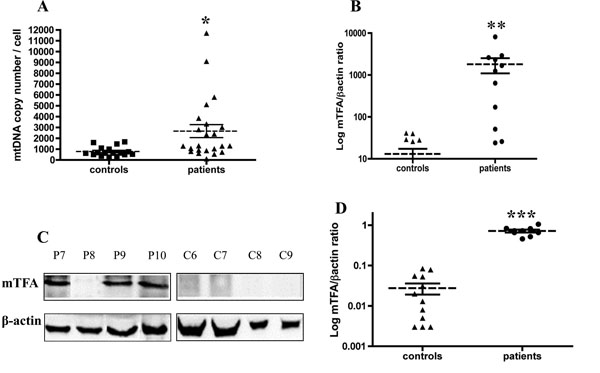 ), a significant 3-fold increase in the mtDNA/nDNA ratio was measured between leukemia patients and controls (t-test, p = 0.012). Values of mtDNA copy number from the control group were in the same range as previously reported in healthy donors with a different method and targed genes [14Gourlain K, Amellal B, Ait Arkoub Z, Dupin N, Katlama C, Calvez V. Quantitative analysis of human mitochondrial DNA using a real-time PCR assay HIV Med 2003; 4: 287-92.].
), a significant 3-fold increase in the mtDNA/nDNA ratio was measured between leukemia patients and controls (t-test, p = 0.012). Values of mtDNA copy number from the control group were in the same range as previously reported in healthy donors with a different method and targed genes [14Gourlain K, Amellal B, Ait Arkoub Z, Dupin N, Katlama C, Calvez V. Quantitative analysis of human mitochondrial DNA using a real-time PCR assay HIV Med 2003; 4: 287-92.].
mTFA Expression
mTFA was described as a major activator of mtDNA transcription and replication. So, we sought to study the expression of this regulatory factor by real-time PCR and western-blot. We observed a significant increase in mTFA mRNAs in leukemia patients compared to controls (t-test, p =0.009) (Fig. 1B ), that was correlated with the increase at the protein level (t-test, p <0.001) (Fig. 1C
), that was correlated with the increase at the protein level (t-test, p <0.001) (Fig. 1C , 1D
, 1D ). The specificity of anti-mTFA antibody was verified using the Ramos and Jurkat cells lines in which we were able to detect a single band at the predicted size (data not shown).
). The specificity of anti-mTFA antibody was verified using the Ramos and Jurkat cells lines in which we were able to detect a single band at the predicted size (data not shown).
Measurement of ROS Production
We measured the superoxide anion production in PBMC isolated from 14/23 leukemia patients and 11/18 healthy donors using the fluorescent probe HE. The more ROS are produced, the more fluorescence is increased (3). As shown in the Fig. (2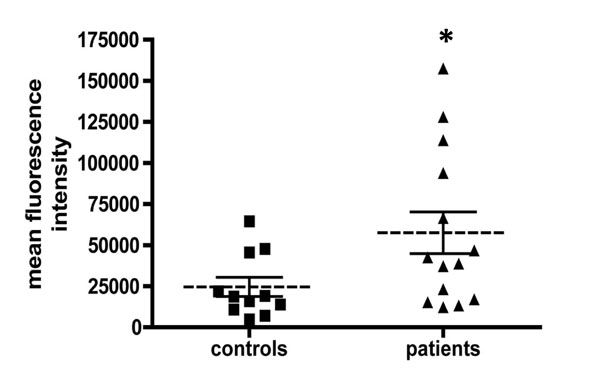 ), the level of ROS production, reflected by the mean fluorescence intensity, was significantly increased by about 2-fold in leukemia patients compared to controls (t-test, p = 0.04).
), the level of ROS production, reflected by the mean fluorescence intensity, was significantly increased by about 2-fold in leukemia patients compared to controls (t-test, p = 0.04).
DISCUSSION
There is an abundant literature about mtDNA mutations and cancers, both in solid tumors and leukemia but the relevance of such mitochondrial abnormalities in tumor genesis is still unclear (see for review [15Lu J, Sharma LK, Bai Y. Implications of mitochondrial DNA mutations and mitochondrial dysfunction in tumorigenesis Cell Res 2009; 19: 802-15.]). These mtDNA point mutations are somatic and detected mainly in the D-loop. Regarding leukemia, mitochondrial mutations were also reported in this region and were suggested to contribute to tumor growth [8Grist SA, Lu X-J, Morley AA. Mitochondrial mutations in acute leukaemia Leukemia 2004; 8: 313-6.]. In contrast, our data don’t support the existence of any link between mtDNA mutations and acute leukemia. Such conclusions have also been recently drawn by Cerezo et al. (2009) [10Cerezo M, Bandelt H-J, Martín-Guerrero I, et al. High mitochondrial DNA Stability in B-Cell chronic lymphocytic leukemia PLoS ONE 2009; 4: e7902-0.] from their study on B-cell CLL. While we detected a single significant difference at the position 343 between patients and healthy controls in PBMC, the epithelial buccal cells of patients, a non-malignant tissue, also displayed the same C nucleotide in the homoplasmic state, this indicates that this mutation was not segregated in tumoral cells. There are different hypothesis to explain discrepancies between our data and those previously published:
Our patients, except N°5, didn’t receive neither chemotherapy nor platelets transfusion at the time of diagnosis that could lead to a false positive detection of polymorphism/mutation [16Meierhofer D, Ebner S, Mayr JA, Jones ND, Kofler B, Sperl W. Platelet transfusion can mimic somatic mtDNA mutations Leukemia 2006; 20: 362-.].
D-loop is known to be a very polymorphic region and when using the rCRS as template, polymorphisms due to haplogroup can be mistaken as mutations. To avoid such pitfall and on the contrary of some studies [17Sharawat SK, Bakhshi R, Vishnubhatla S, Bakhshi S. Mitochondrial D-loop variations in paediatric acute myeloid leukaemia a potential prognostic marker Br J Haematol 2010; 149: 391-8.] we rather compared D-loop sequences obtained from patients with controls coming from the same geographical region.
We also checked that our PCR conditions did not cause amplification of nuclear pseudogenes closely related to mtDNA by using rho° cells.
In our study, we compared tumoral (blood) and non-malignant tissues (epithelial buccal cells) from same patients to determine the role of mtDNA mutation in cancer in contrast to some in which only the tumoral cells were studied [18Kwok CSN, Quah TC, Ariffin H, Tay SKH, Yeoh AEJ. Mitochondrial D-loop polymorphisms and mitochondrial DNA content in childhood acute lymphoblastic leukemia J Pediatr Hematol Oncol 2011; 33: e239-44.].
We found that leukemia cells displayed a higher level of ROS production compared to non-malignant PBMC as recently reported in CML [19Nieborowska-Skorska M, Kopinski PK, Ray R, et al. Rac2-mitochondrial respiratory chain complex III-generated ROS cause genomic instability in chronic myeloid leukemia stem cells and primitive progenitors Blood 2012; 119(18): 4253-63.]. Indeed, alteration of the mitochondrial membrane potential was suggested to disturb electron flow through the mitochondrial respiratory chain and generate elevated level of ROS. While in our study we sequenced only the D-loop region, we can’t rule out the presence of mutations or polymorphisms that would enhance the electron leakage and ROS production. Our data are also in good agreement with the study of Er et al. (2007) [20Er T-K, Tsai S-M, Wu S-H, et al. Antioxidant status and superoxide anion radical generation in acute myeloid leukemia Clin Biochem 2007; 40: 1015-9.] who demonstrated higher superoxide anion production in AML compared to healthy controls. In such conditions, we would have expected to detect common oxidative damage signature mutations (C-T, CC-TT and G-T) of DNA in our patients as reported by Grist et al. (2004) [8Grist SA, Lu X-J, Morley AA. Mitochondrial mutations in acute leukaemia Leukemia 2004; 8: 313-6.]. However, in this study the authors selected a high proportion of patients who had been previously treated and relapsed, furthermore, data of mtDNA sequence analysis were compared to the rCRS. So, the mutations that were reported could be due to chemotherapy treatment or would correspond to polymorphisms. In addition to promote damages to cellular components and DNA mutations, ROS were suggested to play a role in intracellular signaling and to activate pro-proliferative and/ or survival pathways (see for review [21Hole PS, Darley RL, Tonks A. Do reactive oxygen species play a role in myeloid leukemias? Blood 2011; 17: 5816-26.]). ROS would be deleterious in two ways: they would promote DNA instability by promoting DNA breaks and would confer a proliferative advantage in cancer cells. Due to the difficulty to maintain patients’ cells in culture, we didn’t test the role of ROS in proliferation.
ROS were also suggested to regulate the mtDNA copy number as recently demonstrated in nondiabetic hemodialysis patients [22Wang Y-C, Lee W-C, Liao S-C, et al. Mitochondrial DNA copy number correlates with oxidative stress and predicts mortality in nondiabetic hemodialysis patients J Nephrol 2011; 24: 351-8.]. In this study, the authors showed a good and positive correlation between mtDNA content in peripheral blood leukocytes and the plasma thiobarbituric acid-reactive substances level used as an indicator of oxidative stress. In the present study, we observed a significant increase in the mtDNA/nDNA ratio in patients compared to controls. Such an increase was previously reported in blast cells of AML as well as during transformation of chronic granulocytic leukemia [23Boultwood J, Fidler C, Mills KI, et al. Amplification of mitochondrial DNA in acute myeloid leukaemia Br J Haematol 1996; 95: 426-31.]. Aneuploidy is frequently observed in cancers including leukemia cells and could potentialy lead to erroneous nDNA quantification. However this is unlikely in our study since we used the 18S rRNA which is a multicopy gene. mTFA, whose level is known to regulate the mtDNA copy number (see for review [3Kang D, Kim SH, Hamasaki N. Mitochondrial transcription factor A (TFAM): roles in maintenance of mtDNA and cellular functions Mitochondrion 2007; 7: 39-44.]), was increased in patients compared to healthy controls both at the levels of mRNA and protein. Very few data are available about the expression level of mTFA so it’s difficult to compare our data. However, using a non-quantitative RT-PCR mTFA expression was shown to be absent in normal B-lymphocytes from 2 healthy donors while it was greatly expressed in CLL [24Carew JS, Nawrocki ST, Xu RH, et al. Increased mitochondrial biogenesis in primary leukemia cells: the role of endogenous nitric oxide and impact on sensitivity to fludarabine Leukemia 2004; 18: 1934-40.]. So it’s not surprising to observe a strong increase in mTFA expression in patients compared to controls. Moreover, our molecular studies are in good agreement with western-blot experiments since in the control group mTFA was barely detectable while a strong immunoreactivity was measured in patients. We can question about signals that would trigger mTFA overexpression which consequently would induce increase in mtDNA level in leukemia patients. We can speculate that oxidative stress would damage mtDNA and compensatory mechanisms would be initiated to replace altered DNA molecules by new ones via mTFA. Such relationship between ROS and mTFA expression was previously reported in yeast mitochondria subjected to oxidative stress [25Hori A, Yoshida M, Shibata T, Ling F. Reactive oxygen species regulate DNA copy number in isolated yeast mitochondria by triggering recombination-mediated replication Nucleic Acids Res 2009; 37: 749-61.]. More, in addition to its role in mtDNA maintenance, mTFA was recently demonstrated to be localized in nucleus and its over-expression, in the human prostate cancer cell line PC3, was shown to induce cellular proliferation [26Han B, Izumi H, Yasuniwa Y, et al. Human mitochondrial transcription factor A functions in both nuclei and mitochondria and regulates cancer cell growth Biochem Biophys Res Commun 2011; 408: 45-51.].
In conclusion, while our data suggest no evidence for mtDNA instability in leukemogenesis, we observed an increase in oxidative stress, mtDNA and mTFA levels in leukemia patients’ cells. Such mitochondrial alterations could be used a molecular signature of leukemia cells. Further studies would be necessary to determine any link between such abnormalities and cell proliferation.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
FUNDING SOURCE
Les ministères de l’enseignement supérieur et de la recherche Français et Tunisien.
ABBREVIATIONS
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s web site along with the published article
ACKNOWLEDGEMENTS
Fundings come from the Ministères de l’enseignement supérieur et de la recherche Français et Tunisien. Mrs Mouna Saadaoui was supported by le Ministère de l’Enseignement Supérieur et de la Recherche Scientifique de Tunisie. We thank Dr. Balkis Meddeb for providing blood samples of the patients from Aziza Othmana Hospital, Dr. Inès Safra (laboratoire d'hématologie, Institut Pasteur de Tunis, Tunisie) for their advices, Dr. Christian Creveuil (Unité de biostatistique et de recherche clinique, CHU de Caen, France) for its statistical advices, Pr. Xavier Troussard (laboratoire d'hématologie, CHU de Caen, France) for his scientific collaboration and Dr. Florence Truquet (laboratoire d'hématologie, CHU de Caen, France) for her critical reading of the manuscript.



